Abstract
Health care data included in clinical data repositories (CDRs) are increasingly used for quality reporting, business analytics and research; however, extended clinical data from interprofessional practice are seldom included. With the increasing emphasis on care coordination across settings, CDRs need to include data from all clinicians and be harmonized to understand the impact of their collaborative efforts on patient safety, effectiveness and efficiency. This study characterizes the extended clinical data derived from EHR flowsheet data that is available in the University of Minnesota’s CDR and describes a process for creating an ontology that organizes that data so that it is more useful and accessible to researchers. The process is illustrated using a pressure ulcer ontology and compares ease of finding concepts in i2b2 for different data organization approaches. The challenges of the manual process and difficulties combining similar concepts are discussed.
Introduction and Background
Health care data included in clinical data repositories (CDRs) are increasingly used for quality reporting, business analytics and research; however, data generated by clinicians (physicians, nurses, therapists and other providers) from interprofessional practice are seldom standardized in electronic health records (EHRs) nor included in CDRs. With the increasing emphasis on interprofessional practice and care coordination across settings1, CDRs need to include data from all clinicians and be harmonized across settings to support research that leads to a better understanding of the impact of their collaborative efforts on patient safety, effectiveness, and efficiency2. There are a number of current national and regional efforts to share data between CDRs for the purpose of supporting research activities including identification of cohorts for clinical studies and comparative effectiveness of research from multiple sites. These include the eMerge network3, the HMO Research Network (HMORN)4, the Clinical Data Research Networks (CDRNs) making up PCORnet5, MiniSentinel6 for adverse drug reactions and Observational Medical Outcomes Partnership (OMOP)7. All of these networks have generally focused on a relatively restricted set of data. That set includes patient demographics such as birthdate, gender, marital status, and geographic location; diagnoses; procedures; encounters; and sometimes laboratory orders and results as well as medications. However these domains represent only a relatively small fraction of the data characterizing patients and care provided that is captured in the EHR.
Extended clinical data (ECD) are needed to meet the operational and research needs of health system and academics. In this paper, by ECD, we specifically are addressing structured or semi-structured data documented in flowsheets that represent interprofessional patient assessments, patient goals, interventions, and outcomes. Examples of these data include physiological concepts (e.g. pain, pressure ulcers, tissue perfusion, bowel elimination), as well as psychological (e.g. cognitive function, coping), and functional categories (e.g. activities of daily living, self-care, nutrition) along with health related problems (e.g. medication management, fall risk). The structure behind flowsheets changes over time and across service lines to accommodate clinician workflows and updates to EHRs resulting in many variations in observations of the same phenomenon.
There are proposed methodologies for creating an ontology to organize flowsheet data for secondary use that makes sense for operational staff and researchers. The current study focuses on flowsheet data from an instance of Epic Systems Corporation’s (“Epic”) EHR system and builds on previous work by Waitman, Warren, and their colleagues.8,9 Epic flowsheets have a hierarchical architecture that includes templates at the highest level representing a screen view (such as an adult admission assessment) and contains multiple groups of similar observations labeled measures or rows (e.g. vital signs or tobacco use screening). Templates are comprised of groups and/or measures. Groups are collections of measures. Templates and groups provide the context for observations, but the combination results in an exponential growth in the number of template-group-measure combinations to support clinician workflows and EHR upgrades over time. Waitman et al8 used a six-step pruning and clustering technique to condense their flowsheets into higher level templates, groups, and measures for organizing this data in i2b2 (Informatics for Integrating Biology and the Bedside). i2b2 is an informatics framework that simplifies the process of cohort discovery10. This resulted in a considerable reduction in the number of templates, groups, and measures displayed in the navigation hierarchy; however, there continued to be redundancy of concepts represented by measures across groups and templates. Warren et al6 used two focus groups (academic nurse researchers and clinical based nurses who were secondary users of EHR data) and identified 18 templates or high level categories: vital signs, height and weight, intake and output, fall risk, safety, IV therapy, and the remainder represented body systems, psychosocial/educational/cultural/spiritual assessments, and social habit assessments. This approach provided a method of organizing flowsheet data at a high level, but further work was needed to link flowsheet measures to concepts from the EHR data to support quality improvement and research. Our research builds on and goes beyond these studies by creating a more general process for modeling flowsheet data for an entire clinical area (pressure ulcers11).
Purpose of Study
The purpose of this study is to characterize the flowsheet data that is in a CDR and describe a process for developing an ontology that organizes that data to be more useful and accessible to researchers. The process is illustrated using a pressure ulcer ontology as an example.
Method of Development
The University of Minnesota (UMN) collaborates with Fairview Health Services and University of Minnesota Physicians to create and maintain a CDR. The CDR includes over 2 million patients from seven hospitals and 40 clinics. Data are documented in a single Epic EHR instance for the health system and extracted from their Clarity database into the UMN’s CDR. The types of data include patient history and demographics, encounters, procedures, diagnosis, notes, labs, medications, and flowsheets. The data are stored in 153 tables and each of the tables was categorized to belong to one of those groups. The sum of the table record counts in those categories was used to characterize the dataset.
Our discussions with other CTSAs lead us to believe that this is one of the few CDRs that includes flowsheet data. A 200,000 encounter subset of that data was extracted from the CDR for data exploration and to characterize aspects of the flowsheet data related to templates, groups and measures. The hierarchy was abstracted from the data set to create a database of all of the combinations of templates, groups, measures and their value sets. The frequency information for each measure (numbers of times documented, patients, and encounters) was also abstracted. This level of detail was essential to support the ontology development process.
An iterative process for mapping the flowsheet data to a logically organized structure was performed for five clinical areas of interest (prevention of pressure ulcers, falls, catheter associated urinary tract infections, venous thrombosis embolism, and pain management). The process to create a list of concepts and their relationships consisted of several steps. First we generated a list of research questions for the clinical area of interest and identified relevant clinical concepts to extract into an initial concept list. We then searched for the concepts in the extracted hierarchy dataset of templates/groups/measures using terms characterizing the clinical area. When a concept was found, we examined the measures associated with the same groups and templates as the concept to find additional concepts that may be related to the clinical area of interest. The concepts were recorded in a new spreadsheet and all flowsheet measures representing the same concept were mapped to the concept. The research team met weekly to validate findings. This iterative process continued until no new concepts were found. The result of this process was a list of concepts and data captured in the flowsheets for a clinical area.
Concepts were then organized into a logical order. At the highest level, concepts were grouped using a set of categories familiar to clinicians – assessments, goals for problems, interventions, and outcomes. This top-down approach helped form a skeleton for the hierarchy of concepts. Concepts that are clinically similar were grouped together. For example, for the pressure ulcer clinical area, all of the concepts related to Skin Assessments are grouped together. Determining where the concept fit under the higher level concepts followed. Flowsheet measures that had similar descriptions were then grouped together as a single concept based on that measure’s value set. If the value sets were similar, then they likely represented the same concepts. Equally important, if the value sets were different, then they represented different concepts. For example, there are three different measures named “Skin Integrity.” Two of them had value sets with choices like abrasion, blister, body piercing, burn(s), cracked etc., but the third measure’s value set had choices like blanchable erythema, bruising, dark purple area, etc. The first two were concluded to represent the same concept but the third “Skin Integrity” measure was considered a different concept in the ontology.
Initially, spreadsheets were used to document the work. But as the ontologies grew (the pressure ulcer ontology quickly grew to 84 concepts) a more robust ontology development tool, Protégé, was employed. Since the hierarchy kept changing (concepts were moved between subtrees, split and merged), Protégé tracked those changes more easily and accurately. As a final step, the concept lists for pressure ulcers were translated into an i2b2 hierarchical ontology. Determining if the organization of concepts was useful to researchers was the focus. To support this analysis, the original template/group/measure hierarchy was also translated into an i2b2 ontology to enable comparison of the effort needed to navigate to concepts of interest using each organizational approach.
Results
The flowsheet data comprised the largest volume of information (table rows) in the CDR at 34% (Figure 1). That was more than double the second largest volume of data, Orders and Procedures. The flowsheet data consisted of 14,564 measures (each measure is one type of row) in 2,972 groups in 562 templates. There were over 1.2 billion observations stored in the flowsheets. Ninety-five percent (95%) of all of the observations were covered by 2,000 measures; 65% of the measures (9,505) were choice lists. There were 56,965 values within those choice lists.
Figure 1:
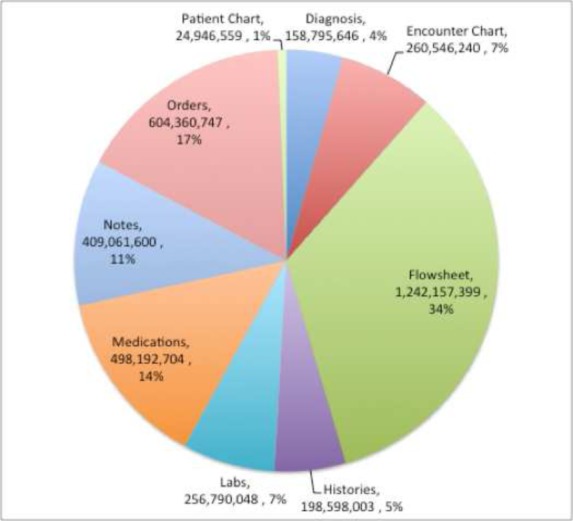
CDR table row proportions
The iterative process of mapping measures to concepts was performed to produce an ontology for pressure ulcers. The mapping contained 84 concepts (from 96 measures) organized into a hierarchy (Figure 2). These concepts appeared on 72 templates. On average, each concept appeared on 12 templates and one concept (Braden Score) appeared on 28 different templates; 6 of the concepts appeared on only a single template. This mapping process clearly demonstrated that measures for similar concepts were distributed across many different templates and groups in the CDR.
Figure 2:
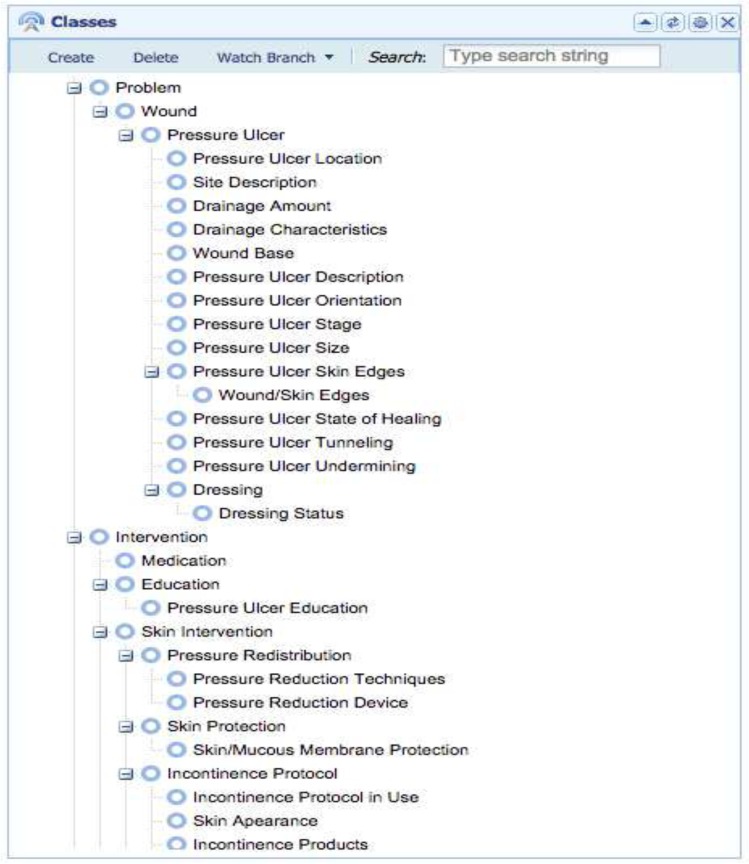
Pressure ulcer ontology
In order to further test the organization of the concepts, i2b2 was used to illustrate how researchers would navigate concepts to answer simple questions, e.g. “How many patients have pressure ulcers?” There were two flowsheet measures that are used to record whether a patient has a pressure ulcer. One of the fields expects “Yes/No” answers. The other has “Suspected/No” as answers. Unfortunately, these measures are named the same in the CDR and they appear on 14 different templates. A researcher using i2b2 would have to follow 14 different paths to find one of the two measures and either put in a “Yes” or “Suspected” (depending on the measure) to find all of the patients with pressure ulcers. Figure 3 shows what the researcher would see in i2b2 if they were using they template/group/measure hierarchy. The i2b2 diagram is only a partial view since it is not possible to open the hierarchy and show all 14 data elements simultaneously. If the researcher used the ontology based navigation, the concept of “Pressure Ulcer Present” would only exist once. Figure 4 shows what the researcher would see in this case. The concept of “Pressure Ulcer Present” is still shown to have two forms. The confirmed form has “Yes/No” answers and the suspected form as “Suspected/No” as answers. However, from a researcher’s point of view, all of the concepts and information needed to specify answers to the question were located near each other in the navigation hierarchy.
Figure 3:
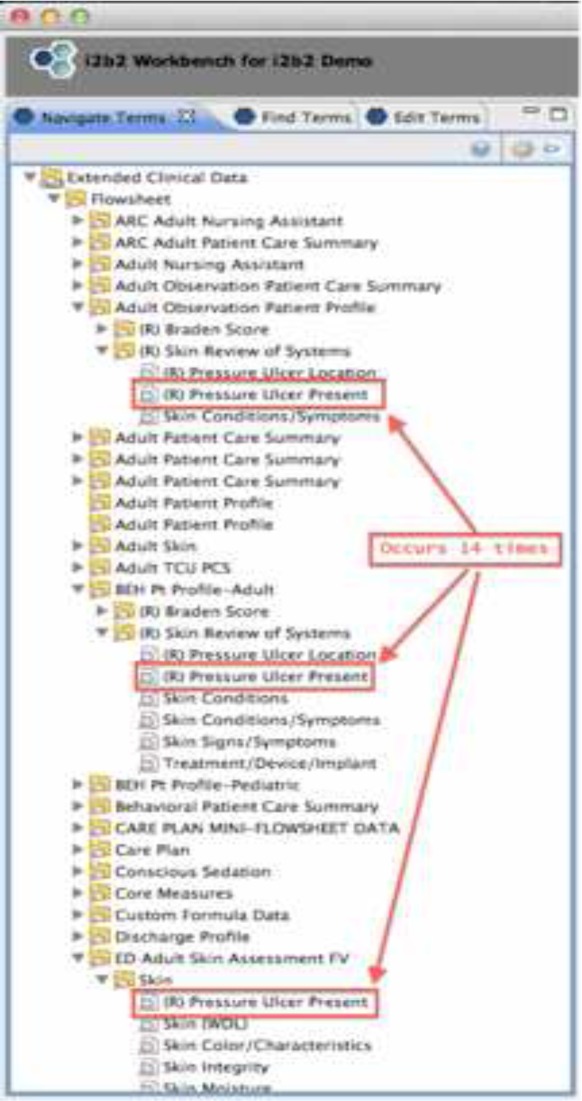
Template based navigation (i2b2)
Figure 4:
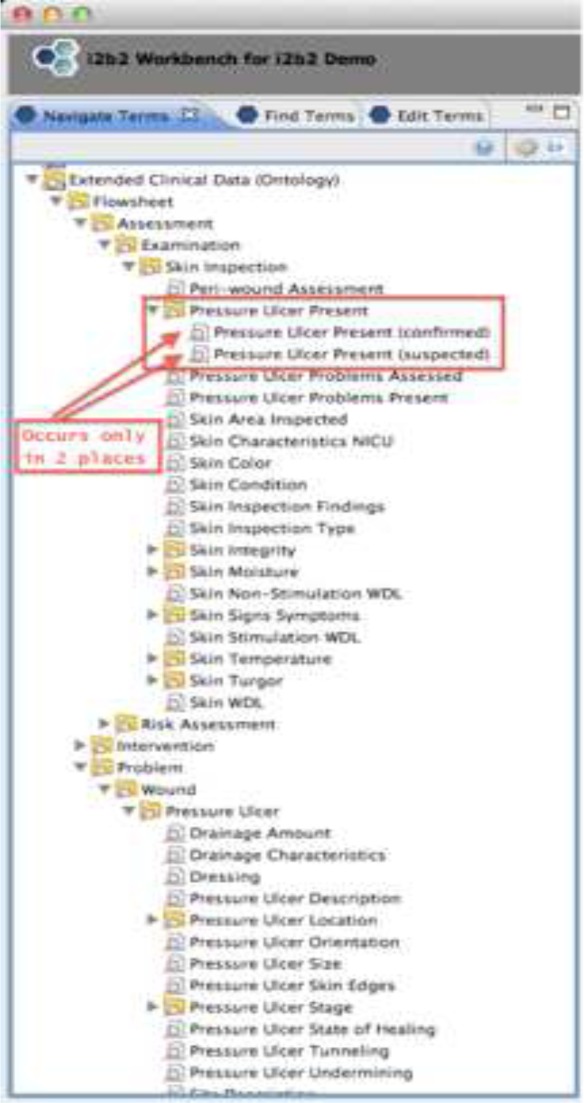
Ontology based navigation (i2b2)
Discussion
The flowsheet data comprised a large proportion of all of the data in the CDR (more than a third of all table rows). This flowsheet data consists of a large number of templates, groups and measures that are customizable at each healthcare organization and even across service lines within a single organization. The information contained in the flowsheets is important to include in research because it provides a more complete and interprofessional view of healthcare concepts and practices. Organizing the flowsheet data into an ontology holds the promise of making searches for information more efficient and accurate from a researcher’s perspective.
One challenge that was encountered was manual mapping. Mapping flowsheet measures to the pressure ulcer ontology manually was a difficult, time consuming process and potentially error prone. This process did produce an ontology of 84 concepts that covered 96 measures to answer research questions. However, a manual approach is not likely to be a practical and scalable way to organize the remaining 14,468 flowsheet measures. Further research on automating the process is needed, though complete automation is probably not attainable.
A second challenge involved determining which flowsheet measures should be included in the overall ontology. While it may not be practical or desired to map all of the flowsheet data into an ontology, it is unclear how mapping priorities should be determined. Waitman, Warren and their colleagues5,6 picked a threshold of only mapping measures that occurred 35 or more times or were used monthly. However, frequency by itself is a limited indicator of importance. For example, low frequency could be indicative of a measure that was just recently implemented or focused on a condition that rarely occurred but was extremely important when it does appear. Frequency can be a way of prioritizing work, but using frequency to dismiss measures could exclude important data. In this study, measures that were deprecated or no longer in use were included. From a researcher’s perspective, deprecated measures, which have been used to document patient care in the past, are equally valuable.
Another challenge was determining when to combine individual flowsheet measures into a single concept. Certainly, if two or more flowsheet measures had the same value set they could be combined. However, if flowsheets had similar but different value sets, it was unclear how to combine them. Combining all of the terms together (union of values) and reconciling the lists by manually mapping the values from the two value sets were considered. It was even more difficult to devise a method for combining a measure that had a list of choices with a measure that accepted free form text. Mapping these value sets to a standard terminology would be a possible solution; however it is unclear how many of these values exist in terminologies like SNOMED CT. For this study, the pressure ulcer ontology had 34 flowsheet measures distributed amongst 12 concepts that could be combined, which is 35% of all of the measures covered by the ontology.
Even after an ontology is produced, mapping the concepts to LOINC and mapping the 56,965 values from the choice lists to SNOMED CT is a daunting task. It is an open question as to where to stop mapping data to standard codes. Further research is needed to determine the importance that researchers place on concepts represented in value sets that therefore would need standardized codes.
Finally, developing the pressure ulcer ontology revealed that some of the concepts necessary to address research questions were not available in the flowsheet data. For example, age, gender, etc. are important to researchers. While these concepts were not included in the ontology, they are accessible to researchers in other parts of the i2b2 hierarchy.
This process has only been conducted on pressure ulcer related flowsheet data from a single healthcare organization. Research is underway to develop ontologies for four additional clinical areas (catheter associated urinary tract infections, pain, venous thromboembolism and falls). Further research is also needed to validate this with another healthcare organization’s flowsheet data. As a result of applying the method of mapping flowsheet measures to clinical concepts, requirements for a software tool to automate the process have been developed. The prototype tool is in test to determine a practical query strategy for finding flowsheet measures, providing a similarity score for matching, and then mapping the measures to concepts. Further requirements for improving the tool are in process.
Conclusion
Extend clinical data derived from flowsheets are a large and important component of health information stored in the EHR and it is increasingly important to include it in an organization’s CDR to provide a more complete picture of care delivery. It is organized in the EHR to make data entry for each clinical workflow efficient. If data is organized in that manner in the CDR, it will be difficult for researchers to find concepts of interest. An ontology based approach for organizing ECD delivers a more useful and efficient navigation process for a researcher. Further research is needed to resolve the challenges of mapping this data to an ontology and to validate this approach in other clinical areas.
Acknowledgments
This was supported by Grant Number 1UL1RR033183 from the National Center for Research Resources (NCRR) of the National Institutes of Health (NIH) to the University of Minnesota Clinical and Translational Science Institute (CTSI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the CTSI or the NIH. The University of Minnesota CTSI is part of a national Clinical and Translational Science Award (CTSA) consortium created to accelerate laboratory discoveries into treatments for patients.
This study was also partially supported by NSF grant: NSF IIS-1344135.
References
- 1.Nandan M, Scott PA. Interprofessional practice and education: Holistic approaches to complex health care challenges. J Allied Health. 2014;43(3):150–156. [PubMed] [Google Scholar]
- 2.Kaushal R, Hripcsak G, Ascheim DD, et al. Changing the research landscape: The New York City clinical data research network. J Am Med Inform Assoc. 2014;21(4):587–590. doi: 10.1136/amiajnl-2014-002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogt TM, Elston-Lafata J, Tolsma DD, Greene SM. The role of research in integrated health care systems: the HMO Research Network. Perm J. 2004;8(4):10–7. doi: 10.7812/tpp/04.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleurence RL, Curtis LH, Califf RM, et al. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrman RE, Benner JS, Brown JS, et al. Developing the Sentinel System—a national resource for evidence development. New England Journal of Medicine. 2011;364(6):498–499. doi: 10.1056/NEJMp1014427. [DOI] [PubMed] [Google Scholar]
- 7.Stang PE, Ryan PB, Racoosin JA, Overhage JM, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Annals of Int Med. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 8.Waitman LR, Warren JJ, Manos EL, Connolly DW. Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data repository to support research and quality improvement. AMIA Annual Symposium Proceedings. 2011;2011:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 9.Warren JJ, Manos EL, Connolly DW, Waitman LR. Ambient Findability: Developing a Flowsheet Ontology for i2B2. Proc 11th Int Congr Nurs Informatics 2012. 2012 Jan;(1):432. [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Choi J, Secalag L, et al. Building an Ontology for Pressure Ulcer Risk Assessment to Allow Data Sharing and Comparisons Across Hospitals; AMIA Annual Symposium Proceedings; 2010. pp. 382–386. [PMC free article] [PubMed] [Google Scholar]


