Abstract
Increasing interest in and experience with electronic health record (EHR)-driven phenotyping has yielded multiple challenges that are at present only partially addressed. Many solutions require the adoption of a single software platform, often with an additional cost of mapping existing patient and phenotypic data to multiple representations. We propose a set of guiding design principles and a modular software architecture to bridge the gap to a standardized phenotype representation, dissemination and execution. Ongoing development leveraging this proposed architecture has shown its ability to address existing limitations.
Introduction
Electronic health record (EHR)-based phenotyping is a rich source of data for clinical and genomic research1. Interest in this area, however, has uncovered many challenges including the quality and semantics of the data collected, as well as the disparate modalities in which the information is stored across institutions and EHR systems2, 3. Significant effort has gone into addressing these challenges, and researchers have an increased understanding of the nuances of EHR data and the importance of multiple information sources such as diagnostic codes coupled with natural language processing (NLP)4, 5.
The process of developing a phenotype algorithm is complex – requiring a diverse team engaged in multiple iterations of hypothesis generation, data exploration, algorithm generation and validation6. The technical tools used during these phases vary across teams and institutions, driven in part by availability, licensing costs, technical infrastructure supported at an institution, and personal preference. For example, initial feasibility queries may be conducted using a system like i2b27 followed by in-depth analysis using statistical software (e.g. SAS) or structured query language (SQL) statements directly against a database. A researcher may document the algorithm by listing steps in Microsoft Word, later converting it to an executable workflow using KNIME (https://www.knime.org/).
Teams creating algorithms need to be able to quickly explore EHR data using tools that they are intimately familiar and comfortable with. However, once an algorithm has been established, the ability to represent and share the algorithm with other institutions in a standardized way currently poses some limitations. While phenotype algorithms are presently published along with scientific publications or in repositories such as the Phenotype KnowledgeBase (PheKB, http://phekb.org), the representation is rarely standardized and executable at other institutions. For example, phenotypes from the electronic Medical Records and Genomics (eMERGE) network8 are represented as textual documents that need to be interpreted by other institutions and adapted for local implementation9.
A solution first requires unambiguous representation of algorithm logic and semantically rich patient data, an execution layer that combines the two to generate sharable and computable results, and finally a repository to share phenotypes and execution results for collaborative research. In this work, we describe our approach towards creating the Phenotype Execution and Modeling Architecture (PhEMA; http://informatics.mayo.edu/phema) to address these challenges.
Background
Several system architectures and implementations are reported in the informatics literature for electronic phenotyping. One example is the i2b2 system, which leverages a service-oriented architecture (SOA) to compose a system of individual “cells”7. Each cell is responsible for a particular function (e.g. terminology management, query execution), and interacts via established application programming interfaces (APIs). While i2b2 is not built on any formal standards, the broad adoption of the platform and its growing community has arguably made its database schema and APIs a “de facto” standard. Many institutions have adopted the i2b2 system, and it benefits from an active community that has extended the system with new functionality via additional cells and other extensions. For example, the Eureka! platform includes a separate cohort authoring interface, which then feeds into an i2b2 instance for additional analysis10. Work has also been done to allow i2b2 to interoperate with the Health Quality Measure Format (HQMF), providing a more standardized option for logic representation11. To date, however, the i2b2 HQMF implementation is not comprehensive enough to be sufficient for more complex phenotype algorithms. In addition, execution in i2b2 is tightly coupled to its patient data model, requiring that an institution map a copy of its clinical data into the i2b2 schema.
Within the realm of EHR-based clinical quality measures (CQMs), HQMF is primarily used to represent quality measure logic. The process of developing quality measures is similar to that of a phenotype, with validated measures being formally defined using the Measure Authoring Tool (MAT; https://www.emeasuretool.cms.gov/). Using the standardized HQMF artifact, popHealth (http://projectpophealth.org/) takes the measure logic and executes it against a set of patients. Furthermore, Bonnie (https://bonnie.healthit.gov/) provides a framework in which quality measure authors may validate their logic. However, there are some limitations. For example, popHealth does not connect to a live data source, requiring patient data to be exported from a clinical repository and then separately imported into the tool. Additionally, while one of the strengths of the eCQM infrastructure is adherence to national standards, they are not sufficiently comprehensive to represent phenotype algorithms12.
Few execution environments exist that can import externally developed algorithm logic and execute against a patient model. The Phenotype Portal (http://phenotypeportal.org/) is one such implementation, which supports HQMF-based algorithms as input and executes against a platform-specific patient model. The benefit of this approach is that it may utilize logic that is authored in any system that can export an HQMF representation (currently limited to the MAT). Other systems, such as the Translational Research Informatics and Data Management Grid (TRIAD)13 and caGRID14, provide standards and APIs to allow interoperability between components and distribution of algorithms but require significant resource to set up and use.
While there are many standalone components and modular architectures that compose phenotyping systems, none to date have fully identified and met all of the needs of EHR-based phenotype algorithm authors.
Meta-Architecture
The concept of a meta-architecture 15 is to provide a high-level strategy for identifying needs of a domain and additional context in order to inform the creation of a formal software architecture. We conducted a review of existing system architectures to perform a gap analysis of strengths and limitations (presented in the Background section), including requirements for setting up the system, extensibility of the base platform, use of published standards and/or APIs, and results from hands-on setup or use of the system. In addition, preliminary results from focus groups with stakeholders at Mayo Clinic and Northwestern University elucidated additional requirements for how a system should operate. This was supplemented with the authors’ collective experience (many of whom are affiliated with eMERGE) in the use of existing tools to map requirements to the following guiding principles:
Apply service oriented architecture (SOA) to develop the system as independent components that interoperate via standard interfaces – given the large number of components (described below), a single system would require full adoption of the entire platform. The decoupled and distributed nature of services16 allows users to use publicly hosted instances (e.g. public terminology server), offsetting setup and maintenance of a local installation of all components. Also, as shown within systems like i2b2, integrating via well-documented, standards and APIs allows users to extend the platform.
Provide integration at the user interface layer as well as with system-to-system interfaces – loosely coupled systems do not always provide an easy flow when moving between them, which can reduce adoption, acceptance and user satisfaction. To address this, we will visually integrate different components when possible, and minimize extra steps needed to use each component. For example, an anticipated workflow is to author and execute a phenotype definition, although authoring and execution would be separate components. Visually integrating these would allow the user to see results from the execution platform within the authoring environment and without having to explicitly go to another window. A successful example of this type of integration is the Eureka! platform’s use of i2b2.10
Embrace similarities with comparable domains (e.g. CQMs and decision support) – many standards and approaches exist to using EHR data, including the realms of decision support and quality reporting, as well as biomedical research. The architecture should focus on the needs of biomedical researchers, but anticipate how it could also support quality reporting and clinical uses. This also would allow adoption of components developed in those domains, and the opportunity to propose enhancements to existing standards in lieu of proposing a new competing standard for logic representation.
Accept that there is a cost to the adoption of any platform, but strive to minimize that cost – the concept of activation energy in biology may be applied to software architecture as well: look for the least amount of energy required to adopt a system. A modular architecture may accomplish this by minimizing the number of components that must be installed, allowing simple implementations that are easy to set up. This also allows a composition of hosted and local systems. In addition, it should account for an institution’s preference to use an existing system and incorporate that into the overall architecture.
Phenotype Execution and Modeling Architecture (PhEMA)
Based on the principles defined in the meta-architecture, we propose PhEMA, which includes the following system components connected where possible via existing standards (see Figure 1):
Library – a repository of phenotype algorithms and supporting metadata, which supports annotation, discovery and reuse of algorithms.
Authoring – a graphical editor to create phenotype definitions using emerging national standards from Health Level 7 (HL7), Center for Medicare and Medicaid Services (CMS), and the Office of the National Coordinator for Health IT (ONC). The representation used will be free of any EHR or platform-specific implementation details.
Clinical Data Repository – the underlying data store for all relevant patient data. This should support recommended best practices in storage, semantics and management of protected health information17. The use of adapters that sit over the underlying data store will allow use of an existing data repository (at the cost of needing to develop an adapter), or adoption of an existing schema (at the cost of mapping to the schema).
Execution – take a phenotype algorithm definition, execute it against the Clinical Data Repository, and report lists of selected patients. Although not necessarily required, it is anticipated that the Execution component will be able to provide an analysis of the algorithm execution, to identify potential errors or bottlenecks (i.e. in which step are 99% of the patients being filtered out).
Validation – environment in which algorithms may be executed against a simulated patient chart to more easily verify the accuracy of the algorithm logic. This supplements the typical practices of executing an algorithm against a full patient population and performing a heuristic assessment if the results seem accurate, or conducting an in-depth chart review.
Data Model Services – the information models that comprise data model elements and phenotype logic require a formal representation. A centralized repository allows all connected systems to identify and respond to new models with minimal or no system changes, with services to access the models in a variety of formats.
Terminology Services – support discovery and use of standardized vocabularies, which are heavily leveraged by phenotypes, and important for cross-institutional portability. This would also include resources such as the Value Set Authority Center (VSAC), which provide curated lists of standard vocabularies groups.
Figure 1.
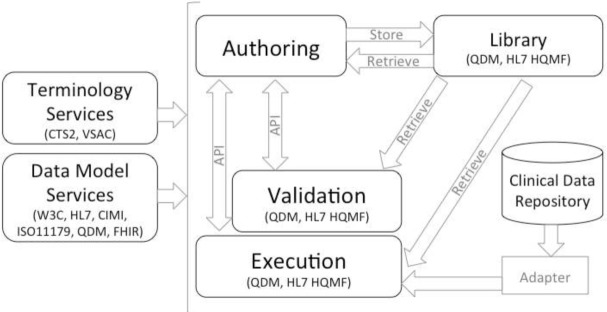
Phenotype Execution and Modeling Architecture (PhEMA)
Many additional components are relevant and beneficial to conduct research (for example, federated execution of phenotype algorithms at multiple institutions), but are not deemed necessary to the core architecture.
Within PhEMA, the Authoring, Execution and Validation components interact with the Library to retrieve and store phenotypes. The Library is intended to also allow a public view of finalized and validated phenotype algorithms, which is currently only supported in the eCQM domain. For the algorithms, we propose using a Quality Data Model (QDM, http://www.healthit.gov/quality-data-model)-based representation. QDM was chosen given its coverage of most logical constructs used in phenotype algorithms, and adoption by the eCQM community. As noted, limitations in this model prevent it from comprehensively representing a phenotype12, which may be addressed by extensions to the model. While this may limit its ability to interoperate with other QDM-based systems, it reaches a necessary balance in adopting existing standards while meeting unmet needs.
In our reference implementation, the Authoring component is a web-based application that queries the Data Model Services for its supported models, allowing it to discover new models as they are introduced (Figure 2). PhEMA’s Data Model Service is a semantic framework that represents QDM elements using the Resource Description Framework (RDF), and provides access via a Linked Open Data API (exposed as REpresentational State Transfer [REST] services). The Authoring component also utilizes Terminology Services to provide access to standard vocabularies using the Common Terminology Services 2 (CTS2, http://informatics.mayo.edu/cts2/) specification. The Authoring component exports QDM-based artifacts, and will establish a bi-directional API with the Validation and Execution systems so a phenotype algorithm may be pushed to and results received from either system, supporting the meta-architecture principle of integration at the user interface and system level.
Figure 2.
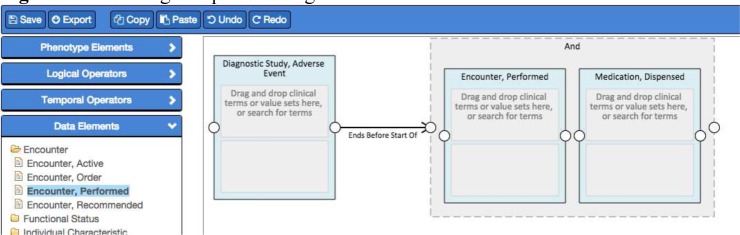
Authoring component integrated with the Data Model Services for the list of Data Elements
The Execution component reads from the Clinical Data Repository for relevant data using the data model implemented in the Phenotype Portal system. This model is based on several standards, and may be mapped to others, such as HL7 FHIR (http://www.hl7.org/implement/standards/fhir/) using an adapter. Effort to date with the Execution component has used workflow-based technologies (e.g. KNIME and Drools), which adds a dimension of computability and portability over direct SQL queries. Ongoing work will connect these with the Data Model Services and Terminology Services to aid with logic execution.
Discussion
We propose PhEMA to support the multiple facets of EHR-based phenotyping. There are notable similarities to other architectures and systems, but also clear differences. For example, PhEMA leverages existing standards as much as possible for the interfaces between its components, while other systems primarily define their own. Also, we propose a comprehensive suite of components, including those not as formally defined in other systems (namely the Library and Validation). These components are necessary to produce standardized phenotype definitions that may be shared with other sites, and to verify their accuracy. The Validation system provides a way to define a gold standard against which the phenotype definition may be verified. This is important to not only provide feedback on correct use of the Authoring system, but to assist in identifying breaking changes if the phenotype is modified.
Software architectures themselves provide a critical blueprint for how a system may be developed, yet are not tangible. Architectures such as PhEMA benefit from a reference implementation for multiple reasons. First, the creation of actual software proves the validity of the proposed architecture. More importantly, it allows institutions to adopt those systems for their own use. It provides concrete source code showing the interoperability between components, aiding future developers in their own work. As described, a reference implementation is currently under development, based on these recommended approaches and standards. Ultimately this will include demonstrating adaptation of an existing system and the creation of a novel component. For example – linking the Execution environment to an existing i2b2 instance and an institutional data warehouse, and developing a new Authoring environment while also demonstrating the ability to process logic created by the MAT. We feel this approach is necessary to fully validate the modularity of the architecture, and to demonstrate the ability to use established systems with other novel components.
While PhEMA provides a reference implementation, the details of each component are ultimately left to the adopter. For example, a Library component may be implemented as a simple database, a shared portal like PheKB.org, or tied to a version control system. The Validation component may be one that uses a simulated patient chart, or may be tied to an actual EHR and chart abstraction system.
The architecture is intended to support the diverse team of phenotype authors, and may be leveraged at any point in the phenotyping process. At a minimum, we envision it will be used once a phenotype is fully defined and validated, and ready to be authored in a standardized manner. Our intent is to facilitate collaborative research by increasing the number of standardized phenotypes that may be shared and executed across institutions.
Conclusion
Leveraging the strengths of existing biomedical research systems, and addressing known limitations in those platforms defined by the needs of the phenotyping community, we propose a modular system architecture to facilitate the full lifecycle of phenotype algorithm authoring. The resulting reference implementation based on this architecture promises a more streamlined set of tools to represent and utilize standardized phenotype algorithms.
Acknowledgments
This work has been supported in part by funding from PhEMA (R01 GM105688) and eMERGE (U01 HG006379, U01 HG006378 and U01 HG006388).
References
- 1.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12(6):417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 2.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS Computational Biology. 2012;8(12):e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen LV. The electronic health record for translational research. J Cardiovasc Transl Res. 2014;7(6):607–14. doi: 10.1007/s12265-014-9579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen LV, Thompson WK, Pacheco JA, Kho AN, Carrell DS, Pathak J, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform. 2014 doi: 10.1016/j.jbi.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Ann Symp. 2007:548–52. [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genetics in Medicine. 2013;15(10):761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Ann Symp. 2011:274–83. [PMC free article] [PubMed] [Google Scholar]
- 10.Post AR, Kurc T, Willard R, Rathod H, Mansour M, Pai AK, et al. Temporal abstraction-based clinical phenotyping with Eureka! AMIA Ann Symp. 2013:1160–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Klann GJ, Murphy NS. Computing Health Quality Measures Using Informatics for Integrating Biology and the Bedside. J Med Internet Res. 2013;15(4):e75. doi: 10.2196/jmir.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson WK, Rasmussen LV, Pacheco JA, Peissig PL, Denny JC, Kho AN, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Ann Symp. 2012:911–20. [PMC free article] [PubMed] [Google Scholar]
- 13.Payne P, Ervin D, Dhaval R, Borlawsky T, Lai A. TRIAD: The Translational Research Informatics and Data Management Grid. Appl Clin Inform. 2011;2(3):331–44. doi: 10.4338/ACI-2011-02-RA-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15(2):138–49. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malan R, Bredemeyer D. Meta-Architecture. 2004. [August 20, 2014]. Available from: http://www.bredemeyer.com/ArchitectingProcess/MetaArchitecture.htm.
- 16.Erl T. SOA : principles of service design. Upper Saddle River, NJ: Prentice Hall; 2008. [Google Scholar]
- 17.Huser V, Cimino JJ. Desiderata for healthcare integrated data repositories based on architectural comparison of three public repositories. AMIA Ann Symp. 2013:648–56. [PMC free article] [PubMed] [Google Scholar]


