Abstract
Personalized predictive models are customized for an individual patient and trained using information from similar patients. Compared to global models trained on all patients, they have the potential to produce more accurate risk scores and capture more relevant risk factors for individual patients. This paper presents an approach for building personalized predictive models and generating personalized risk factor profiles. A locally supervised metric learning (LSML) similarity measure is trained for diabetes onset and used to find clinically similar patients. Personalized risk profiles are created by analyzing the parameters of the trained personalized logistic regression models. A 15,000 patient data set, derived from electronic health records, is used to evaluate the approach. The predictive results show that the personalized models can outperform the global model. Cluster analysis of the risk profiles show groups of patients with similar risk factors, differences in the top risk factors for different groups of patients and differences between the individual and global risk factors.
Introduction
Predictive modeling is one of the most popular and important methodologies used in clinical and healthcare research today and has been successfully applied to a number of use cases including the early detection of disease onset and the greater individualization of care [1–4]. The conventional approach in predictive modeling is to build a single “global” predictive model using all the available training data which is then used to compute risk scores for individual patients and to identify population wide risk factors. Recent work in the area of personalized medicine show that the patient population is heterogeneous, that each patient has unique characteristics and that it is important to have targeted, patient specific predictions, recommendations and treatments [5–7] From the perspective of personalized medicine, a “one size fits all” global predictive model may not be the best approach for individual patients. A static global model captures information that is important for the entire training patient population but may miss less popular information that is important for individual patients. An alternative approach is to build a patient-specific or “personalized” predictive model for each patient. The model would be customized for the individual patient since it is built using information from the patient and from clinically similar patients. Because they are dynamically trained for specific patients, personalized predictive models can leverage the most relevant patient information and have the potential to generate more accurate risk scores and to identify more relevant and informative patient-specific risk factors.
Personalized models in the context of healthcare applications have recently been investigated. A comparative study of several global, local and personalized modeling approaches applied to a number of bioinformatics classification tasks is presented in [8]. A personalized modeling framework that leverages evolutionary optimization techniques is proposed in [9]. In these studies, k-nearest neighbor (KNN), support vector machine (SVM) and connectionist classification models were explored and static distance measures (e.g., Euclidean, Manhattan and Mahalanobis) were used to find the nearest neighbors. They found that across a range of different bioinformatics classification tasks, personalized models can provide improved accuracy over global and local models. A patient-specific model averaging approach for learning Bayesian network models using all available training data is described in [10]. They found that including models with local structure is better than only using models with global structure. Other more distantly related work include extensions of the KNN classifier to include more sophisticated class probability estimators [11] and the application of collaborative filtering approaches to disease risk prediction [12].
This study extends the investigation and analysis of personalized predictive models along a number of important dimensions, including: 1) using a trainable similarity metric to find clinically similar patients, 2) creating personalized risk factor profiles by analyzing the parameters of the trained personalized models and 3) clustering the risk factor profiles to facilitate an analysis of the characteristics and distribution of the patient specific risk factors.
Methods
Study Subjects
A 15,038 patient cohort was constructed from an anonymous longitudinal medical claims database consisting of four years of data covering over 300,000 patients. 7,519 patients with a diabetes diagnosis in the last two years but not in the first two years were identified as incident cases. Each case was paired with a matched control patient based on age (+/− 5 years), gender and primary care physician resulting in 7,519 control patients without any diabetes diagnosis in all four years. The patients’ diagnosis information, medication orders, medical procedures and laboratory tests from the first two years of data were used in the study.
Feature Construction from Longitudinal Patient Data
A feature vector representation for each patient is generated based on the Patient’s longitudinal data. This data can be viewed as multiple event sequences over time (e.g. a patient can have multiple diagnoses of hypertension at different dates). To convert such event sequences into feature variables, an observation window (e.g. the first two years) is specified. Then all events of the same feature within the window are aggregated into a single or small set of values. The aggregation function can produce simple feature values like counts and averages or complex feature values that take into account temporal information (e.g., trend and temporal variation). In this study, only basic aggregation functions are used: count for categorical variables (diagnoses, medications and procedures) and mean for numeric variables (lab tests). This results in over 8,500 unique feature variables. To reduce the size of the feature space, an initial global feature selection is performed using the information gain measure [13] to select the top features for each feature type for subsequent use: 50 diagnoses, 50 procedures, 15 medications and 15 lab tests for a total of 130 features.
Personalized Predictive Modeling
Personalized predictive modeling involves the following processing steps:
- Patient Similarity Computation
- ○ Receive a new test patient.
- ○ Identify a cohort of K similar patients from the training set using a patient similarity measure.
- Feature Filtering
- ○ Select a subset of the features using information from the test patient and the similar patient cohort.
- Predictive Modeling
- ○ Train a personalized predictive model using the similar patient cohort.
- ○ Compute a risk score for the new test patient using the trained personalized predictive model.
- Risk Factor Profile Computation
- ○ Analyze the trained personalized predictive model to create a personalized risk factor profile.
Patient Similarity Computation
A number of different similarity measures can be used to identify the cohort of patients from the training set that are most clinically similar to the test patient. In this study, a trainable similarity measure called Locally Supervised Metric Learning (LSML) that is customizable for a specific disease target condition is used [14]. The distance metric is defined as , where xi and xj are the patient feature vectors for patients i and j respectively and W is a transformation matrix that is estimated from the training data. LSML is formulated as an average neighborhood margin maximization problem and tries to find a weight matrix W where the local class discriminability is maximized. A trainable metric is important because different clinical scenarios will likely require different patient similarity measures. For example, two patients that are similar to each other with respect to one disease target, e.g., diabetes, may not be similar at all for a different disease target such as lung cancer. The use of static similarity measures, e.g., Euclidean or Mahalanobis, for all target conditions may not be optimal. In this study, the LSML similarity measure is trained for the diabetes disease onset target and then used to find the most clinically similar case and control patients for a given test patient. The selection is done without considering the case-control status of the training patients so the resulting similar patient cohort may not maintain the global case-control balance. It is possible to select the similar case and control patients separately and then combine them into one cohort if a specific case-control balance is desired. A comparison of similar patient selection using LSML, Euclidean distance and random selection is presented in the Results section.
Feature Filtering
Using only the K most similar patients from the training set can reduce the amount of data available for training the personalized predictive model. Reducing the dimensionality of the feature vectors by selecting a subset of the initial features can help compensate for this. A number of approaches can be used to do this including performing cross-validated feature selection on the similar patient training cohort using an information gain or Fisher score metric [13]. In this study, a simple filtering heuristic is used: the selected features consist of the union of the features that occur in the test patient feature vector and all features that occur in two or more feature vectors from the K most similar patients. The goal of the filtering is to ensure that only features that can impact the test patient are included.
Predictive Modeling
For each test patient, a logistic regression (LR) predictive model was dynamically trained using data from the selected case and control patients that are most similar to the test patient based on the LSML similarity measure. The personalized predictive model was then used to compute a score (the risk of diabetes disease onset) for that test patient. Predictive modeling experiments were performed using 10-fold cross validation and performance was measured using the standard AUC (area under the ROC curve) metric. AUC and 95% confidence intervals (CIs) are reported.
Risk Factor Profile Computation
After training, the parameters in the predictive model are analyzed to identify the important risk factors captured by the model and used to create a “risk factor profile” for the patient(s) represented by the model. For the logistic regression model, the beta coefficient for each feature captures the change in the log odds for a unit change in that feature. In addition to the value of the coefficient, the significance of the coefficient can be assessed by computing the Wald statistic and the corresponding P-value. The important risk factors are the features with statistically significant, large magnitude coefficients. The beta coefficient values of these selected features can then be used to create the risk factor profile. For the global predictive model, only a single “population wide” risk factor profile can be derived. For the personalized predictive models, a risk factor profile is derived for each patient resulting in a large number of profiles. In this case, it is useful to examine the risk profiles individually as well as the distribution of the risk profiles across the patient population. Exploring and comparing the individual profiles allows one to pinpoint the risk factor differences among the patients. Examining the distribution of the profiles provides a global view of their behavior and relationships. One scalable approach that can support both individual comparisons and global distributional analysis is to perform hierarchical clustering on the risk profiles. An analysis of the clustering results can provide insight into the characteristics and distribution of the profiles. One can assess the degree of similarity and difference of the risk factors for different patients. In addition, it may be possible to discover any structural relationships in the patient population with respect to common risk factors identified by the personalized models.
Results
Prediction Performance
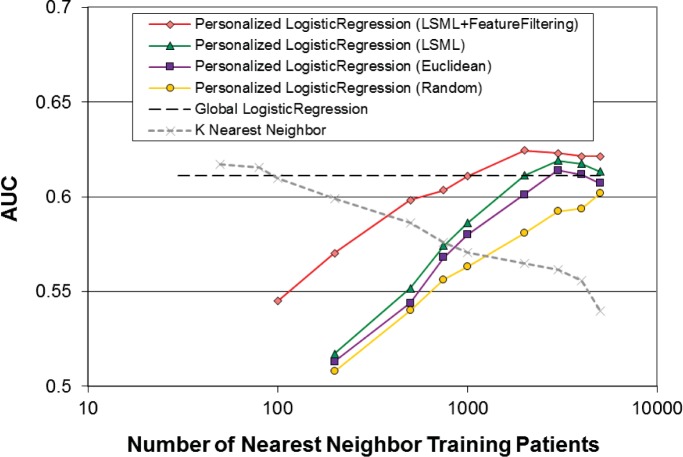
Performance of the personalized logistic regression model in terms of AUC as a function of the number of nearest neighbor training patients is shown in Figure 1. There are four curves corresponding to four different configurations. In addition, the performance of the global logistic regression model (–) and the performance of a K nearest neighbor (KNN) classifier (×) are shown for reference. First, as a baseline, K randomly selected patients are used for training the Personalized model (◦) Performance steadily increases towards the global model performance as the number of training patients increases. This behavior is expected because for parametric models such as logistic regression, there needs to be sufficient data for the model parameters to be properly trained. Second, instead of selecting patients randomly, the Euclidean distance metric is used to select the K most similar Patients for training (□) For a fixed number of training patients, similarity based selection is consistently better than random selection. Also, performance starts to level off after about 3000 training patients, suggesting that there is little to gain from using more dissimilar patients for training. Third, the LSML similarity metric is used to select the K most similar patients for training (Δ). Performance using a custom trained similarity measure is better than using a static measure for all values of K. Fourth, the dimensionality of the feature vectors is reduced using the filtering approach described earlier (⋄) This reduces the training data requirements on the model and result in significant performance improvements, especially for smaller values of K. Again, there is a diminishing return for using more dissimilar training patients as performance levels off for values of K larger than 2000. Performance of the personalized models is comparable to the global model (AUC: 0.611, 95%CI: 0.605–0.617) at K=1000 and better than the global model for larger values of K (AUC: 0.624, 95%CI: 0.617–0.631 at K=2000). The K most similar patients (selected via LSML) can also be used directly for classification using a k-nearest neighbor (KNN) classifier. The behavior of the exemplar-based KNN classifier contrasts with that of the parametric logistic regression classifier. KNN performance (×) is comparable to the global model for small values of K (AUC: 0.612, 95%CI: 0.606–0.618 at K=50) but drops steadily as K increases.
Figure 1:
Performance of the personalized logistic regression model in terms of AUC as a function of the number of nearest neighbor training patients for different classifier configurations.
Risk Factor Profiles
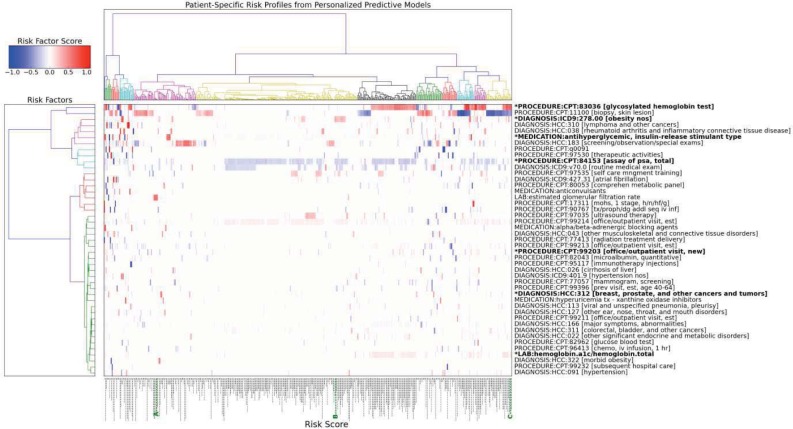
To facilitate the analysis of the characteristics and distribution of the patient specific risk factors, agglomerative hierarchical clustering (using a Euclidean distance measure) is performed on the personalized risk factor profiles. Figure 2 is a hierarchical heat map plot showing the top risk factors identified by the personalized predictive models for 500 randomly selected patients. The patient specific risk factor profiles (i.e., the columns) are clustered along the horizontal axis. The individual risk factors (i.e., the rows) are clustered along the vertical axis. The color in the heat map corresponds to the risk factor score values (i.e., beta coefficient values) in the patient risk profiles: red is high while blue is low. Analysis of the risk factor profile clusters shows that some patients share very similar risk factors and are grouped together in the same cluster whereas other patients have very different and almost non-overlapping risk factors and belong to groups that are far apart in the cluster tree. The patient specific risk scores are plotted as vertical bars along the bottom of the horizontal axis; the longer the bar, the higher the risk score. Patients with certain risk factor profiles have consistently higher risk scores. For example, patients with high values For “PROCEDURE:CPT:83086 [glycosylated hemoglobin test]” and “LAB:hemoglobin.a1c/hemoglobin.total” in their risk profiles have much higher risk scores than those with low values for these factors. Patients with similar risk scores can have very different risk factors. For example, the three case patients, highlighted in green as A, B and C on the bottom axis of Figure 2, all have risk scores around 0.75 but the top risk factors for each patient are different: “LAB: estimated glomerular filtration rate” For patient A, “DIAGNOSIS:ICD9:278.00 [obesity nos]” for patient B, and “PROCEDURE:CPT:83036 [glycosylated hemoglobin test]” and “LAB:hemoglobin.a1c/hemoglobin.total” for patient C. The personalized risk factor for each patient can also differ from the risk factors captured by the global model (indicated by highlighted risk factor names with a * prefix). Indeed, a large number of risk factors not captured by the global model are identified in the personalized models as useful predictors. Finally, the risk factor clusters along the vertical axis can be used to identify groups of risk factors that have high co-occurrence rates across the patient risk factor profiles. For example, “PROCEDURE:CPT:84153 [assay of psa, total]” and “DIAGNOSIS:ICD9:v70.0 [routine medical exam]” frequently occur together.
Figure 2:
Hierarchical heat map plot showing the top risk factors for diabetes onset identified by the personalized predictive models for 500 randomly selected patients. Patient specific risk factor profiles (the columns) are clustered along the horizontal axis. Risk factors (the rows) are clustered along the vertical axis. Risk factors captured by the global model are highlighted and have a * prefix in the name. The risk score for each patient is plotted as a vertical bar along the bottom.
Discussion and Conclusion
Patient specific personalized predictive models trained using a smaller set of data from patients that are clinically similar to the query patient can perform better than a global predictive model trained using all the training data. Unlike statically trained global models, personalized models are trained dynamically and can leverage the most relevant information available in the patient record. Personalized predictive models can be analyzed to identify risk factors that are important for the individual patient and used to create personalized risk factor profiles. Cluster analysis of the risk profiles show groups of patients with similar risk factors, differences in the top risk factors for different groups of patients and differences between the individual and global risk factors. Once identified, the patient specific risk factors may be leveraged to support better targeted therapies, customized treatment plans and other personalized medicine applications.
There are many possible directions for future work. First, the approach and methods need to be validated on larger patient data sets and additional disease targets. Next, more sophisticated methods can be developed and used for a number of the processing steps. This includes expanding the feature construction processing to include temporal features, exploring the impact of balanced vs. unbalanced selection of similar case-control training patients, improving the robustness of the feature selection filtering by using cross validation methods, extending the predictive modeling to include additional classification algorithms that have interpretable models, such as Bayesian classifiers and decision trees, and developing risk factor profile extraction approaches for these additional classifiers. More in depth analysis of the characteristics and distribution of the risk factors captured by the personalized models is also needed. This can include exploring risk factors that could be masked by excluding the well-known risk factors from the model a priori. Finally, the potential of combining information captured by the global and personalized models as well as by different types of classifiers should also be explored.
References
- 1.Amarasingham R, Moore BJ, Tabak YP, Drazner MH, Clark CA, Zhang S, et al. An automated model to identify heart failure patients at risk for 30-day readmission or death using electronic medical record data. Med Care. 2010 Nov;48(11):981–8. doi: 10.1097/MLR.0b013e3181ef60d9. [DOI] [PubMed] [Google Scholar]
- 2.Pittman J, Huang E, Dressman H, Horng C-F, Cheng SH, Tsou M-H, et al. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8431–6. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012 Jun;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 4.Vickers AJ. Prediction models in cancer care. CA Cancer J Clin [Internet] 2011. Jun 23, [cited 2013 Nov 25]; Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3189416/ [DOI] [PMC free article] [PubMed]
- 5.President’s Council of Advisors on Science and Technology Priorities for Personalized Medicine. [Internet] 2008. Sep, Available from: www.whitehouse.gov/files/documents/ostp/PCAST/pcast_report_v2.pdf.
- 6.Overby CL, Pathak J, Gottesman O, Haerian K, Perotte A, Murphy S, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc JAMIA. 2013 Jul 9; doi: 10.1136/amiajnl-2013-001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyderman R. Personalized health care: from theory to practice. Biotechnol J. 2012 Aug;7(8):973–9. doi: 10.1002/biot.201100297. [DOI] [PubMed] [Google Scholar]
- 8.Kasabov N. Global, local and personalised modeling and pattern discovery in bioinformatics: An integrated approach. Pattern Recogn Lett. 2007 Apr;28(6):673–85. [Google Scholar]
- 9.Kasabov N, Hu Y. Integrated optimisation method for personalised modelling and case studies for medical decision support. Int J Funct Inform Pers Med. 2010 Mar 1;3(3):236–56. [Google Scholar]
- 10.Visweswaran S, Angus DC, Hsieh M, Weissfeld L, Yealy D, Cooper GF. Learning Patient-Specific Predictive Models from Clinical Data. J Biomed Inform. 2010 Oct;43(5):669–85. doi: 10.1016/j.jbi.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Z, Hsu W, Liu Z, Lee ML. PROCEEDINGS OF THE 6TH PAKDD. Springer; 2002. SNNB: A Selective Neighborhood based Naive Bayes for Lazy Learning; pp. 104–14. [Google Scholar]
- 12.Chawla NV, Davis DA. Bringing big data to personalized healthcare: a patient-centered framework. J Gen Intern Med. 2013 Sep;28(Suppl 3):S660–5. doi: 10.1007/s11606-013-2455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell TM. Machine Learning. 1st ed. McGraw-Hill Science/Engineering/Math; 1997. p. 432. [Google Scholar]
- 14.Wang F, Sun J, Li T, Anerousis N. Two Heads Better Than One: Metric+Active Learning and its Applications for IT Service Classification; Ninth IEEE International Conference on Data Mining, 2009, ICDM ’09; 2009. pp. 1022–7. [Google Scholar]