Abstract
Medication in for ma lion is one of [he most important clinical data types in electronic medical records (EMR) This study developed an NLP application (PredMED) to extract full prescriptions and their relevant components from a large corpus of unstructured ambulatory office visit clinical notes and the corresponding structured medication reconciliation (MED REC) data in the EMR. PredMED achieved an 84.4% F-score on office visit encounter notes and 95.0% on MED„REC data, outperforming two available medication extraction systems. To assess the potential for using automatically extracted prescriptions in the medication reconciliation task, we manually analyzed discrepancies between prescriptions found in clinical encounter notes and in matching MED_REC data for sample patient encounters.
Introduction
Medication information is one of the most important clinical data types in electronic medical records (EMR). A complete understanding of a patient’s medication status is critical for healthcare safety and quality. It is also useful in detecting drug-related pathology or changes in clinical signs that may be the result of drug therapy1. Since 2005, medication reconciliation (MR) has been part of the Joint Commission’s National Patient Safety Goals2. The goal of MR is to obtain and maintain accurate and complete medication information for a patient, which will be used in the course of patient care to ensure safe and effective medication use3, However, poor communication of medication information at transition points as well as unstructured medication descriptions recorded in free-text clinical notes challenge the reconciliation process.
Clinical notes contain rich medical information, such as the patient’s symptoms, current medication prescriptions, examination findings, lab/x-ray results, etc. In recent years, many systems have leveraged natural language processing (NLP) technologies to extract information embedded in clinical notes, including Medical Language Extraction and Encoding System (MedLEE)4: Apache clinical Text Analysis and Knowledge Extraction Systems (cTAKES)5; and Health Information Text Extraction (HITEx)6, 7. In 2009, the 3rd i2b2 workshop on NLP challenges focused on medication identification, including medication name, dose, administration route, frequency, duration and reason in clinical discharge summaries8. Vanderbilt University used an automated medication extraction system (MedEx)9 to accurately extract medication name, strength, route, form, dosage, duration, and frequency. Mayo’s Medication Extraction and Normalization system (MedXN) was recently developed to extract similar medication information and normalize it to the most appropriate RxNorm concept unique identifier10.
This study develops an NLP application to extract prescription information from a large corpus of unstructured ambulatory office visit clinical notes and to investigate its value for supplementing the structured medication reconciliation (MED_REC) data in the EMR. For this effort, prescription information includes medications and devices (e.g., syringes). This work is part of a larger project, called PredMED (Predictive Modeling for Early Detection)11. We conjecture that extracted prescriptions can also enable automated medication reconciliation by comparing prescription medication information found in encounter clinical notes with prescriptions listed in MED_REC entries for the same patients.
Materials and Methods
An NLP application was developed and validated for identifying prescriptions in both office visit encounter notes and in MED_REC; then it was used to further investigate discrepancies between those two sources. For medication prescriptions, seven component types were extracted, shown in Table 1. For syringe prescriptions, two types were extracted, including MedSize which indicates gauge, capacity and/or length, and MedForm (see Table 2). For other device prescriptions, relevant annotations were extracted (e.g., ONE TOUCH ULTRA DEVI, with “one touch ultra” as MedName and “DEVI” as MedForm).
Table 1:
Medication prescription component types, including definitions and examples.
| Type | Definition | Example (bold, italic) |
|---|---|---|
| MedName | Medication name, including prescribed medications, over the counter medications, biological substances, and medication classes suggested by doctors. |
Olcium 600 MG PO TABS Coumadin 2.5 MG PO TABS Aspirin 81 MGPOTABS |
| MedStrength | The strength of a single medication. | Lopressor 50 MG PO TABS |
| MedRoute | The route of drug administration. | Lopressor 50 MG PO TABS |
| MedForm | The physical appearance of the medication. | Lopressor 50 MG PO TABS |
| MedDosaee | The dose intake amount of each medication. | one tablet daily |
| MedFrequency | The frequency of medication intake suggested or required. | every 6 hours as needed |
| MedDuration | The time period over which the medication is administered. | × 3days |
Table 2:
Syringe prescription annotation types, including definitions and examples.
| Type | Definition | Examples |
|---|---|---|
| Med Size | Capacity: the amount of medication the syringe can contain. | 1 cc, 0.5 ml, 3cc |
| Gauge: the size of needle. | 18G, 21G, 26G, 29 G | |
| Length: the length of needle. | 1-1/2″. 5/16″ | |
| MedForm | The device prescription category of syringe/needle. | M1SC, KIT |
Source of Data
Data for this study were obtained from the Geisinger Health System (GHS) primary care practice EMRs. The dataset consisted of the full encounter records for 6355 incident primary care HF patients diagnosed between 2003 and 2010. We randomly selected 187,829 office visit encounter notes and the corresponding matched 1,264,730 MED_REC records in the structured EMR, as the training corpus to develop the NLP application. The encounter notes and MED_REC records were matched based on the combination of patient id and contact date. In order to evaluate the performance, gold standard test sets were created for a small corpus of 50 "Office Visit" encounter notes randomly selected from the dataset as well as a corpus of the 346 matching MED_REC entries. Two doctors individually annotated die small corpora using Knowtator, a general-purpose text annotation tool12. Initial inter-annotator agreement (IAA) was computed. Then, the two doctors adjudicated their differences, by consensus, to produce the annotations which served as the final gold standards.
Tools
IBM Content Analytics Studio 3.013 (ICA), an Eclipse-based development environment for building custom text analyzers in various languages, was used for basic text processing and for developing dictionaries and grammars. The resulting analysis resources were inserted into an Unstructured Information Management Architecture (U1MA) pipeline14, 15, which also provided text pre-processing and additional text analysis engines for segment type identification, semantic constraints, and output formatting. A concordance program16 was used for linguistic analysis of the syntax and contexts of prescriptions in the entire training corpus. This knowledge was incorporated into the development of dictionaries, grammar rules, and constraints for the NLP application.
Prescription Analysis Pipeline
Figure 1 depicts the high-level prescription analysis pipelines. Basic text processing includes sentence and paragraph boundary detection, tokenization, dictionary look-up, and part-of-speech lagging in ICA. Word-error correction and section header detection were also performed. Relevant dictionaries, grammars and rule sets were built to recognize words and phrases used in prescriptions (see Table 1 and Table 2).
Figure 1:
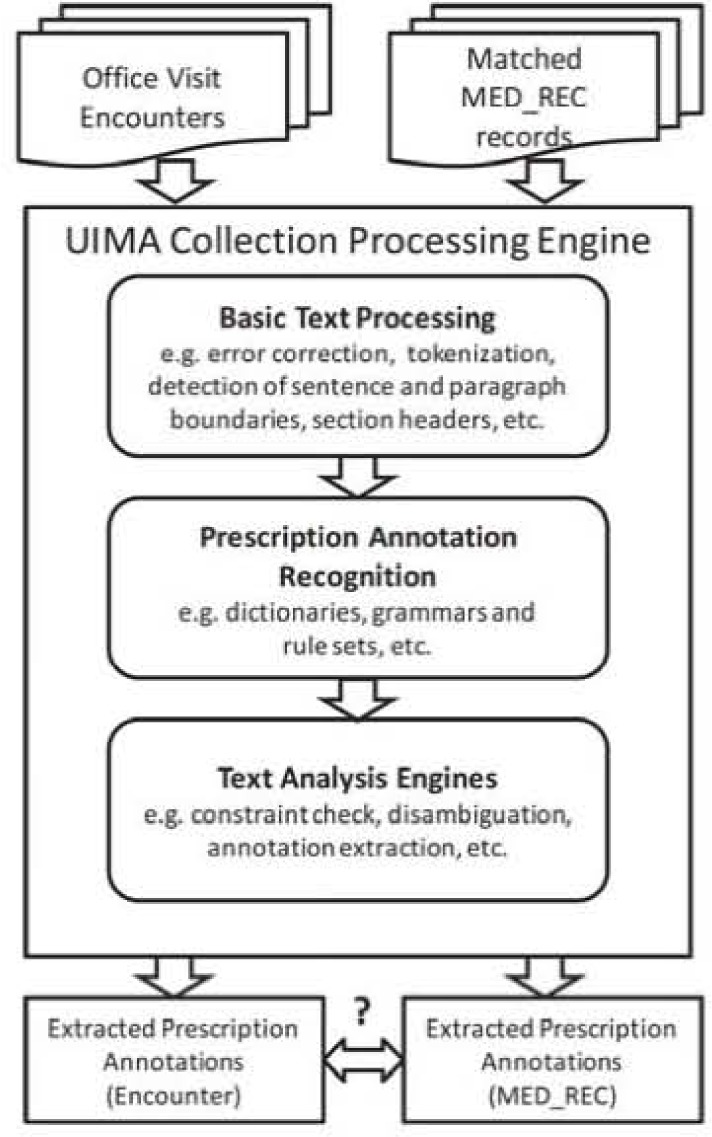
Analysis Pipelines.
To identify MedNames, we used a dictionary built from RxNorm17 by combining term types: single ingredient (IN, e.g., “acetazolamide”), multiple ingredients (MIN. e.g., “acetic acid” / “hydrocortisone”), precise ingredient (PIN. e.g., “acetazolamide sodium”) and brand name (BN, e.g., “acid gone”). Pharmacy primary-class (e.g., “diuretics”), sub-classes (e.g., “loop diuretics”) and generic names (e.g., “domperidone”) from structured Geisinger EMR records were also imported as look-up dictionaries for McdName. An Addenda dictionary contained further MedNames encountered during training that were not in any of our other sources. A dictionary of Prescription Verbs allowed us to recognize verbs (e.g., “take”, “discontinue”, “SIG:”, “Rx:”) that typically occur with prescriptions. Further dictionaries were built for identifying DosageForms (e.g., “AERO”, “caps”); syringe CapacityUnits (e.g., “ml”); MedRoutes (e.g., “OR”, “by mouth”); Latin frequencies (e.g., “b.i.d.”); TimeUnits (e.g., “hour”, “week”); TimeOfDay (e.g., “morning”, “mealtime”); etc. These dictionaries were built manually, based on standard lexical resources together with extensive exploration of concordances over the Geisinger encounter note corpus.
We created parsing rules to recognize candidate MedDescriptions (for MED_REC) and Prescriptions (for encounter notes). The top-level rules, which are 1CA aggregate rules (i.e., they operate over an entire sentence, skipping extraneous tokens) are: Med Description ➔ MedPart? MedName MedPart{l,4} and Prescription ➔ RxPart* MedName RxPart*. MedPart can he any of MedStrength, MedRoute. MedForm, or MedSize. RxPart can be any of MedDosage, MedFrequency, MedDuration, PrescriptionVerb, or a MedPart. Example candidates are shown in Figure 2.
Figure 2:

Examples of candidate annotations and their components.
Once the Prescription candidates are recognized, a UIMA text analysis engine (TAE) performs disambiguation and applies constraints before final annotations are created. For example, a MedName will be ignored if no RxPart is found in the same Prescription. Similarly, an RxPart without a valid MedName will also be ignored. To disambiguate ambiguous lower-case MedRoutes (e.g., “to”, “in”, or “or”), we use a “preferred route” attribute in the MedNames dictionary to determine whether an apparent route is just an ordinary word. Based on observation of many prescriptions, a heuristic ignores a Prescription if there are twelve or more ordinary words between RxParts. The TAE enforces similar, but simpler, constraints and disambiguation when annotating the parts of MedDescriptions.
Evaluation and Discrepancy Analysis
The performance of this prescription pipeline (PredMED) on individual annotations and the whole Prescription was evaluated on the gold standards developed with the 50 office visit encounter notes as well as the 346 matched MED_REC entries. Measurement metrics of precision, recall and F-score were utilized. (Precision = TP/(TP+FP): Recall = TP/(TP+FN); F-score = {2 × Precision × Recall)/(Precision + Recall); where “TP” is true positives, “FP” is false positives: “FN” is false negatives.) We also applied MedEx9 and MedXN10, two openly available UIMA-based medication extraction systems, to the test corpora and compared their performance with PredMED’s using Chi-square or Fisher’s exact test. Differences with P<0.05 were considered significant. To investigate prescription discrepancies between office visit encounter notes and matched MED_REC records, the extractions were matched on MedName, MedStrength, MedRoute. and MedForm (or, for devices, on MedName. MedSize, and MedForm).
Results
Manual Annotation Statistics
Table 3 shows the IAA of two doctors’ initial annotations of 50 office visit encounter notes and the corresponding 346 MEDREC entries. The overall agreement was greater for MED_REC entries than for encounter notes (96.8% vs. 91.7%). The relatively simple and consistent description of medications in structured MED_REC. without narrative mentions of medication dosage, duration and frequency, explains the better agreement.
Table 3:
IAA for initial expert annotations of 50 office visit encounter notes and 346 matched MED_REC entries, before adjudication.
| Annotation Types | Encounter Notes | MED_REC |
|---|---|---|
| All Types | 91.7% | 96.8% |
| MedName | 92.3% | 98.2% |
| MedStrength | 94.1% | 95.4% |
| MedRoute | 87.9% | 98.2% |
| MedForm | 98.1% | 96.9% |
| MedSize | 53.9% | 35.3% |
| MedDosage | 94.0% | – |
| MedDuration | 56.3% | – |
| MedFrequency | 85.4% | – |
| Prescription | 93.2% | – |
Performance Evaluations
Table 4 shows the evaluation results of PredMED on 50 office visit encounter notes in terms of precision (P). recall (R) and F-score (F). Prescription, MedName, MedStrength, MedRoute, MedForm, and MedFrequency achieved F-scores above 80%. MedDosage and MedDuration had high precision (>90%) but low recall. Table 5 shows the evaluation results of PredMED on MED_REC. Four annotation types - MedName. MedStrength, MedRoute, MedForm achieved high F-score (>90%). The low performance on MedSize and MedDuration reflects the complexity of the language with which they are expressed, which also is consistent with the low initial IAA between doctors - MedSize: 53.9%, MedDuration: 56.3%.
Table 4:
Performance of PredMED on 50 office visit encounter notes and comparisons to the performance of MedEx and MedXN (* indicates significantly different performance when compared to PredMED: bold represents performance inferior to PredMED; italics represents performance superior Lo PredMRD).
| Annotation Types | Total # | PredMED | MedEx | MedXN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P(%) | R(%) | F(%) | P(%) | R(%) | F(%) | P(%) | R(%) | F(%) | ||
| AH Types | 2848 | 85.8 | 83.0 | 84.4 | 79.6* | 64.8* | 71.5* | 81.6* | 75.3* | 783* |
| Prescription | 464 | 78.2 | 83.4 | 80.8 | – | – | – | – | – | – |
| MedName | 475 | 78.4 | 81.7 | 80.0 | 63.8* | 90.3* | 74.8* | 82.8 | 85.1 | 83.9 |
| MedStrength | 343 | 91.2 | 90.4 | 90.8 | 87.6 | 86.3 | 86.9 | 89.6 | 93.3 | 91.4 |
| MedRoute | 436 | 83.7 | 91.5 | 87.4 | 90.1 | 43.6* | 58.7* | 98.4* | 57.6* | 72.7* |
| MedForm | 355 | 96.5 | 91.8 | 94.1 | 89.5* | 54.9* | 68.1* | 48.4* | 58.9* | 53.1* |
| MedSize | 8 | 50.0 | 12.5 | 20.0 | – | – | – | – | – | – |
| MedDosage | 357 | 92.5 | 65.3 | 76.5 | 97.9* | 52.9* | 68.7* | 99.3* | 75.6* | 85.9* |
| MedDuration | 21 | 93.3 | 66.7 | 77.8 | 13.2* | 23.8* | 17.0* | 88.9 | 76.2 | 82.1 |
| MedFrequency | 389 | 89.7 | 78.7 | 83.8 | 89.1 | 60.7* | 72.2* | 86.2 | 82.0 | 84.1 |
Table 5:
Performance of PredMED on MED_REC and comparisons to the performance of MedEx and MedXN. (* indicates significantly different performance when compared lo PredMED: bold represents performance inferior to PredMED: italic represents performance superior to PredMED).
| Annotation Types | Total # | PredMED | MedEx | MedXN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P(%) | R(%) | F(%) | P(%) | R(%) | F(%) | P(%) | R(%) | F(%) | ||
| All Types | 1379 | 98.8 | 91.5 | 95.0 | 97.9 | 83.5* | 90.1* | 98.9 | 81.9* | 89.6* |
| MedName | 354 | 98.5 | 91.5 | 94.9 | 95.9 | 93.2 | 94.6 | 98.2 | 89.8 | 93.8 |
| MedStrength | 337 | 100.0 | 87.2 | 93.2 | 98.7 | 88.1 | 93.1 | 99.7 | 90.5 | 94.9 |
| MedRoute | 332 | 98.7 | 94.0 | 96.3 | 100.0 | 87.1* | 93.1 | 100.0 | 81.9* | 90.1* |
| MedForm | 343 | 98.2 | 93.3 | 95.7 | 97.0 | 65.6* | 78.3* | 97.4 | 65.3* | 78.2* |
| MedSize | 13 | 50.0 | 38.5 | 43.5 | – | – | – | – | – | – |
Performance Comparison
Table 4 shows the performance comparisons between MedEx/MedXN and PredMED on office visit encounter notes. Med Ex and MedXN do not extract Prescription and MedSize. PredMED had a significantly greater overall F-score (84.4%) compared to MedEx (71.5%) and MedXN (78.3%). compared to MedEx. PredMED had greater F-scores on MedName, MedRoute. MedForm, MedDosage, MedDuration and MedFrequency; and similar performance on MedStrength. Compared to MedXN, PredMED had greater F-scores on MedRoute, MedForm.
Table 5 shows the performance comparisons on MED_REC. PredMED again achieved a significantly greater F-score on all types: 95.0% vs. MedEx (90.1%) and MedXN (89.6%). Compared to MedEx, PredMED had a significantly greater F-score on MedForm. and similar F-score on MedName. MedStrength and MedRoute. Compared to MedXN. PredMED had a significantly greater F-score on MedRoute. MedForm, and similar F-score on MedName. MedStrength. It is worth noting that the performance observed here for MedEx and MedXN is lower than that described in other publications9, 10, We believe that to be due to (a) our focus on extracting full prescriptions and not just isolated components and (b) the more complex syntax of prescriptions in me GHS encounter notes, as compared to discharge summaries.
Discussion and Conclusions
PredMED demonstrates significantly better prescription extraction performance (F-score: 84.4% in office visit encounter notes: 95.0% in MED_REC) compared to the other two extractors: MedEx (71.5%; 90.1%) and MedXN (78.3%; 89.6%). Due to the simple, consistent language of the medication descriptions in MED_REC, all three pipelines have similar F-score on MedName and MedStrength and similar precision on MedRoute and MedForm.
PredMED significantly outperformed MedEx and MedXN in recall of MedRoute and MedForm. Error analysis shows that the major reason is that MedEx and MedXN failed to capture several MedRoutes (e.g., “OR”/”by mouth” [oral], “IN” [inhalation], “EX” [external], “IJ” [injection], “OF” [ophthalmic], “NA” [nasal], etc.) and MedForms (e.g., “AERO” [aerosol], “AERS” [aerosol solution], “CPDR” (capsule delayed release], “CREA” [cream], “TB24” [tablet extended release 24 hour], etc.). MedXN’s low precision on MedForm was mainly caused by misanalysing the MedForm word in MedDosage contexts; for example, in “Lasix 40 MG OR TABS, one tab by mouth daily”, the phrase “one tab” should be Med Dosage and “tab” should not be MedForm.
MedDosage was a challenge for all three systems. PredMED interpreted “one a day” as MedFrequency, rather than annotating “one” as MedDosage. Similarly, MedEX often missed the MedDosage number, as in “Effexor XR 75MG OR CP24, 1 po q am”. MedXN misrecognized MedDosages as MedStrengths, as in “PREDNISONE 10 MG OR TABS. 40mg po q d for 3 days”.
PredMED and MedXN performed similarly on MedDuration. Their relatively low recall was due to inability to identify variant duration expressions in narratives (e.g., “since Friday”, “chronically”, etc.). MedEx’s low recall was traced to its failure to recognize expressions like “for Number TimeUnits” and “x Number TimeUnits”. And MedEx’s inability to disambiguate other expressions including Number+ TimeUnits contributed to its low precision (e.g., “Disp: 1 month”, “a 73 year old female”). Finally, MedEx did not recognize certain Latin expressions of MedFrequency, such as “qd” or“q pm”.
Error analysis further reveals that spurious sentence boundaries were inserted into the clinical notes during Extraction, Transformation and Loading (ETL). This caused PredMED to miss prescriptions where the MedName appeared in a different sentence than the Prescription components, thereby reducing recall. Separately, precision suffered because of the false positives that resulted from including a number of common words (e.g., “ICE”, “valve”) in the MedNames dictionary.
Comparison of medications documented on the same date from two sources (i.e., MED_REC entries and clinical notes) illustrates the need for medication reconciliation. As an example. Figure 3 shows the medication discrepancies found for a single patient: 21 out of 25 were mentioned in both sources, while of the remaining four, two appeared only in MED_REC and two were found only in the encounter note. In an application setting, these discrepancies would be presented to a healthcare professional for reconciliation and appropriate follow-up. Note that this example is based on a single time point – the encounter date. In an actual reconciliation application, we’ll need to create and exploit a more general timeline of prescription information for the patient. Of course, prescription timelines will require full semantic analysis of prescription mentions, including the semantics of prescription verbs, such as “start”, “stop”, “reduce”, “discontinue”, etc.
Figure 3:
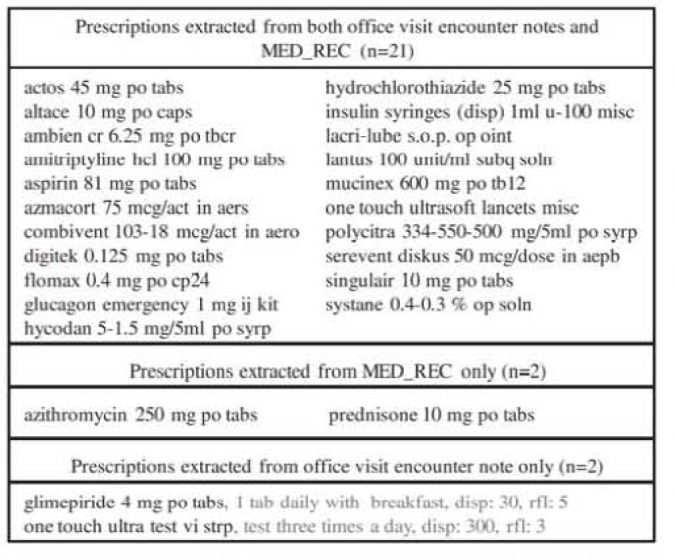
Example of matched and unmatched prescriptions extracted from two sources.
The current extraction pipeline needs to be improved in the following areas: disambiguating MedName: improving recall on MedDosage, MedDuration, and MedFrequency: extracting new component types, such as MedNeeessity; and using the best ideas from other extractors such as MedEx and MedXN. One of the most important improvements will be normalization, which will include normalizing MedNames (as is done by MedXN), MedStrength, MedDosage. and other components. An example of this need arose in the case of another patient, for whom the encounter note contained “coreg 12.5 mg po tabs, take one tablet two times daily” while “carvedilol 25 mg po tabs” was mentioned in the corresponding MED_REC. In this case, the generic name (“carvedilol”) and the brand name (“coreg”) should be normalized to the same medication and the strength and dosage should be shown to match, using a simple calculation.
In the future, we’d like to prototype an application based on these observations. We envision a system which can automatically extract prescriptions from different sources, identify potential medication discrepancies and present the evidence to a healthcare professional for assessment, reconciliation, and action,
Acknowledgments
This work was supported in part by NIH grant number 1R0IHL116832-01. The authors would like to sincerely thank Dr. Amos Cahan and Dr. Xinxin (Katie) Zhu, from IBM Research, for annotating the office visit encounter notes and the MED REC medication descriptions, creating gold standards for this study; Mr. Harry Stavropoulos, from IBM Research, for providing database support; and Ms. Zahra Daar from the Geisinger Health System, for facilitating the collection and exchange of research data.
References
- 1.Fitzgerald RJ. Medication errors: the importance of an accurate drug history. Br J Clin Pharmacol. 2009;67(6):671–5. doi: 10.1111/j.1365-2125.2009.03424.x. Epub 2009/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Joint Commission National Patient Safety Goals Effective. Jan 1, 2014.
- 3.American Pharmacists A, American Society of Health-System P. Steeb D, Webster L. Improving care transitions: optimizing medication reconciliation. J Am Pharm Assoc (2003) 2012;52(4):e43–52. doi: 10.1331/JAPhA.2012.12527. Epub 2012/07/25. [DOI] [PubMed] [Google Scholar]
- 4.Friedman C, Hripcsak G, DuMouchel W, Johnson SB, Clayton PD. Natural language processing in an operational clinical information system. Natural Language Engineering. 1995;1(01):83–108. [Google Scholar]
- 5.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. Epub 2010/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Health Information Text Extraction (HITEx) https://www.i2b2.org/software/projects/hitex/hitex_manua1.html.
- 7.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, comorbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. Epub 2006/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514–8. doi: 10.1136/jamia.2010.003947. Epub 2010/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. Epub 2010/01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd RJ, Steinhubl SR, Sun J, Ebadollahi S, Stewart WF. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int J Med Inform. 2014;83(12):983–92. doi: 10.1016/j.ijmedinf.2012.12.005. Epub 2013/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogren PV. Knowtator: A Protege plug-in for annotated corpus construction; Proceedings of the 2006 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology; 2006. pp. 273–5. [Google Scholar]
- 13.Release Notes for IBM Content Analytics Studio 3.0 http://www-01.ibm.comlsupoort/docview.wss?uid=swg27024048#descnmion.
- 14.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Natural Language Engineering. 2004;10(3–4):327–48. [Google Scholar]
- 15.Apache UIMA https://uima.apache.org/
- 16.TextSTAT - Simple Text Analysis Tool http://neon.niederlandistik.fu-berlin.de/static/textstat/TextSTAT-Doku-EN.html.
- 17.Unified Medical Language System - RxNorm http://www.nlm.nih.gov/research/umls/rxnorm/


