Abstract
Peripheral arterial disease (PAD) results from an obstruction of blood flow in the arteries other than the heart, most commonly the arteries that supply the legs. The complexity of the known signaling pathways involved in PAD, including various growth factor pathways and their cross talks, suggests that analyses of high-throughput experimental data could lead to a new level of understanding of the disease as well as novel and heretofore unanticipated potential targets. Such bioinformatic analyses have not been systematically performed for PAD. We constructed global protein-protein interaction networks of angiogenesis (Angiome), immune response (Immunome), and arteriogenesis (Arteriome) using our previously developed algorithm GeneHits. The term “PADPIN” refers to the angiome, immunome, and arteriome in PAD. Here we analyze four microarray gene expression datasets from ischemic and nonischemic gastrocnemius muscles at day 3 posthindlimb ischemia (HLI) in two genetically different C57BL/6 and BALB/c mouse strains that display differential susceptibility to HLI to identify potential targets and signaling pathways in angiogenesis, immune, and arteriogenesis networks. We hypothesize that identification of the differentially expressed genes in ischemic and nonischemic muscles between the strains that recovers better (C57BL/6) vs. the strain that recovers more poorly (BALB/c) will help for the prediction of target genes in PAD. Our bioinformatics analysis identified several genes that are differentially expressed between the two mouse strains with known functions in PAD including TLR4, THBS1, and PRKAA2 and several genes with unknown functions in PAD including EphA4, TSPAN7, SLC22A4, and EIF2a.
Keywords: bioinformatics, systems biology, angiogenesis, microarray, thrombospondin 1
peripheral arterial disease (PAD) affects ∼8 million to 12 million people in the United States and over 200 million worldwide, especially those over the age of 50 (17). PAD results from atherosclerosis that causes an obstruction of blood flow in the peripheral arteries, most commonly the arteries that perfuse the legs. With the goal to increase blood flow around blockages, clinical trials using drugs and gene delivery aimed at stimulating vascular growth and remodeling have been performed for more than a decade, but these trials have been met with limited success (1, 41, 71).
Vascular growth and remodeling during physiological or pathological conditions in adults are primarily mediated by arteriogenesis and angiogenesis programs (36, 69). Arteriogenesis is a vascular remodeling process that leads to increase the diameter of pre-existing conduit vessels (∼25–100 μm), i.e., arterioles or small arteries, triggered by shear stress and inflammation at the site of blood vessel occlusion. Angiogenesis is a vascular remodeling process that leads to the formation of microvessels (<20 μm) by sprouting or intussusception. Induction of arteriogenesis is needed for the supply of bulk blood flow at the proximal end in ischemic cardiovascular diseases like PAD. In addition, induction of angiogenesis is critical for efficient perfusion of the ischemic tissue in PAD. In addition, inflammation is known to be one of the common driving forces behind both arteriogenesis and angiogenesis during ischemic tissue injuries (2, 56). This indicates the extensive molecular interactions in the inflammatory environment modulating arteriogenesis and angiogenesis in PAD. In this study, we refer to the “immune response” as inflammation.
In mice, hindlimb ischemia (HLI) induces arteriogenesis in abductor region of thigh and angiogenesis in gastrocnemius and tibialis anterior region of the leg. While the majority of the collateral large vessel network (≥100 μm) occurs in the abductor region of the thigh, arteriogenesis of medium collaterals (∼25–50 μm) can still occur in the gastrocnemius and tibialis anterior region. Recent reports have shown that while arteriogenesis is needed to compensate for the bulk supply of the blood flow to the ischemic limb, angiogenesis is critical for efficient perfusion recovery of the mice from HLI (44). This strongly suggests that in addition to arteriogenesis, angiogenesis is critical for perfusion recovery from PAD. Since gastrocnemius muscle constitutes a potential reservoir for angiogenic signaling and to some extent arteriogenic signaling, we chose to use gastrocnemius muscle from C57BL/6 and BALB/c mice (nonischemic and ischemic) to differentiate the gene expression patterns in angiogenic, immune, and arteriogenic signaling networks. Global gene expression analysis suggests that inflammatory and immune responses play a major role in PAD (18), and the genes involved in the three biological processes of angiogenesis, immune response, and arteriogenesis are the potential reservoir for predictions of novel targets in PAD.
Our study primarily builds on previously published animal models describing the apparent susceptibility of two genetically different mouse strains (C57BL/6 and BALB/c) (8, 16, 19, 57). It is well known that C57BL/6 and BALB/c mice differ dramatically in their ability to recover from HLI; however, genetic changes that occur in the ischemic hindlimb between these two strains that could determine the perfusion recovery are not known. In our previous experiments we have observed that microRNA-93 (miR-93) is one of the critical microRNAs that is upregulated in C57BL6 ischemic muscle but not in BALB/c (27). We hypothesized that one of the apparent reasons for the inability for BALB/c to recovery from HLI is due to lower miR-93 levels in ischemic muscle. We confirmed our hypothesis by gain-of-function experiments (injecting miR-93 mimics into BALB/c HLI mice) and by loss-of-function experiments (knocking down miR-93 by injecting antagomir-93 in C57BL6 HLI mice). Scrambled miR-93 served as control in gain-of-function experiments in BALB/c HLI mice and scrambled antagomir-93 served as controls in loss-of-function experiments in C57BL6 mice. In our subsequent experiments we determined to identify and delineate the molecular events in the initial stages of disease progression between C57BL6 and BALB/c HLI mice that can play key roles in the better recovery of C57BL6 mice and worse recovery of BALB/c mice. We collected total gastrocnemius muscle from scrambled miR-93-treated BALB/c mice at day 3 post-HLI (similar to BALB/c 3 days post-HLI) or scrambled antagomir-93-treated C57BL6 mice day 3 post-HLI (similar to C57BL6 3 days post-HLI). RNA microarrays were performed from the RNA extracted from these 3-day post-HLI gastrocnemius tissues. Data obtained from the microarray were used to identify the molecular events that control the perfusion recovery between mouse strains.
Available microarray datasets in PAD patients include gene expression analysis of peripheral blood mononuclear cells (42) and human femoral arteries (14, 18). However, these cells or tissues are very different from the ischemic skeletal muscles that are the site of ischemia and the stimulus for vascular remodeling and growth. The only available gene transcriptional data comparing the ischemic vs. nonischemic gastrocnemius muscles from the two mouse strains at an informative early time point is described in Ref. 27. The main goal of this study is to determine the differential gene expression changes in the ischemic skeletal muscle between these two strains at an earlier time point to identify the potential molecules that could be responsible for better recovery in C57BL/6 mice and/or for worse recovery in BALB/c mice. Nonischemic muscle from both strains served as the baseline controls for this study. We have previously shown that at day 3 post-HLI the capillary density and perfusion recovery between these two mouse strains are comparable, which makes it a critical time point to investigate the upstream signaling changes between the two strains (43). By day 7 several changes in blood flow and perfusion recovery between these two mouse strains become significantly different, thereby making time points other than day 3 not relevant for the study.
The failure of several strategies to improve blood flow in PAD suggests a critical need to identify the potential targets that can improve perfusion recovery in PAD. High-throughput data have been successfully used for other diseases, most prominently in cancer, to identify important novel targets. To integrate hundreds of angiogenesis-related molecules and infer angiogenesis-annotated genes and proteins, we have developed a machine learning algorithm to construct the angiome, a global protein-protein interaction network (PIN) relevant to angiogenesis (12). Such analyses for PAD-related angiogenesis, immune response, and arteriogenesis have not been performed. The primary goal of this study is to use the “omic” approach to identify important set of genes and signaling pathways in PAD, with the eventual goal of identifying new therapeutic targets. We used large-scale high-throughput gene expression datasets from ischemic and nonischemic muscles from the two mouse strains to construct PADPIN, the global PIN of angiogenesis, inflammation, and arteriogenesis in PAD.
METHODS
Animal model of HLI and perfusion imaging.
Unilateral hindlimb ischemia was induced in 8–12 wk old male C57BL/6 (n = 11) and BALB/c (n = 12) mice by surgical ligation and excision of the femoral artery above the inguinal ligament of its bifurcation at saphenous and popliteal arteries as previously described (39). All the animal protocols in our experiments were reviewed and approved by the University of Virginia Institutional Animal Care and Use Committee. Laser Doppler perfusion imaging was performed immediately on ischemic and nonischemic feet of the mice immediately postoperation. Mice were anesthetized by intraperitoneal injection of ketamine (90 mg/kg) and xylazine (10 mg/kg) throughout the surgical and postoperative imaging process. Laser Doppler imaging to measure the blood flow was measured at day 3, day 7, day 14, day 21, and day 28 time intervals post-HLI (39).
Animal treatments, tissue isolation, and mRNA array.
Animal treatments, tissue isolation, and mRNA array were performed as previously described (27). In brief, scrambled antagomir-93 sequences were synthesized according to the nucleotide modifications as previously described (37). Oligonucleotide sequences for scrambled antagomir-93 (6) are 5′-AAGGCAAGCUGACCCUGAAGUU-3′. Oligonucleotides were dissolved in PBS and injected retro-orbitally at a dose of 8 mg/kg body wt, 30 min before HLI (6, 23). Scrambled miR-93 mimic (cat. #4464058; Ambion, Austin, TX) was also dissolved in PBS, and a total dose of 300 μM was delivered to each mouse by intramuscular injection in two sites (100 μM in 25 μl at each site) of the gastrocnemius muscle and one site in the tibialis anterior muscle (100 μM in 25 μl) 30 min before HLI (21).
For mouse mRNA arrays between ischemic and nonischemic tissue, gastrocnemius muscle from BALB/cJ mice (n = 3 per group) was harvested at day 3 after HLI. Total RNA was extracted by the TRIzol total transcriptome isolation protocol. After quality control, RNAs were separated into 50 μg aliquots, and arrays were performed with the GeneChip mouse genome 430 2.0 array (Affymetrix, Santa Clara, CA).
Antibodies.
Bcl2 (cat. #2772), Bax (cat. #2876), and Caspase-3 (cat. #9661) antibodies were purchased from Cell Signaling and Technology (Danvers, MA). Actin antibody was purchased from Sigma (cat. #A2103; St. Louis, MO).
Western blots.
Tissue lysates from scrambled-antagomir-93-treated ischemic C57BL/6 mice and scrambled-miR-93 mimic-treated BALB/c mice [used in the microarray analysis (27)] were resolved on SDS-PAGE and immunoblotted for Bcl2, Bax, and Caspase-3.
Densitometry and statistics.
Densitometry of the immunoblots was performed by NIH ImageJ 1.4 analysis, and the values were plotted on Graph Pad Prism 6.0 for statistical analysis and bar graph presentation. P < 0.05 was considered statistically significant for western blot analysis.
Construction of PIN of immune response and arteriogenesis.
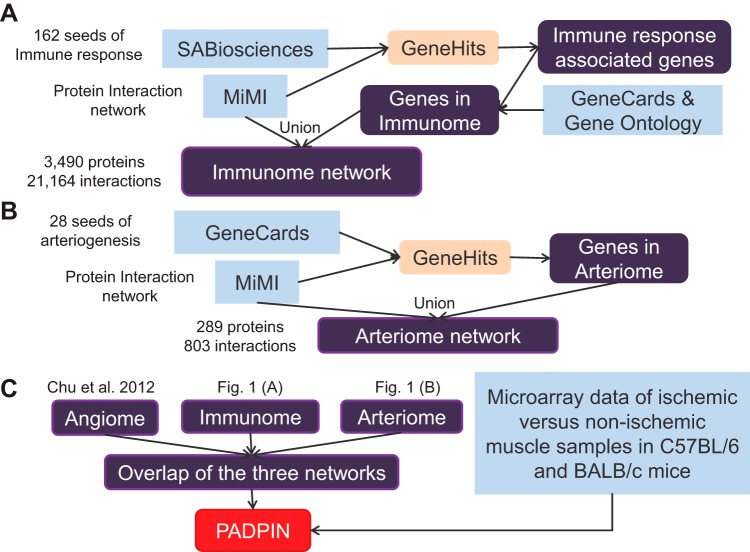
The flowchart of constructing the PADPIN is shown in Fig. 1. We have constructed a gene search engine GeneHits described in Ref. 12. Using GeneHits, we have constructed the angiome, the global PIN of angiogenesis, using three resources SABiosciences, Gene Ontology (GO), and GeneCards (54). The three databases are the resources for the proteins in PADPIN. The Michigan Molecular Interactions (MiMI) database (68) integrates 11 protein interaction data sources (BIND, CCSB, DIP, GRID, HPRD, IntAct, KEGG, MDC, MINT, PubMed, and Reactome). We included all 11 databases from MiMI plug-in 3.0.1 on Cytoscape 2.8 (63), which are the resources for interactions in PADPIN. The angiome comprises 1,233 proteins and 5,726 interactions; the flowchart for constructing the angiome is shown in Ref. 11. Following the same protocol described in Refs. 11 and 12, we used 162 immune response-associated proteins from SABiosciences as the query genes in GeneHits, together with the list of genes by searching the keyword “immune” in the GeneCards and GO “GO:0006955: immune system” to construct the immunome, the global PIN of immune response (Fig. 1A). There are no GO categories or SABiosciences annotations for arteriogenesis. To construct the global PIN of arteriogenesis (arteriome), we searched the keyword “arteriogenic” and “arteriogenesis” in GeneCards, which provides 28 query proteins (Fig. 1B). We used Cytoscape to plot the PINs (63).
Fig. 1.
Flowchart of constructing the immunome, arteriome, and protein-protein interaction networks of peripheral artery disease (PADPIN). Flowchart of construction of the immunome (A), arteriome (B), and PADPIN (C).
Mouse microarray data analysis.
Table 1 summarizes the four microarray datasets from mouse models of PAD (27). The gene expression in ischemic and nonischemic gastrocnemius muscle from C57BL/6 and BALB/c mice is from Ref. 27. We created a preclinical model performing unilateral femoral artery ligation; the 3-day postischemia time point was selected as the one where perfusion recovery was comparable between the strains. The recovery of blood flow after HLI at later time points in BALB/c mice is much poorer than in C57BL/6 mice (16). Global comparisons of microarray samples between the two different mouse strains and between ischemic and nonischemic muscle samples were not investigated in Ref. 27; this was accomplished in the present work. We used GenePattern (51) to compute the fold change, P value, and z-score. The datasets for each experimental condition in Table 1 include three samples. We set up the number of permutations as 0 to calculate asymptotic P values by the independent two-sided t-test in GenePattern (51). We generated the volcano plot of log2 (fold change ratio) vs. −log10 (P values) with Matlab. All the microarray datasets and simulations are freely provided on request.
Table 1.
Summary of four microarray studies from PAD mouse model
| Samples of mRNA Extraction | Samples, n | Description of Mouse Models |
|---|---|---|
| Ischemic gastrocnemius muscle samples from C57BL/6 mice | 3 | C57BL/6 mice recover remarkably well from HLI |
| Ischemic gastrocnemius muscle samples from BALB/c mice | 3 | recovery after HLI in BALB/c is much poorer than C57BL/6 mice |
| Nonischemic gastrocnemius muscle samples from C57BL/6 mice | 3 | C57BL/6 mice recover remarkably well from HLI |
| Nonischemic gastrocnemius muscle samples from BALB/c mice | 3 | recovery after HLI in BALB/c is much poorer than C57BL/6 mice |
Model is from Ref. 27. HLI, hindlimb ischemia.
Construction of the PIN of PAD and bioinformatics analysis.
We used the union of the three PINs (angiome, immunome, and arteriome) as the pool of genes in angiogenesis, inflammation, and arteriogenesis, respectively. The PIN of peripheral arterial disease (PADPIN) refers to the subsets of the union of three PINs, filtered by the microarray data of ischemic vs. nonischemic muscle samples in C57BL/6 and BALB/c mouse, respectively (Fig. 1C). GO provides the resource and terms of biological processes, cellular components, and molecular function in many different species (3). We used ToppGene https://toppgene.cchmc.org/ (9) to analyze GO molecular functions and biological processes of the differentially expressed genes in the mouse microarray data (27). We used gene set enrichment analysis (GSEA) to determine whether an a priori defined set of genes, e.g., corresponding to the proteins in angiome, immunome, and arteriome, shows statistically significant differences between the two phenotypes (66). The suggested default value for the threshold of false discovery rate (FDR) by GSEA is 25%.
Coexpressed genes in other muscle tissue samples.
We compare the differentially expressed genes as the query to retrieve coexpressed genes in other microarray data from muscle samples. The algorithm is based on SEEK (Search-based Exploration of Expression compendium) http://seek.princeton.edu/ (73). SEEK gathers 5,210 microarray datasets and RNA-Seq platforms from various tissues such as cancer cells, skeletal muscle, and endothelial cells. We choose “muscle (noncancer)” in SEEK to refine the results with the microarray data from other muscle samples. The weight of each microarray dataset is determined by the number of coexpressed genes in that dataset, so the dataset with higher weight represents more relevance to the query genes. The P value is computed by random coexpressed edges from a random set of genes of the same size.
RESULTS
Differences in perfusion recovery and cell death between two mouse strains at day 3 post-HLI.
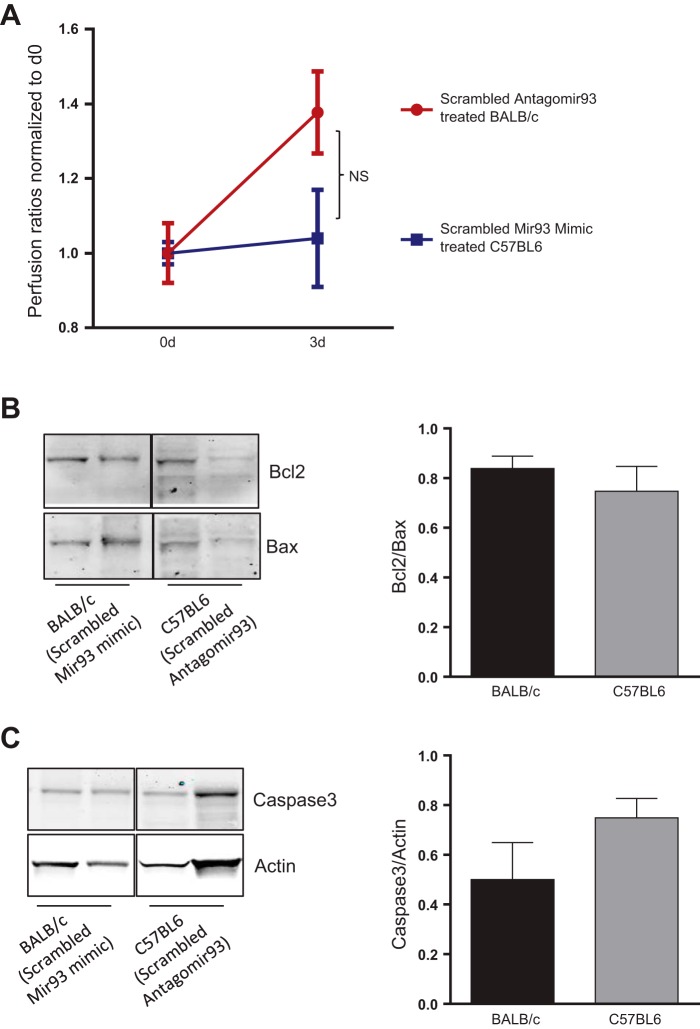
Though there are well-established differences in outcomes between inbred mouse strains, the time course following surgical HLI can be informative. We have used the strategy to select time points following HLI when outcomes are similar (27). To compare array data we wanted to verify that outcomes between the C57BL/6 and BALB/c were similar at the selected time point. Indeed, no significant differences in the extent of perfusion recovery between scrambled miR-93 mimic-treated BALB/c (day 0: 23.43 ± 1.97, day 3: 32.28 ± 2.8 means ± SE) and scrambled antagomir-93-treated C57BL/6 (day 0: 28.48 ± 0.99, day 3: 29.67 ± 3.73 means ± SE) at day 3 post-HLI were observed (P = 0.52). Furthermore, there were no significant differences in Bcl2/Bax ratios (P = 0.49) or activation of caspase-3 (P = 0.17) between scrambled miR-93 mimic-treated BALB/c and scrambled antagomir-93-treated C57BL/6 gastrocnemius muscle samples day 3 post-HLI (Fig. 2).
Fig. 2.
Measurement of perfusion ratio and Western blots. A: perfusion ratios of scrambled antagomir-93-treated C57BL/6 and scrambled microRNA (miR)-93 mimic-treated BALB/c hinlimb ischemia (HLI) at day 3; n = 11–12 per each group. B: Western blot analysis of Bcl2/Bax ratios in scrambled antagomir-93-treated C57BL/6 and scrambled miR-93 mimic-treated BALB/c HLI muscle lysates at day 3. C: Western blot analysis of activated caspase-3 in scrambled antagomir-93-treated C57BL/6 and scrambled miR-93 mimic-treated BALB/c HLI muscle lysates at day 3; n = 5 per each group. No significant differences in perfusion ratios, Bcl2/bax ratios, or activated caspase-3 levels were observed between both strains at day 3 post-HLI.
Construction of the angiome, immunome, and arteriome.
We have used our machine learning algorithm GeneHits to construct the global PIN of angiogenesis (angiome) comprising 1,233 proteins and 5,726 interactions (12). In this study, we constructed the global PIN of immune response and arteriogenesis as immunome and arteriome, respectively (Fig. 1, A and B). The immunome comprises 3,490 proteins and 21,164 interactions (Supplemental Table S1).1 We refer to immune response as “inflammation” in this study. The number of genes in the immunome may be more than the genes annotated as inflammation; however, this large set of genes in the immunome allows us to have a large pool of genes related to inflammation and that overlap with angiogenesis and arteriogenesis in PAD. The arteriome comprises 289 proteins and 803 interactions (Supplemental Table S2). We compared the three sets in a Venn diagram (Fig. 3) and list the overlapped genes among the three sets in Supplemental Table S3. The high overlap of genes between angiome and immunome might reflect the regulation by endothelial cells of various biological processes, including both vascular and immune response (15).
Fig. 3.
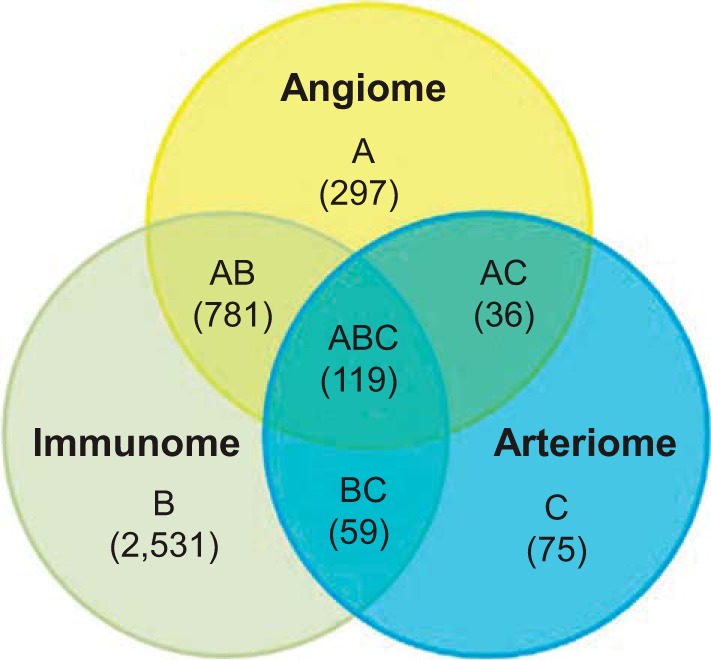
Venn diagram for angiome, immunome, and arteriome. The numbers in the Venn diagram indicate the numbers of proteins in each protein-protein interaction network (PIN).
Differentially expressed genes in mouse PAD model.
We then systematically identified differentially expressed genes from the four microarray datasets in Table 1 by GenePattern 3.6.1 (51). First, we compared the gene expression transcripts from ischemic vs. nonischemic gastrocnemius muscle from C57BL/6 mice, as an example of a strain with a favorable/desirable response. Then we compared the gene transcripts from ischemic vs. nonischemic gastrocnemius muscle from BALB/c mice, as an example of a strain with a nonfavorable/undesirable response. Next, we compared the differential gene expression of ischemic samples between C57BL/6 and BALB/c mice. Finally, we compared the differential gene expression of nonischemic samples between C57BL/6 and BALB/c mice.
We sorted all 21,747 genes by fold change in each experimental condition of the four comparisons (Supplemental Table S4). The gene name, z-score, P value, fold change, and mean and SD of all the genes in each experimental condition of the four comparisons are provided in Supplemental Table S4. We constructed a volcano plot of log2 (fold change ratio) vs. −log10 (P values) from the differentially expressed genes between ischemic vs. nonischemic C57BL/6 in Fig. 4. We identified the differentially expressed genes that are included in the angiome, immunome, and arteriome, with the absolute fold change ranked as top 5% of all the genes and P value <0.05; the list of differentially expressed genes between ischemic vs. nonischemic gastrocnemius muscle in C57BL/6 and BALB/c mice is provided in Supplemental Table S5. Many of the differentially expressed genes in Supplemental Table S5 are in the immunome, i.e., within the areas B, AB, BC, or ABC in the Venn diagram in Fig. 3. We present the top five up- and downregulated genes with the highest absolute fold change under the statistical significance P value <0.05 in Table 2, A and B, respectively. For example, the anti-angiogenic protein thrombospondin-1 (THBS1) is identified by its upregulated fold change as 1.66 with the P value 0.0418. Notably, the next four proteins on the list belong to the immunome, but not to angiome or arteriome. Among the downregulated genes, the top one that belongs to both angiome and immunome is MAP2K4, a known signal transduction molecule. We also list the top five up- and downregulated genes between ischemic and nonischemic BALB/c mice in Table 2, C and D, respectively. For example, toll-like receptor 4 (TLR4) is identified with the upregulation fold change 1.614 and P value 0.004. It is noteworthy that EPHA4 (Ephrin receptor A4) is in the angiome/immunome/arteriome intersection.
Fig. 4.
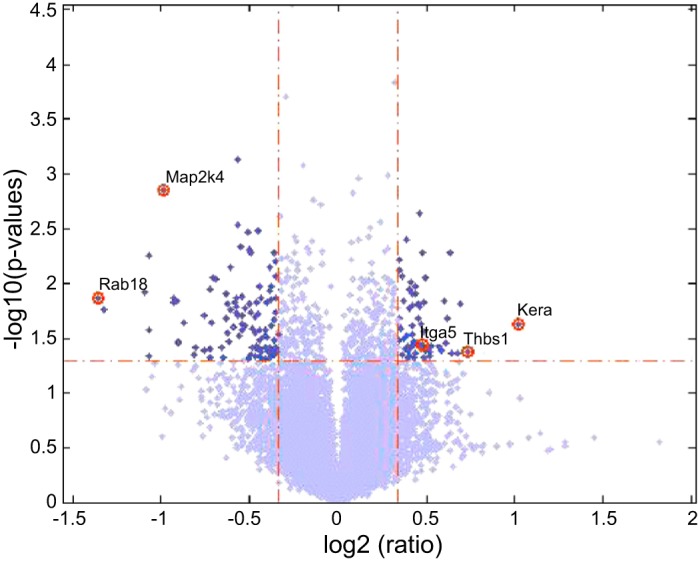
Volcano plot of the microarray data. The volcano plot of log2 (fold change ratio) as x-axis vs. −log10 (P values) as y-axis from the differentially expressed genes between ischemic vs. nonischemic C57BL/6.
Table 2.
Top five up- and downregulated genes in ischemic vs. nonischemic C57BL/6 and BALB/c mice
| Gene Name | Description | P Value | Fold Change | Venn Diagram (Fig. 3) |
|---|---|---|---|---|
| A: Upregulated genes ischemic vs. nonischemic C57BL/6 mice | ||||
| THBS1 | thrombospondin 1 | 0.042 | 1.660 | AB (angiome + immunome) |
| RAPGEF6 | Rap guanine nucleotide exchange factor (GEF) 6 | 0.035 | 1.523 | B (immunome) |
| TPP2 | tripeptidyl peptidase II | 0.040 | 1.502 | B (immunome) |
| UBE2D2 | ubiquitin-conjugating enzyme E2D 2 | 0.037 | 1.437 | B (immunome) |
| ATR | ataxia telangiectasia and Rad3 related | 0.046 | 1.432 | B (immunome) |
| B: Downregulated genes ischemic vs. nonischemic C57BL/6 mice | ||||
| TSPAN7 | tetraspanin 7 | 0.045 | −2.100 | B (immunome) |
| MAP2K4 | mitogen-activated protein kinase kinase 4 | 0.001 | −1.984 | AB (angiome + immunome) |
| KIF1C | kinesin family member 1C | 0.034 | −1.866 | B (immunome) |
| IL15RA | interleukin 15 receptor, alpha | 0.023 | −1.823 | B (immunome) |
| RBL2 | retinoblastoma-like 2 (p130) | 0.045 | −1.761 | B (immunome) |
| C: Upregulated genes ischemic vs. nonischemic BALB/c mice | ||||
| TLR4 | toll-like receptor 4 | 0.004 | 1.614 | B (immunome) |
| KCTD12 | potassium channel tetramerization domain containing 12 | 0.026 | 1.560 | C (arteriome) |
| SLC2A3 | solute carrier family 2 (facilitated glucose transporter), member 3 | 0.009 | 1.526 | AB (angiome + immunome) |
| EPHA4 | EPH receptor A4 | 0.012 | 1.521 | ABC (angiome + immunome + arteriome) |
| BAG4 | BCL2-associated athanogene 4 | 0.029 | 1.517 | B (immunome) |
| D: downregulated genes ischemic vs. nonischemic BALB/c mice | ||||
| SLC22A4 | solute carrier family 22 (organic cation/zwitterion transporter), member 4 | 0.024 | −1.528 | B (immunome) |
| TPP2 | tripeptidyl peptidase II | 0.037 | −1.43 | B (immunome) |
| FBXW11 | F-box and WD repeat domain containing 11 | 0.005 | −1.408 | B (immunome) |
| NR2C1 | nuclear receptor subfamily 2, group C, member 1 | 0.018 | −1.391 | B (immunome) |
| KSR1 | kinase suppressor of ras 1 | 0.041 | −1.366 | B (immunome) |
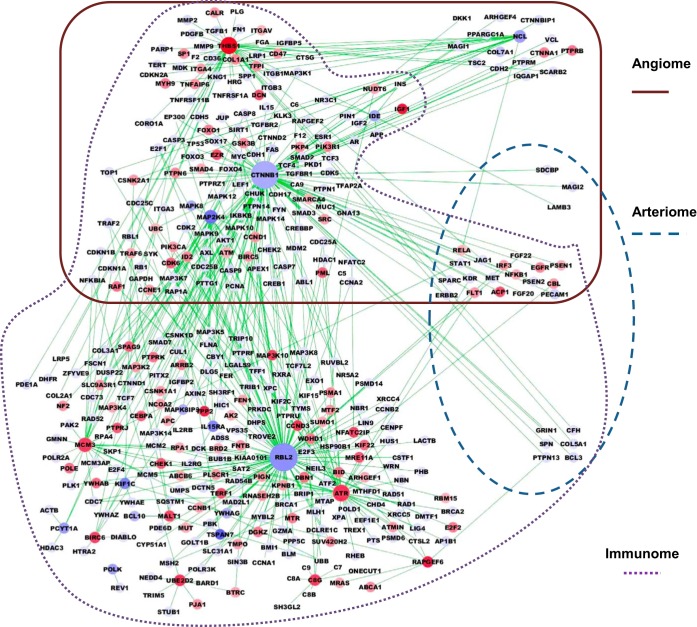
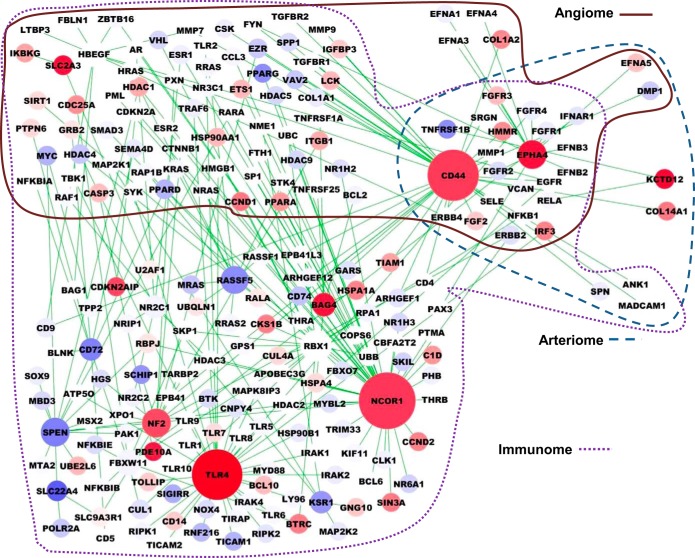
To visualize the PADPIN, we selected the top 10 upregulated and top 10 downregulated genes with the highest absolute fold change under the statistical significance P value < 0.05 between ischemic vs. nonischemic gastrocnemius muscle in C57BL/6 and BALB/c mice, respectively (Supplemental Table S5). We selected the proteins in the angiome, immunome, and arteriome that are linked to the 20 proteins with the most differentially expressed gene expression in C57BL/6 and BALB/c mouse to construct the representative subnetwork of PADPIN in Figs. 5 and 6, respectively. The color of nodes from red to blue represents the gene expression level from high to low. The size of nodes from large to small represents the degree of nodes, i.e., the number of interactions, from high to low. Several proteins that are linked to the proteins (i.e., genes) in Supplemental Table S5 but absent in this table are shown in the figures, such as CD47 linked to THBS1 in angiome (Fig. 5) and TLR7 linked to TLR4 in immunome (Fig. 6). The total number of proteins in Fig. 5 (C57BL/6) is 395, which is more than the number of proteins in Fig. 6 (BALB/c) as 215; this difference is due to the interactions in the angiome, immunome, and arteriome. The numbers of proteins in the PADPIN of C57BL/6 (Fig. 5) in the Venn diagram A, B, C, AB, BC, AC, and ABC (Fig. 3) are 15, 207, 0, 146, 6, 3, and 18, respectively. The numbers of proteins in the PADPIN of BALB/c (Fig. 6) in the Venn diagram A, B, C, AB, BC, AC, and ABC (Fig. 3) are 4, 112, 2, 70, 3, 2, and 22, respectively. The ratio of the number of proteins annotated in angiome and immunome to the total 395 proteins (C57BL/6, Fig. 5) is 46% (= 182/395) and 95% (= 377/395) in PADPIN of BALB/c, and 45 and 96% in PADPIN of C57BL/6, respectively. This ratio might reflect the fact that the number of proteins in immunome is more than in angiome, and both PINs are larger than arteriome. The distinct visualization of PADPIN between the two PADPINs in Figs. 5 and 6 clearly shows the diverse molecules involved in ischemic vs. nonischemic tissues between the two mouse strains; we will specifically compare the gene expression between the two mouse strains in the following paragraph.
Fig. 5.
Subnetwork of PADPIN for C57BL/6 mouse data. PIN generated by the top 10 up- and top 10 downregulated genes in ischemic vs. nonischemic muscles in C57BL/6 mouse comprises 395 proteins and 428 interactions. The color of nodes from red to blue represents the gene expression level from high to low. The size of nodes from large to small represents the degree of nodes, i.e., the number of interactions, from high to low.
Fig. 6.
Subnetwork of PADPIN for BALB/c mouse data. PIN generated by the top 10 up- and top 10 downregulated genes in ischemic vs nonischemic muscles in BALB/c mouse comprises 215 and 225 interactions. Same notation as in Fig. 5.
Differentially expressed genes between two inbred mouse strains.
The BALB/c mice show much poorer perfusion recovery after HLI following ligation of the femoral artery. We further compare the differential gene expression between C57BL/6 vs. BALB/c mice of ischemic and nonischemic muscles, respectively, with absolute fold change value ranked as top 5% of all the genes and P value < 0.05 in Supplemental Table S6. There are 197 and 175 differentially expressed genes between the two mouse strains in ischemic and nonischemic muscles, respectively. Even under normal conditions in nonischemic muscles, the 69 up- and 106 downregulated gene expressions in Supplemental Table S6 strongly suggest the highly diverse gene expression profiles between the two mouse strains. We show the top five up- and downregulated genes between C57BL/6 vs. BALB/c mice of ischemic muscles in Table 3, A and B, and nonischemic muscle in Table 3, C and D, respectively. We used Toppgene (9) for the gene list enrichment analysis of these differentially expressed genes between the two strains (Supplemental Table S7). Supplemental Table S7 shows that the statistically significant GOs and pathway analysis between two mouse strains in ischemic tissues include cell proliferation (GO:0008283, P value = 8.73E-18), positive regulation of immune system process (GO:0002684, P value = 1.15E-09), negative regulation of apoptotic process (GO:0043066, P value = 1.29E-09), mTOR signaling pathway (P value = 8.08E-08), insulin signaling (P value = 2.29E-07), and PI3K-Akt signaling pathway (P value = 3.08E-06). The statistically significant GOs and pathway analysis between two mouse strains in nonischemic tissues include cell proliferation (GO:0008283, P value = 9.08E-19), apoptotic process (GO:0006915, P value = 1.90E-11), vasculature development (GO:0001944, P value = 4.73E-11), pathways in cancer (P value = 4.49E-07), insulin signaling (P value = 6.75E-07), and PI3K-Akt signaling pathway (P value = 2.02E-05). The GO and pathway analysis show the different regulation of cell proliferation, apoptosis, vasculature, and immune responses between the C57BL/6 and BALB/c mice.
Table 3.
Top five up- and downregulated genes in C57BL/6 vs. BALB/c ischemic and nonischemic muscle samples
| Gene Name | Description | P Value | Fold Change | Venn Diagram (Fig. 3) |
|---|---|---|---|---|
| A: Upregulated genes C57BL/6 vs. BALB/c ischemic muscle samples | ||||
| PRKAA2 | protein kinase, AMP-activated, alpha 2 catalytic subunit | 0.006 | 43.245 | A (angiome) |
| FGFBP1 | fibroblast growth factor binding protein 1 | 0.011 | 40.621 | AC (angiome + arteriome) |
| MBD2 | methyl-CpG binding domain protein 2 | 0.003 | 40.568 | B (immunome) |
| PTTG1 | pituitary tumor-transforming 1 | 9.56E-04 | 33.334 | AB (angiome + immunome) |
| RAC57BL/6B | RAC57BL/6B, member RAS oncogene family | 0.026 | 33.078 | B (immunome) |
| B: Downregulated genes C57BL/6 vs. BALB/c ischemic muscle samples | ||||
| EIF2S1 | eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa | 8.00E-04 | −32.648 | B (immunome) |
| GBP1 | guanylate binding protein 1, interferon-inducible | 0.03 | −15.790 | B (immunome) |
| PLAU | plasminogen activator, urokinase | 0.001 | −12.673 | AB (angiome + immunome) |
| ATP1A2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | 0.003 | −10.270 | B (immunome) |
| SPON1 | spondin 1, extracellular matrix protein | 0.007 | −4.244 | AB (angiome+immunome) |
| C: Upregulated genes C57BL/6 vs. BALB/c nonischemic muscle samples | ||||
| PRKAA2 | protein kinase, AMP-activated, alpha 2 catalytic subunit | 0.012 | 73.065 | A (angiome) |
| MBD2 | methyl-CpG binding domain protein 2 | 0.003 | 50.154 | B (immunome) |
| FGFBP1 | fibroblast growth factor binding protein 1 | 0.002 | 43.532 | AC (angiome+arteriome) |
| PTTG1 | pituitary tumor-transforming 1 | 0.001 | 28.310 | AB (angiome+immunome) |
| RAC57BL/6B | RAC57BL/6B, member RAS oncogene family | 0.007 | 27.804 | B (immunome) |
| D: Downregulated genes C57BL/6 vs. BALB/c nonischemic muscle samples | ||||
| EIF2S1 | eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa | 0.041 | −22.259 | B (immunome) |
| GBP1 | guanylate binding protein 1, interferon-inducible | 0.041 | −15.149 | B (immunome) |
| PLAU | plasminogen activator, urokinase | 0.002 | −14.248 | AB (angiome + immunome) |
| DMP1 | dentin matrix acidic phosphoprotein 1 | 0.042 | −12.464 | AC (angiome + arteriome) |
| ATP1A2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | 7.29E-04 | −10.203 | B (immunome) |
We applied GSEA (66) to determine whether an a priori defined set of genes shows statistically the significant differences between two phenotypes, e.g., C57BL/6 and BALB/c mice; the Kolmogorov-Smirnov test was used to determine the significance of the enrichment. The significant FDR q-value could indicate that the proteins in the angiome, immunome, or arteriome are disproportionally ranked at either the head or tail of the ranked list. By comparing the gene transcripts between C57BL/6 with BALB/c ischemic muscles, we computed the q-value as 0.085, 0.089, and 0.276 in angiome, immunome, and arteriome, respectively. The set of angiogenesis and immune response-related proteins was significantly perturbed in ischemic tissues between the two mouse strains by the computed q-values, but the set of arteriogenesis-related proteins was not. The q-values for the nonischemic muscles between the two mouse strains are 0.305, 0.313, and 0.591 in angiome, immunome, and arteriome, respectively. Neither the angiogenesis-, immunity-, nor arteriogenesis-related proteins were significantly perturbed in nonischemic tissues between the two mice strains.
Comparison with other microarray datasets from muscle samples.
The microarray data in our analysis are from the gastrocnemius muscles (27), which contain the skeletal myocytes, fibroblasts, inflammatory cells, and endothelial cells. This heterogeneous bulk tissue reflects the complexity of different cells in the mouse PAD model, which is the limit of our bioinformatics analysis. To overcome this difficulty, we compared the coexpressed genes with the microarray data from other muscle samples. We used the list of up- and downregulated genes between ischemic and nonischemic tissues of the two mouse strains (Supplemental Table S5) as the input query to SEEK http://seek.princeton.edu/. We compared the differentially expressed genes in our PAD model with other microarray datasets from muscle tissues by the coexpression genes of the query. This analysis can help identify cell-specific genes and compare with other microarray data from muscle samples, which are not specific to PAD.
The output of query-weighted dataset of the SEEK analysis is shown in Supplementary Table S9, which is ranked by the query relevance. The output of the coexpressed genes is in Supplementary Table S10, which is ranked by the coexpression score. We found that three most relevant datasets are GSE21212 (5), GSE13791 (13), and GSE13736 (65) based on endothelial cells and fibroblasts. The coexpressed genes in the endothelial cell-specific dataset (GSE13736) (65) support our analytical results that our bioinformatics results from the gastrocnemius muscle samples reveal angiogenesis-related genes.
DISCUSSION
The major goal of this bioinformatics study was to identify potential targets to treat PAD by comparing the sets of genes (between better recovery and worse recovery mouse strains) that can play critical roles in modulating angiogenesis, arteriogenesis, and inflammation. We observed a high level of overlap between angiome and immunome (Fig. 3), indicating extensive cross talks between systemic immune responses and vasculature in ischemic hindlimb. We observed that there are 69 up- and 106 downregulated genes at basal levels (nonischemic) in BALB/c and C57BL/6 mice, respectively (Supplemental Table S6). This suggests highly diverse gene expression changes in the hindlimb between the two mouse strains. It is well known that genetic differences between C57BL/6 and BALB/c play critical roles in their recovery from ischemic hindlimb injury (16, 19, 57). Differences in these genes at their basal level can strongly predispose these mouse strains either to adapt and to recover better or to become more susceptible and recover more poorly from ischemic hindlimb injury. The contribution of these genes in the apparent ability of C57BL/6 vs. BALB/c to adapt and recover from acute ischemic leg injury by promoting arteriogenesis or angiogenesis or by modulating immune responses needs to be further investigated.
The driving stimuli of arteriogenesis include shear stress and inflammation, whereas angiogenesis is driven by the hypoxic environment that develops due to the occlusion in PAD (1). The process of pre-existing network of arterioles to form functional collaterals to bypass vascular obstruction is referred to as arteriogenesis, which is distinguished from angiogenesis (69). The hemodynamic factors and biochemical signaling in the arteriolar network that contribute to functional collaterals are not yet clear (58). A dysfunctional arteriolar network has been observed in diabetic peripheral artery obstructive disease with critical limb ischemia. The inability of the arteriolar network to promote an effective VEGF-driven arterial wall neoangiogensis leads to increased arterial damage (48). These reports clearly indicate that the blood flow disturbances at the site of occlusion can lead to significant changes in gene expression as well as in the phenotypical changes in the vessel wall. More work is needed to understand the signaling events that can separate arteriogeneic signaling events from angiogenic signaling events in ischemic diseases.
Differences between the two selected mouse strains.
BALB/c strain has fewer native collaterals in hindlimb than C57BL/6, which are associated with greater reduction in perfusion immediately after femoral ligation, slower recovery of perfusion, and worse ischemia (7). However, perfusion scores and capillary density are similar between the two strains at day 3 post-HLI (56). A clear demarcation in the extent of perfusion recovery between the two strains was apparent by day 7. In our current study, choosing day 3 post-HLI samples is very critical. By day 7 post-HLI, several dramatic changes in blood flow and cell death occur between these two strains, making it difficult to delineate the network interactions that are associated with changes in blood flow and cell death signaling cascades. In addition to baseline differences in collaterals, cell death signaling pathways and immune cell interactions might be different between C57BL/6 and BALB/c mice. To overcome this limitation, we chose the mice that have similar perfusion scores and cell death evident by no differences in Bcl2/Bax ratios or caspase-3 activation at day 3 post-HLI for microarray analysis (27) (Fig. 2). This allows us to perform microarray analysis between two mouse stains that are relatively similar in perfusion flow recovery and tissue necrosis/cell death post-HLI and make our microarray data analysis between these two mouse strains more robust. Hence, we expect that even though both strains differ largely at baseline in collaterals, using the samples at day 3 post-HLI along with their respective nonischemic hindlimb controls greatly increase the strength of the data.
Using different methodologies, investigators have shown the causative genetic variations to angiogenesis and arteriogenesis between the mouse strains to be linked to Lsq1 and Candq1 (now called determinant of collateral extent) quantitative trait loci on chromosome 7 (16, 59). The current study is the first to compare the differentially expressed genes between two mouse strains of ischemic and nonischemic muscle samples. We notice that the seven specific genes in Candq1 (Rabep2, Sh2b1, Cln3, Apobr, Il27, MapK3, and Ppp4c), which are related to endothelial cells (59), are not listed as differentially expressed genes in Supplemental Table S6. We identified differentially expressed genes between the two mouse strains based on the microarray data analysis, and we correlated them with the differences in collateral blood flow.
Target identification for therapeutic angiogenesis.
One of the limitations for the study is the unavailability of the gene expression data from muscles of human PAD patients; the available data are from diseased arteries or blood cells. The microarray data in Ref. 18 collected 30 femoral arteries, including 11 intermediate lesions, 14 advanced lesions, and 5 normal femoral arteries. The samples of microarray data in Ref. 14 were harvested from the thromboendarterectomy specimens of common femoral artery in 20 patients with PAD. The peripheral blood mononuclear cells were isolated by layering the blood samples over histopaque (42). To overcome these limitations, we compared the differentially expressed genes between ischemic vs. nonischemic C57BL/6 and BALB/c mice with the three microarray datasets in PAD patients (14, 18, 42). We notice that all these tissues are very different from gastrocnemius muscles used in Hazarika et al. (27). While limitations exist in all animal models, factors that adversely limit the extent of perfusion recovery or increase the degree of tissue loss in humans have parallel effects in mouse models of PAD. We performed bioinformatics analysis to explore these datasets and compare differential gene expression between ischemic and nonischemic C57BL/6 and BALB/c mice with three PAD patients' microarray sets; this analysis gave us important biological insights to predict new targets of therapeutic angiogenesis.
In some cases, gene expression changes in the muscle can also be detected in peripheral blood cells. We compared the list of differentially expressed genes from mice with peripheral blood mononuclear cells from human PAD patients (42) using Gene Expression Omnibus ID accession number GSE27034. Table 4 lists the fold change values for genes in angiome and immunome from each study. We observed the general similarity of upregulated genes between mouse and human samples, but some changes have opposite directions of fold change. The diverse microarray protocol, platforms, and tissue resources may limit the comparability in Table 4. We also notice that the fold change (PAD/control) of TLR4 in Croner et al. (14) is 40.57, which is much higher than any other microarray data.
Table 4.
Comparison of the fold change of genome-wide differentially expressed genes between ischemic vs. nonischemic muscles from mice (Supplemental Table S5) with samples from human PAD patients
| Tissue |
|||||
|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Current Study: Mouse Gastrocnemius Muscles | Fu (18): Human Femoral Arteries | Croner (14): Human Common Femoral Artery | Masud (42): Human PBMCs |
| A: Upregulated genes of ischemic vs. nonischemic C57BL/6 mice (Supplemental Table S5) | |||||
| BTG1 | B-cell translocation gene 1, anti-proliferative | 1.337564 | 2.022 | ||
| MYO1F | myosin IF | 1.313371 | 2.558 | ||
| THBS1 | thrombospondin 1 | 1.659983 | 4.575 | 2.12 | |
| TLR4 | toll-like receptor 4 | 1.36603 | 40.571 | 1.82 | |
| B: Downregulated genes of ischemic vs. nonischemic C57BL/6 mice (Supplemental Table S5) | |||||
| CYCS | cytochrome c, somatic | −1.36222 | −1.484 | ||
| OGT | O-linked N-acetylglucosamine (GlcNAc) transferase | −1.30688 | −1.440 | ||
| C: Upregulated genes of ischemic vs. nonischemic BALB/c mice (Supplemental Table S5) | |||||
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma | 1.290306 | −2.509 | ||
| COL1A2 | collagen, type I, alpha 2 | 1.269932 | 2.342 | ||
| EBF1 | early B cell factor 1 | 1.334283 | −1.5 | ||
| KCTD12 | potassium channel tetramerization domain containing 12 | 1.560756 | 1.623 | ||
| LAMB1 | laminin B1 | 1.30851 | 3.575 | ||
| SLC2A3 | solute carrier family 2 (facilitated glucose transporter), member 3 | 1.526328 | 5.753 | ||
| TLR4 | toll-like receptor 4 | 1.614745 | 40.571 | 1.82 | |
| D: Downregulated genes of ischemic vs. nonischemic BALB/c mice (Supplemental Table S5) | |||||
| HBEGF | heparin-binding EGF-like growth factor | −1.33892 | 2.032 | ||
PAD, peripheral artery disease; PBMC, peripheral blood mononuclear cell.
In Table 4, we notice that THBS1 is upregulated in the ischemic vs. nonischemic muscle samples of C57BL/6 mice (27), human femoral arteries (18), and human peripheral blood mononuclear cells (42). A previous study of Isenberg et al. (32) shows that the whole tissue cAMP levels in THBS1 null mice are elevated. Yao et al. (72) show that THBS1 can limit cAMP-mediated events in vascular smooth muscle cells (VSMC) and arterial rings. In PAD patients, the basal muscle dialysate protein level of THBS1 with exercise and passive movement was higher than healthy control subjects (28). Inhibition of THBS1/CD47 signaling has also shown to promote tissue recovery and survival rate in aged and diet-induced vasculopathy in experimental animal models of PAD (30). We predict that the inhibition of THBS1 is a potential therapeutic angiogenesis target for PAD patients. Nitric oxide (NO) modulates blood flow and tissue perfusion by relaxing and dilating the arteries. THBS1, through its cell surface receptor CD47, limits the availability of NO and thus decreases tissue blood flow and perfusion (53). Blockage of THBS1-CD47 signaling pathways could alleviate tissue ischemia in the animal models, such as enhancement of ischemic tissue survival in a porcine model (33) and prevention of necrosis of full thickness skin grafts (31). THBS1 has also been proposed as the potential biomarker of PAD (62). Thus, we propose that THBS1 and its receptors CD47 or CD36 are the therapeutic targets for PAD.
Following the identification of THBS1 as a potential novel target in PAD, we examine other molecules that are associated with THBS1 (Supplemental Table S8). None of the four genes CD47 (leukocyte surface antigen CD47), F2 (prothrombin), SDC4 (syndecan-4), and CD36 (platelet glycoprotein 4) under the GO: 0070053 thrombospondin receptor activity is up- or downregulated. Binding of THBS1 to CD47 inhibits NO signaling by inhibiting the cGMP synthesis (29). We found the 93 genes involved in NO pathway (Supplemental Table S8), based on GO:0007263 NO-mediated signal transduction and SABiosiences NO signaling pathway http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-062A.html. In addition to THBS1, nitric oxide synthase 1 exhibits upregulation with the fold change 5.173 and P value = 0.038 between the ischemic tissues of C57BL/6 and BALB/c mice. We did not find any other genes involved in the NO pathway between ischemic vs. nonischemic tissues in the two mouse strains. THBS1 is also a major activator of transforming growth factor-β1, which mediates cell proliferation, wound healing, and the immune response (40). We found 429 genes in GO:0008283 cell proliferation (Supplemental Table S8). The upregulated genes in ischemic vs. nonischemic muscle samples of C57BL/6 mice involved in cell proliferation include BIRC6, WWTR1, and HIPK2. The downregulated genes in ischemic vs. nonischemic muscle samples of C57BL/6 mice include CLCF1, PTK2, DLG1, FGFR1, and MDM2.
Tetraspanin-7 is the top gene that is downregulated in ischemic C57BL/6 muscle compared with nonischemic. Tetraspanin-7 belongs to tetraspanin family and has been shown to be involved in HIV-1 infection (55). It has also been shown to interact with PrPC (24). The role of Tetraspanin-7 in vascular biology or in PAD is not known. In addition to THBS1, we notice that TLR4 is listed in the top 5% of upregulated genes in Supplemental Table S5 (fold change = 1.366, P value = 0.036) in ischemic vs. nonischemic C57BL/6 mice (Supplemental Table S5) and the top gene that is upregulated in ischemic BALB/c muscle compared with nonischemic, which are also listed in Croner et al. (14) and Masud et al. (42). TLR4 plays the pivotal role in inflammation and neuronal apoptosis of cerebral ischemia, and inhibition of TLR4 can reduce cerebral ischemia-induced shear stress (67). TLR4 was overexpressed in the serum samples from PAD patients after treatment with anti-β2-glycoprotein I (ABGPI) antibodies (70). SLC22A4 [solute carrier family 22 (organic cation/zwitterion transporter), member 4] is the top gene that is downregulated in BALB/c ischemic muscle compared with nonischemic. The role of SLC22A4 in vascular biology or ischemic diseases is not yet clear (61). PRKAA2a (AMPKa2) is the top gene that was upregulated in the C57BL/6 vs. BALB/c ischemic and nonischemic muscle comparison. PRKAA2a has been shown to have a great impact as a metabolic sensor in skeletal muscle (45). Exercise intensity has been shown to be correlated with the increasing activity of PRKAA2a in human skeletal muscle (10). Furthermore, activation of PRKAA2a significantly improved the exercise performance and vascular insufficiency in high-fat diet-fed mice (4). PRKAA2a gene variants are associated with total cholesterol and serum lipoproteins in normal female Caucasians (64). Interestingly, increased cAMP levels were well known to activate PRKAA2a (47), and platelet-derived TSP-1 has been demonstrated to block PGE2-induced cAMP2 levels (52). Furthermore, skeletal muscle, VSMC, and cardiac tissue from TSP-1 null mice have significantly higher basal cAMP levels (32, 72). Hence, it can be hypothesized that increased TSP-1 levels can decrease cAMP levels, which can decrease PRKAA2a activation and the angiogenic state in ischemic muscle (34, 60). These reports point toward more complex TSP-1, PRKAA2a and cAMP interactions that are occurring between immune cells and vasculature in ischemic hind limb. EIF2S1 (eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa) is the top gene that is down-regulated between C57BL/6 vs. BALB/c ischemic and nonischemic muscle. Phosphorylation of EIF2S1 has been shown to play prominent roles in cerebral ischemia (20). The functional role of EIF2a in PAD is not yet clear.
Interestingly, EphA4 was the only gene at the intersection of arteriome, angiome, and immunome in Table 2. The Ephrin family is a large family of receptor tyrosine kinases that play critical roles in angiogenic remodeling of blood and lymphatic vessels (38), axonal guidance and cortical development (46), and tumor angiogenesis (49). EphA4-deficient mice exhibits abnormal blood-spinal barrier characteristics, displaying a loss of tight association of astrocytes with blood vessels and leaky blood-spinal barrier (22). EphA4 is expressed on both small and large blood vessels correlating with our angiome and arteriome intersection in the bioinformatics analysis. Furthermore, EphA4 has also been shown to modulate adhesion of monocytes to endothelial cells in human atherosclerosis plaques (35). EphA4 constitutively associates with αIIbβ3 integrin in platelets, and activated platelets upregulate the surface expression of EphA4 (50). Taken together, the functional and expression profiles of EphA4 closely match the arteriome, angiome, and immunome intersection in our bioinformatic analysis. Since it has been shown that blocking EphA4 decreases angiogenesis (25), understanding the biochemical signaling events between proangiogenic EphA4 and antiangiogenic THBS1 in activated platelets in the pathologies of thrombosis and atherosclerosis in PAD is important and interesting. The bidirectional cross talk of platelets and vascular wall in the ischemic environment involving EphA4 and THBS1 is intriguing.
Tissues from the gastrocnemius muscles.
The sample tissue in our microarray data is from the gastrocnemius muscle instead of abductor muscle. This might be one of the major reasons that we obtained a limited number of proteins related to arteriogenesis in the microarray analysis and PADPIN (Fig. 5 and 6). The main reason for choosing the gastrocnemius muscle in the microarray is that the abductor region of thigh is well known to be very heterogeneous, even within each mouse, making the predictions variable and limited. It has been shown that lack of angiogenesis in the distal part of the leg, i.e., gastrocnemius and tibialis anterior, will not result in efficient perfusion recovery (26, 44). Therefore, using gastrocnemius muscle in this study has the advantage of identifying the gene changes in the distal muscle that are critical for perfusion recovery and will provide more clinically relevant information to promote perfusion recovery in PAD.
RNA isolated from gastrocnemius muscle is overwhelmingly from skeletal muscle fibers, fibroblasts, smooth muscle cells, and immune cells infiltrating the ischemic tissue. It is difficult to separate cell-specific molecular changes with our previous methodology (27). We do not know the contributions of different cells and their impact on the microarray data in the bulk human tissue. Laser microdissection could provide a solution, but in most cases it is not feasible. We provide the computational simulations using SEEK (73) in Supplementary Tables S9 and S10, which show that we were able to isolate the signals from endothelial cells in gastrocnemius muscles. The use of different tissues, e.g., the thigh and peripheral blood mononuclear cells, to identify the molecular changes should be very valuable and informative in future studies.
Conclusions
By using the machine learning tools, protein-protein interaction databases, microarray study, and GO analysis, we constructed global PIN of angiogenesis, immune response, and arteriogenesis; identified novel genes; and predicted therapeutic targets that play critical roles in peripheral arterial disease. This study provides bioinformatics predictions for future experimental validation in peripheral arterial disease, including applications to computational drug repositioning.
GRANTS
The research was supported by National Heart, Lung, and Blood Institute Grants HL-101200 and R21 HL-122721.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: L.-H.C., C.G.V., and A.S.P. conception and design of research; L.-H.C. analyzed data; L.-H.C., C.G.V., B.H.A., and A.S.P. interpreted results of experiments; L.-H.C. and C.G.V. prepared figures; L.-H.C. drafted manuscript; L.-H.C., C.G.V., B.H.A., J.S.B., and A.S.P. edited and revised manuscript; L.-H.C., C.G.V., B.H.A., J.S.B., and A.S.P. approved final version of manuscript; C.G.V. performed experiments.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Annex BH. Therapeutic angiogenesis for critical limb ischaemia. Nat Rev Cardiol 10: 387–396, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo AG, Iruela-Arispe ML. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc Res 86: 226–235, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltgalvis KA, White K, Li W, Claypool MD, Lang W, Alcantara R, Singh BK, Friera AM, McLaughlin J, Hansen D, McCaughey K, Nguyen H, Smith IJ, Godinez G, Shaw SJ, Goff D, Singh R, Markovtsov V, Sun TQ, Jenkins Y, Uy G, Li Y, Pan A, Gururaja T, Lau D, Park G, Hitoshi Y, Payan DG, Kinsella TM. Exercise performance and peripheral vascular insufficiency improve with AMPK activation in high-fat diet-fed mice. Am J Physiol Heart Circ Physiol 306: H1128–H1145, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhasin M, Yuan L, Keskin DB, Otu HH, Libermann TA, Oettgen P. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics 11: 342, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324: 1710–1713, 2009. [DOI] [PubMed] [Google Scholar]
- 7.Chalothorn D, Faber JE. Strain-dependent variation in collateral circulatory function in mouse hindlimb. Physiol Genomics 42: 469–479, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalothorn D, Moore SM, Zhang H, Sunnarborg SW, Lee DC, Faber JE. Heparin-binding epidermal growth factor-like growth factor, collateral vessel development, and angiogenesis in skeletal muscle ischemia. Arterioscler Thromb Vasc Biol 25: 1884–1890, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ZP, Stephens TJ, Murthy S, Canny BJ, Hargreaves M, Witters LA, Kemp BE, McConell GK. Effect of exercise intensity on skeletal muscle AMPK signaling in humans. Diabetes 52: 2205–2212, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Chu LH, Lee E, Bader JS, Popel AS. Angiogenesis interactome and time course microarray data reveal the distinct activation patterns in endothelial cells. PLoS One 9: e110871, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu LH, Rivera CG, Popel AS, Bader JS. Constructing the angiome: a global angiogenesis protein interaction network. Physiol Genomics 44: 915–924, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costales JA, Daily JP, Burleigh BA. Cytokine-dependent and-independent gene expression changes and cell cycle block revealed in Trypanosoma cruzi-infected host cells by comparative mRNA profiling. BMC Genomics 10: 252, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croner RS, Balzer K, Schellerer V, Muller V, Schlabrakowsi A, Sturzl M, Naschberger E, Lang W. Molecular characterization of peripheral arterial disease in proximal extremity arteries. J Surg Res 178: 1046–1058, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Danese S, Dejana E, Fiocchi C. Immune regulation by microvascular endothelial cells: directing innate and adaptive immunity, coagulation, and inflammation. J Immunol 178: 6017–6022, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Dokun AO, Keum S, Hazarika S, Li Y, Lamonte GM, Wheeler F, Marchuk DA, Annex BH. A quantitative trait locus (LSq-1) on mouse chromosome 7 is linked to the absence of tissue loss after surgical hindlimb ischemia. Circulation 117: 1207–1215, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowkes FG, Rudan D, Rudan I, Aboyans V, Denenberg JO, McDermott MM, Norman PE, Sampson UK, Williams LJ, Mensah GA, Criqui MH. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet 382: 1329–1340, 2013. [DOI] [PubMed] [Google Scholar]
- 18.Fu S, Zhao H, Shi J, Abzhanov A, Crawford K, Ohno-Machado L, Zhou J, Du Y, Kuo WP, Zhang J, Jiang M, Jin JG. Peripheral arterial occlusive disease: global gene expression analyses suggest a major role for immune and inflammatory responses. BMC Genomics 9: 369, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukino K, Sata M, Seko Y, Hirata Y, Nagai R. Genetic background influences therapeutic effectiveness of VEGF. Biochem Biophys Res Commun 310: 143–147, 2003. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Bonilla L, Cid C, Alcazar A, Burda J, Ayuso I, Salinas M. Regulatory proteins of eukaryotic initiation factor 2-alpha subunit (eIF2 alpha) phosphatase, under ischemic reperfusion and tolerance. J Neurochem 103: 1368–1380, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol 192: 69–81, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldshmit Y, Galea MP, Bartlett PF, Turnley AM. EphA4 regulates central nervous system vascular formation. J Comp Neurol 497: 864–875, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JP, Hoefer I, Pasterkamp G, Bode C, Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 123: 999–1009, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Guo M, Huang T, Cui Y, Pan B, Shen A, Sun Y, Yi Y, Wang Y, Xiao G, Sun G. PrPC interacts with tetraspanin-7 through bovine PrP154-182 containing alpha-helix 1. Biochem Biophys Res Commun 365: 154–157, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Xu Y, Yang Y, Xi J, Tian W, Duggineni S, Huang Z, An J. Discovery and characterization of a novel cyclic peptide that effectively inhibits ephrin binding to the EphA4 receptor and displays anti-angiogenesis activity. PLoS One 8: e80183, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazarika S, Angelo M, Li Y, Aldrich AJ, Odronic SI, Yan Z, Stamler JS, Annex BH. Myocyte specific overexpression of myoglobin impairs angiogenesis after hind-limb ischemia. Arterioscler Thromb Vasc Biol 28: 2144–2150, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazarika S, Farber CR, Dokun AO, Pitsillides AN, Wang T, Lye RJ, Annex BH. MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation 127: 1818–1828, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoier B, Walker M, Passos M, Walker PJ, Green A, Bangsbo J, Askew CD, Hellsten Y. Angiogenic response to passive movement and active exercise in individuals with peripheral arterial disease. J Appl Physiol 115: 1777–1787, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isenberg JS, Frazier WA, Roberts DD. Thrombospondin-1: a physiological regulator of nitric oxide signaling. Cell Mol Life Sci 65: 728–742, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isenberg JS, Hyodo F, Pappan LK, Abu-Asab M, Tsokos M, Krishna MC, Frazier WA, Roberts DD. Blocking thrombospondin-1/CD47 signaling alleviates deleterious effects of aging on tissue responses to ischemia. Arterioscler Thromb Vasc Biol 27: 2582–2588, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Isenberg JS, Pappan LK, Romeo MJ, Abu-Asab M, Tsokos M, Wink DA, Frazier WA, Roberts DD. Blockade of thrombospondin-1-CD47 interactions prevents necrosis of full thickness skin grafts. Ann Surg 247: 180–190, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isenberg JS, Qin Y, Maxhimer JB, Sipes JM, Despres D, Schnermann J, Frazier WA, Roberts DD. Thrombospondin-1 and CD47 regulate blood pressure and cardiac responses to vasoactive stress. Matrix Biol 28: 110–119, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isenberg JS, Romeo MJ, Maxhimer JB, Smedley J, Frazier WA, Roberts DD. Gene silencing of CD47 and antibody ligation of thrombospondin-1 enhance ischemic tissue survival in a porcine model: implications for human disease. Ann Surg 247: 860–868, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izumi Y, Shiota M, Kusakabe H, Hikita Y, Nakao T, Nakamura Y, Muro T, Miura K, Yoshiyama M, Iwao H. Pravastatin accelerates ischemia-induced angiogenesis through AMP-activated protein kinase. Hypertens Res 32: 675–679, 2009. [DOI] [PubMed] [Google Scholar]
- 35.Jellinghaus S, Poitz DM, Ende G, Augstein A, Weinert S, Stutz B, Braun-Dullaeus RC, Pasquale EB, Strasser RH. Ephrin-A1/EphA4-mediated adhesion of monocytes to endothelial cells. Biochim Biophys Acta 1833: 2201–2211, 2013. [DOI] [PubMed] [Google Scholar]
- 36.Kim H, Cho HJ, Kim SW, Liu B, Choi YJ, Lee J, Sohn YD, Lee MY, Houge MA, Yoon YS. CD31+ cells represent highly angiogenic and vasculogenic cells in bone marrow: novel role of nonendothelial CD31+ cells in neovascularization and their therapeutic effects on ischemic vascular disease. Circ Res 107: 602–614, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689, 2005. [DOI] [PubMed] [Google Scholar]
- 38.Kuijper S, Turner CJ, Adams RH. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med 17: 145–151, 2007. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Hazarika S, Xie D, Pippen AM, Kontos CD, Annex BH. In mice with type 2 diabetes, a vascular endothelial growth factor (VEGF)-activating transcription factor modulates VEGF signaling and induces therapeutic angiogenesis after hindlimb ischemia. Diabetes 56: 656–665, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Dee Z, Pidcock K, Gutierrez LS. Thrombospondin-1: multiple paths to inflammation. Med Inf (Lond) 2011: 296069, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mac Gabhann F, Qutub AA, Annex BH, Popel AS. Systems biology of pro-angiogenic therapies targeting the VEGF system. Wiley Interdiscip Rev Syst Biol Med 2: 694–707, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masud R, Shameer K, Dhar A, Ding K, Kullo IJ. Gene expression profiling of peripheral blood mononuclear cells in the setting of peripheral arterial disease. J Clin Bioinforma 2: 6, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClung JM, McCord TJ, Keum S, Johnson S, Annex BH, Marchuk DA, Kontos CD. Skeletal muscle-specific genetic determinants contribute to the differential strain-dependent effects of hindlimb ischemia in mice. Am J Pathol 180: 2156–2169, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meisner JK, Song J, Annex BH, Price RJ. Myoglobin overexpression inhibits reperfusion in the ischemic mouse hindlimb through impaired angiogenesis but not arteriogenesis. Am J Pathol 183: 1710–1718, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mu J, Brozinick JT Jr, Valladares O, Bucan M, Birnbaum MJ. A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol Cell 7: 1085–1094, 2001. [DOI] [PubMed] [Google Scholar]
- 46.North HA, Clifford MA, Donoghue MJ. ‘Til Eph do us part’: intercellular signaling via Eph receptors and ephrin ligands guides cerebral cortical development from birth through maturation. Cerebr Cortex 23: 1765–1773, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omar B, Zmuda-Trzebiatowska E, Manganiello V, Goransson O, Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal 21: 760–766, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orrico C, Pasquinelli G, Foroni L, Muscara D, Tazzari PL, Ricci F, Buzzi M, Baldi E, Muccini N, Gargiulo M, Stella A. Dysfunctional vasa vasorum in diabetic peripheral artery obstructive disease with critical lower limb ischaemia. Eur J Vasc Endovasc Surg 40: 365–374, 2010. [DOI] [PubMed] [Google Scholar]
- 49.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 10: 165–180, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prevost N, Woulfe DS, Jiang H, Stalker TJ, Marchese P, Ruggeri ZM, Brass LF. Eph kinases and ephrins support thrombus growth and stability by regulating integrin outside-in signaling in platelets. Proc Natl Acad Sci USA 102: 9820–9825, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 38: 500–501, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Roberts W, Magwenzi S, Aburima A, Naseem KM. Thrombospondin-1 induces platelet activation through CD36-dependent inhibition of the cAMP/protein kinase A signaling cascade. Blood 116: 4297–4306, 2010. [DOI] [PubMed] [Google Scholar]
- 53.Rogers NM, Sharifi-Sanjani M, Csányi G, Pagano PJ, Isenberg JS. Thrombospondin-1 and CD47 regulation of cardiac, pulmonary and vascular responses in health and disease. Matrix Biol 37: 92–101, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010: baq020, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol 82: 1021–1033, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaper W, Scholz D. Factors regulating arteriogenesis. Arterioscler Thromb Vasc Biol 23: 1143–1151, 2003. [DOI] [PubMed] [Google Scholar]
- 57.Scholz D, Ziegelhoeffer T, Helisch A, Wagner S, Friedrich C, Podzuweit T, Schaper W. Contribution of arteriogenesis and angiogenesis to postocclusive hindlimb perfusion in mice. J Mol Cell Cardiol 34: 775–787, 2002. [DOI] [PubMed] [Google Scholar]
- 58.Scotland RS, Vallance PJ, Ahluwalia A. Endogenous factors involved in regulation of tone of arterial vasa vasorum: implications for conduit vessel physiology. Cardiovasc Res 46: 403–411, 2000. [DOI] [PubMed] [Google Scholar]
- 59.Sealock R, Zhang H, Lucitti JL, Moore SM, Faber JE. Congenic fine-mapping identifies a major causal locus for variation in the native collateral circulation and ischemic injury in brain and lower extremity. Circ Res 114: 660–671, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibata R, Ouchi N, Kihara S, Sato K, Funahashi T, Walsh K. Adiponectin stimulates angiogenesis in response to tissue ischemia through stimulation of amp-activated protein kinase signaling. J Biol Chem 279: 28670–28674, 2004. [DOI] [PubMed] [Google Scholar]
- 61.Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, Kawamoto M, Johns SJ, DeYoung J, Carlson E, Ferrin TE, Herskowitz I, Giacomini KM. PharmGKB update: III. Genetic variants of SLC22A1, solute carrier family 22 (organic cation transporter), member 1. Pharmacol Rev 56: 161, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Smadja DM, d'Audigier C, Bieche I, Evrard S, Mauge L, Dias JV, Labreuche J, Laurendeau I, Marsac B, Dizier B, Wagner-Ballon O, Boisson-Vidal C, Morandi V, Duong-Van-Huyen JP, Bruneval P, Dignat-George F, Emmerich J, Gaussem P. Thrombospondin-1 is a plasmatic marker of peripheral arterial disease that modulates endothelial progenitor cell angiogenic properties. Arterioscler Thromb Vasc Biol 31: 551–559, 2011. [DOI] [PubMed] [Google Scholar]
- 63.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spencer-Jones NJ, Ge D, Snieder H, Perks U, Swaminathan R, Spector TD, Carter ND, O'Dell SD. AMP-kinase alpha2 subunit gene PRKAA2 variants are associated with total cholesterol, low-density lipoprotein-cholesterol and high-density lipoprotein-cholesterol in normal women. J Med Genet 43: 936–942, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stark L, Matussek A, Strindhall J, Geffers R, Buer J, Kihlström E, Monnecke S, Löfgren S, Lindgren PE. Staphylococcus aureus isolates from blood and anterior nares induce similar innate immune responses in endothelial cells. APMIS 117: 814–824, 2009. [DOI] [PubMed] [Google Scholar]
- 66.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki Y, Hattori K, Hamanaka J, Murase T, Egashira Y, Mishiro K, Ishiguro M, Tsuruma K, Hirose Y, Tanaka H, Yoshimura S, Shimazawa M, Inagaki N, Nagasawa H, Iwama T, Hara H. Pharmacological inhibition of TLR4-NOX4 signal protects against neuronal death in transient focal ischemia. Sci Rep 2: 896, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tarcea VG, Weymouth T, Ade A, Bookvich A, Gao J, Mahavisno V, Wright Z, Chapman A, Jayapandian M, Ozgur A, Tian Y, Cavalcoli J, Mirel B, Patel J, Radev D, Athey B, States D, Jagadish HV. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res 37: D642–D646, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Troidl K, Schaper W. Arteriogenesis versus angiogenesis in peripheral artery disease. Diabetes Metab Res Rev 28, Suppl 1: 27–29, 2012. [DOI] [PubMed] [Google Scholar]
- 70.Varela C, de Haro J, Bleda S, Rodriguez-Padilla J, Ferruelo A, Acin F. Circulating anti-β-glycoprotein I antibodies of peripheral arterial disease patients trigger a genomic overexpression of Toll-like receptor 4 in endothelial cells. J Vasc Surg 61: 1041–1049, 2015. [DOI] [PubMed] [Google Scholar]
- 71.Vuorio T, Jauhiainen S, Yla-Herttuala S. Pro- and anti-angiogenic therapy and atherosclerosis with special emphasis on vascular endothelial growth factors. Exp Opin Biol Ther 12: 79–92, 2012. [DOI] [PubMed] [Google Scholar]
- 72.Yao M, Roberts DD, Isenberg JS. Thrombospondin-1 inhibition of vascular smooth muscle cell responses occurs via modulation of both cAMP and cGMP. Pharmacol Res 63: 13–22, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Q, Wong AK, Krishnan A, Aure MR, Tadych A, Zhang R, Corney DC, Greene CS, Bongo LA, Kristensen VN, Charikar M, Li K, Troyanskaya OG. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat Meth 12: 211–214, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.