Abstract
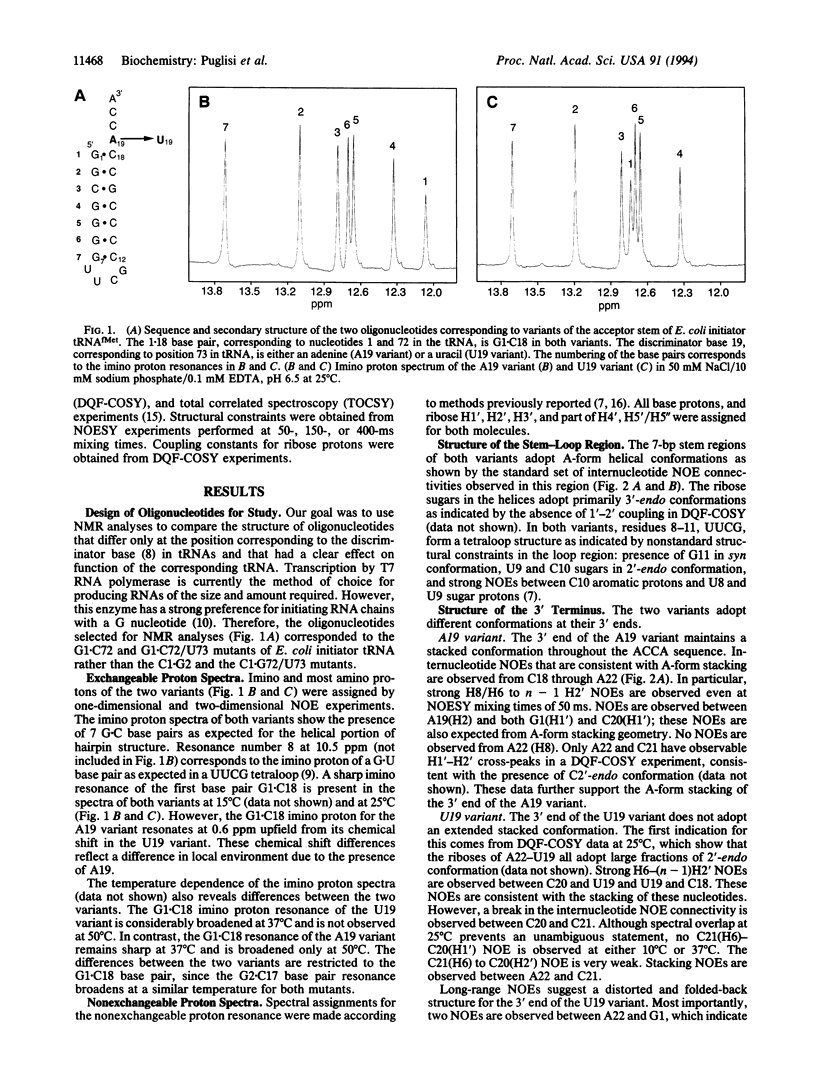
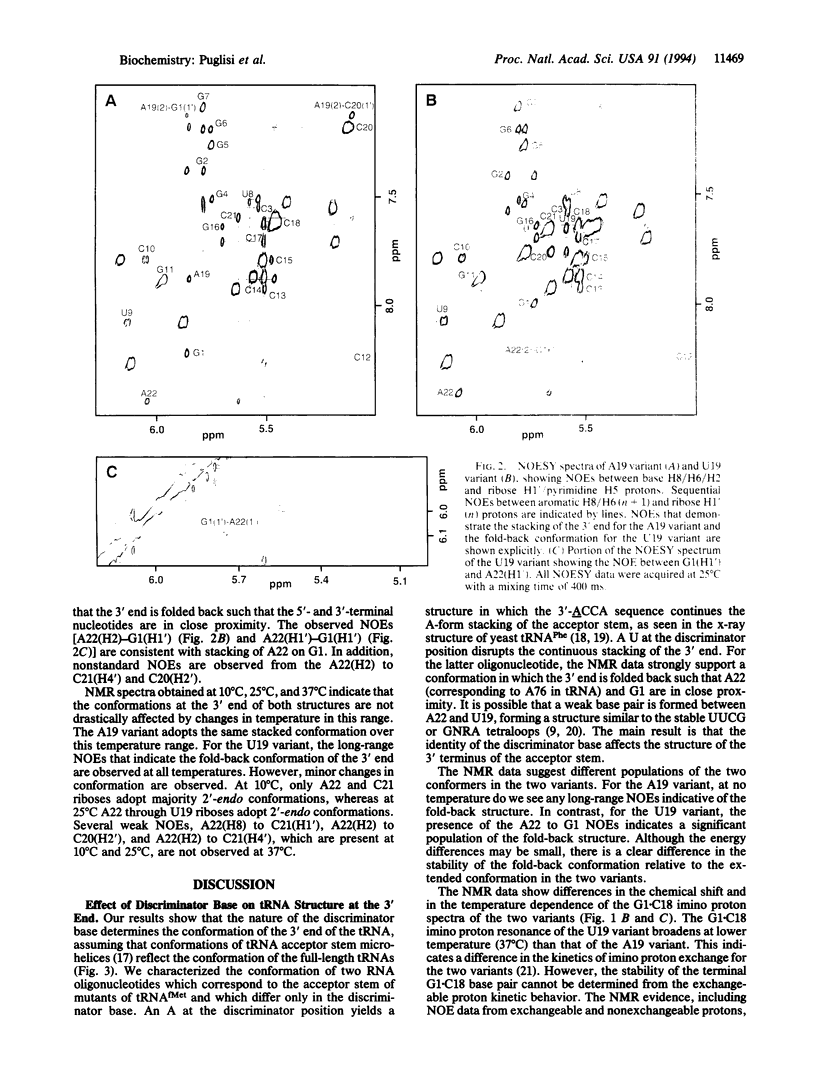
An important step in initiation of protein synthesis in Escherichia coli is the specific formylation of the initiator methionyl-tRNA (Met-tRNA) by Met-tRNA transformylase. The determinants for formylation are clustered mostly in the acceptor stem of the initiator tRNA. Here we use NMR spectroscopy to characterize the conformation of two RNA microhelices, which correspond to the acceptor stem of mutants of E. coli initiator tRNA and which differ only at the position corresponding to the "discriminator base" in tRNAs. One of the mutant tRNAs is an extremely poor substrate for Met-tRNA transformylase, whereas the other one is a much better substrate. We show that one microhelix forms a structure in which its 3'-ACCA sequence extends the stacking of the acceptor stem. The other microhelix forms a structure in which its 3'-UCCA sequence folds back such that the 3'-terminal A22 is in close proximity to G1. These results highlight the importance of the discriminator base in determining tRNA conformation at the 3' end. They also suggest a correlation between tRNA structure at the 3' end and its recognition by Met-tRNA transformylase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breitschopf K., Gross H. J. The exchange of the discriminator base A73 for G is alone sufficient to convert human tRNA(Leu) into a serine-acceptor in vitro. EMBO J. 1994 Jul 1;13(13):3166–3169. doi: 10.1002/j.1460-2075.1994.tb06615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Seno T., Söll G. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci U S A. 1972 Oct;69(10):3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube S. K., Marcker K. A., Clark B. F., Cory S. Nucleotide sequence of N-formyl-methionyl-transfer RNA. Nature. 1968 Apr 20;218(5138):232–233. doi: 10.1038/218232a0. [DOI] [PubMed] [Google Scholar]
- Dutka S., Meinnel T., Lazennec C., Mechulam Y., Blanquet S. Role of the 1-72 base pair in tRNAs for the activity of Escherichia coli peptidyl-tRNA hydrolase. Nucleic Acids Res. 1993 Aug 25;21(17):4025–4030. doi: 10.1093/nar/21.17.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson M. R., Mandal N., RajBhandary U. L. Relationship between the structure and function of Escherichia coli initiator tRNA. Biochimie. 1993;75(12):1051–1060. doi: 10.1016/0300-9084(93)90004-c. [DOI] [PubMed] [Google Scholar]
- Ferguson B. Q., Yang D. C. Topographic modeling of free and methionyl-tRNA synthetase bound tRNAfMet by singlet-singlet energy transfer: bending of the 3'-terminal arm in tRNAfMet. Biochemistry. 1986 Oct 21;25(21):6572–6578. doi: 10.1021/bi00369a035. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Guillon J. M., Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J Mol Biol. 1992 Mar 20;224(2):359–367. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Komatsoulis G. A., Abelson J. Recognition of tRNA(Cys) by Escherichia coli cysteinyl-tRNA synthetase. Biochemistry. 1993 Jul 27;32(29):7435–7444. doi: 10.1021/bi00080a014. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kössel H., RajBhandary U. L. Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J Mol Biol. 1968 Aug 14;35(3):539–560. doi: 10.1016/s0022-2836(68)80013-0. [DOI] [PubMed] [Google Scholar]
- Lee C. P., Dyson M. R., Mandal N., Varshney U., Bahramian B., RajBhandary U. L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Mandal N., Dyson M. R., RajBhandary U. L. The discriminator base influences tRNA structure at the end of the acceptor stem and possibly its interaction with proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7149–7152. doi: 10.1073/pnas.90.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Seong B. L., RajBhandary U. L. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991 Sep 25;266(27):18012–18017. [PubMed] [Google Scholar]
- Limmer S., Hofmann H. P., Ott G., Sprinzl M. The 3'-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6199–6202. doi: 10.1073/pnas.90.13.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H. Transfer RNA identity. FASEB J. 1993 Jan;7(1):72–78. doi: 10.1096/fasebj.7.1.8422977. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992 Jul 20;226(2):399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- Pscheidt R. H., Wells B. D. Different conformations of the 3' termini of initiator and elongator transfer ribonucleic acids. An EPR study. J Biol Chem. 1986 Jun 5;261(16):7253–7256. [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RajBhandary U. L. Initiator transfer RNAs. J Bacteriol. 1994 Feb;176(3):547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Schimmel P., Giegé R., Moras D., Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. The structural basis for the resistance of Escherichia coli formylmethionyl transfer ribonucleic acid to cleavage by Escherichia coli peptidyl transfer ribonucleic acid hydrolase. J Biol Chem. 1975 Jan 25;250(2):542–547. [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Asahara H., Tamura K., Hasegawa T., Himeno H. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J Mol Evol. 1992 Nov;35(5):436–443. doi: 10.1007/BF00171822. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Kierzek R., Turner D. H. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987 Jul 14;26(14):4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Varani G., Tinoco I., Jr RNA structure and NMR spectroscopy. Q Rev Biophys. 1991 Nov;24(4):479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]




