Abstract
The secondary use of electronic health records (EHR) represents unprecedented opportunities for biomedical discovery. Central to this goal is, EHR-phenotyping, also known as cohort identification, which remains a significant challenge. Complex phenotypes often require multivariate and multi-scale analyses, ultimately leading to manually created phenotype definitions. We present Ontology-driven Reports-based Phenotyping from Unique Signatures (ORPheUS), an automated approach to EHR-phenotyping. To do this we identify unique signatures of abnormal clinical pathology reports that correspond to pre-defined medical terms from biomedical ontologies. By using only the clinical pathology, or “lab”, reports we are able to mitigate clinical biases enabling researchers to explore other dimensions of the EHR. We used ORPheUS to generate signatures for 858 diseases and validated against reference cohorts for Type 2 Diabetes Mellitus (T2DM) and Atrial Fibrillation (AF). Our results suggest that our approach, using solely clinical pathology reports, is an effective as a primary screening tool for automated clinical phenotyping.
Introduction & Background
Electronic health records (EHR) capture an increasing variety and amount of clinical data leading to initiatives that are leveraging this potential for knowledge discovery. From adverse event and medical error detection for patient safety1,2 to case-control studies3, those new tools often rely on the researchers’ ability to isolate accurate cohorts of patients with a given phenotype. In this context, the term phenotyping has been used to describe automated and manual methods for identifying these patient cohorts in the EHR4. Advancement of automated phenotyping algorithms is a major roadblock in the field4. Several nationwide efforts, such as eMERGE5 and SHARPn6, have developed selection algorithms for high-throughput phenotype extractions. Those algorithms often comprise of a series of arithmetic and logical operations that are applied to the clinical data. The data types used in these algorithms are heterogeneous and may vary between institutions necessitating continual re-evaluation7. There is an opportunity in phenotyping to apply statistical learning methods, like Association Rule Mining (ARM), for modeling selection algorithms8 or the use of tensor factorization of medications and diagnoses to identify patients9. Other approaches have focused on certain types of clinical data like the diagnoses codes, which often are ICD-9-CM codes. Machine learning techniques trained on these data have been able to classify patients even when data are missing by using inductive logical programming10. The exclusive use of a particular clinical data type (e.g., medications or clinical pathology reports) is advantageous because it allows the exploration other the other data types in the selected cohort while minimizing bias to the extent possible. In particular, ICD-9-CM codes have been widely used for phenotyping and, in some cases, enhanced by additional information, such patient-reported data11. However, ICD-9-CM are primarily used for billing purposes and not for differential diagnosis, introducing complicated biases12. Clinical pathology is the medical subfield that deals with the analysis of bodily fluids for diagnosis and prognosis and clinical pathology reports, commonly called “lab reports,” may be more reliable than ICD-9 codes for EHR phenotyping, while maintaining the same level of standardization.
We present Ontology-driven Reports-based Phenotyping with Unique Signatures (ORPheUS), a knowledge-based phenotyping method that generates a unique clinical pathology signature for each term of a given ontology (i.e. each disease phenotype). Each “phenotype signature” is comprised of a set of abnormal laboratory tests (ATs). Our approach relies on only one type of clinical data – the clinical pathology reports – to minimize biases and increase interoperability. In total we generated clinical pathology signatures for 858 distinct diseases. We validated three of these signatures against reference patient cohorts using definitions from PheKB.org. We evaluated for precision and recall as well as the recovery of known co-morbidities. In each case we found that ORPheUS significantly outperforms the null model, with the T2DM signature recovering 17.2% of diabetics at 81.4% precision (F1 score=0.28).
Methods
Clinical Data Sources
The New York Presbyterian/Columbia University Medical Center (NYP/CUMC) clinical data warehouse contains about 470 million laboratory values from clinical pathology reports from more than 1.3 million patients over the last decade. We selected 177 of the most commonly ordered tests performed from blood, urine, plasma, and cerebrospinal fluid. We restricted our cohort of study to patients over 18 years old at order time with specified sex and at least one of these 177 laboratory tests. It narrowed our study to 767,389 patients with 172,518,869 values total. We preprocessed these data to assert if those reports were normal, abnormal, high, or low accounting for the patients’ age and sex, and according to our normal ranges database (Yahi, et al, in preparation).
Annotating abnormal laboratory tests with ontology terms
ORPheUS uses abnormal laboratory tests (ATs). We associated each AT to the medical terms from a given ontology through statistical enrichment analysis. We created the initial set of annotations by defining a search term by concatenating the name of the laboratory test with its non-normal status (i.e., “blood glucose low”, “blood glucose high”, etc.). Then we searched for each of these terms in the medical search engine UpToDate (www.uptodate.com) and gathered the titles of the first three pages of results. Once regrouped in a text file, these titles were annotated with the Annotator API by the NCBO (www.bioontology.org)13 and counted the number of times an ontology term would appear. We attributed 10 points for an exact match and 8 points for a synonym match. This is a one-time process associate ATs to clinical ontology terms and it is not repeated for the following steps of the phenotyping. We looped through all the terms of the ontology to associate each medical term with the ATs associated with its semantic descendants. We performed a Fisher’s exact test and a permutation analysis on these annotations sets to identify the ATs significantly associated to each ontology term, assessing significance using a FDR <= 0.05. Therefore, each ontology term (e.g., “Diabetes mellitus”), we have a set of significant ATs. We call this set of ATs the phenotype signature.
Selecting cohorts of patients for reference standard
We applied phenotype selection algorithms available on PheKB (www.phekb.org) to construct a reference standard. We therefore identified case cohorts for Atrial Fibrillation (AF)14 and Type 2 Diabetes Mellitus (T2DM)15,16. The data required by these algorithms consists of ICD-9 codes, CPT-4 codes, drug prescriptions, and clinical notes. We tested the performance of ORPheUS on these reference groups of patients.
Phenotyping with ORPheUS
We identified the presence of the phenotype signatures, complete (i.e., all the ATs of the signature are found in the patient’s clinical history) or partial (i.e. a subset of the ATs in the signature), in a patient’s clinical pathology records. For each patient, we look for the presence of any of the ATs belonging to the signature in his medical record to consider this patient as a potential candidate. We referenced laboratory tests with a universal code system named Logical Observation Identifiers Names and Codes (LOINC)17 and we used these codes to match ATs. We sorted those candidates by the number of distinct ATs of the target signature they had without any constraint in time. We designated by true positive (TP) the patients at the intersection of each of these prediction sets and its reference cohort of patients. To assess statistical significance, we compared the precision of the predictions from the signatures to a randomly selected cohort of the same size. For each group of candidates with N distinct ATs, we compared the precision of the prediction against the precision of a randomly selected cohort of the same size relative to all the patients with at least N distinct clinical pathology reports. We performed this random selection 20 times for each category. To compute the recall, we proceeded the same way except that the predictions were evaluated against the complete cohort of reference patients.
Results
Signatures
We annotated 351 abnormal laboratory test (ATs) with terms from the Human Disease Ontology (DOID)18. We then identified those ATs that were specific to each term to generate 858 signatures. The average signature contained 10.8 ± 14 ATs. The minimum number of ATs in a signature was 1 (for 95 signatures), and the maximum 50 (DOID:1579 Respiratory system disease). We did not construct a signature for parent term, “Disease,” in the ontology. Diabetes Mellitus with 14 distinct ATs is a little above the average of signatures (Table 1 – Signature for Diabetes Mellitus). Congenital heart disease presents 16 ATs and Mycoardial infarction 14 (Table 2 and 3).
Table 1.
– Signature of Diabetes Mellitus (DOID:9351)
| Diabetes Mellitus (DOID:9351)
| |
|---|---|
| Clinical Pathology Report | Status |
| Glucose in Serum or Plasma | High/Low |
| Fasting glucose in Serum or Plasma | High/Low |
| Glucose in Blood | High/Low |
| Glucose in Serum or Plasma post challenge | High/Low |
| Hemoglobin A1c/Hemoglobin.total in Blood by HPLC | High/Low |
| Glucose in Blood (Meter) | High/Low |
| Hemoglobin A1c/Hemoglobin.total in Blood | Low |
| Hemoglobin in Blood | High |
Table 2.
– Signature Congenital heart disease (DOID:1682)
| congenital heart disease (DOID:1682)
| |
|---|---|
| Clinical Pathology Report | Status |
| Carbon dioxide, total in Arterial blood | High/Low |
| Carbon dioxide, total in Serum or Plasma | High |
| Estradiol (E2) in Serum or Plasma | High |
| Thyroxine (T4) free in Serum or Plasma | High |
| Calcium.ionized in Arterial blood | High |
| Erythrocyte mean corpuscular volume by Automated count | Low |
| Oxygen saturation in Arterial blood | High/Low |
| Oxygen saturation Calculated from oxygen partial pressure in Blood | High |
| Oxygen saturation in Venous blood | High/Low |
| Oxygen [Partial pressure] in Arterial blood | High/Low |
| Oxygen [Partial pressure] in Venous blood | Low |
| Thyroxine (T4) in Serum or Plasma | High |
Table 3.
– Signature of myocardial infarction (DOID:5844)
| myocardial infarction (DOID:5844)
| |||
|---|---|---|---|
| Clinical Pathology Report | Status | Clinical Pathology Report | Status |
| Basophils [#/volume] in Blood | High | Platelet mean volume in Blood | High |
| Eosinophil [#/volume] in Blood | High | INR in Platelet poor plasma by Coagulation assay | High |
| Eosinophils [#/volume] in Blood by Manual count | High | Carbon dioxide [Partial pressure] in Arterial blood | High |
| Fibrinogen in Platelet poor plasma by Coagulation assay | High | Platelets in Blood | High |
| Hematocrit of Blood by Automated count | High | Potassium in Arterial blood | High |
| Hematocrit of Blood | Low | Sirolimus in Blood | High |
| International Normalized Ratio POC | High | Thrombin time in Platelet poor plasma by Coagulation assay | High |
Phenotyping performances
We computed the precision and recall curves for the Diabetes Mellitus in 83,246 patients with T2DM as determined by the reference standard. We observed that of the 14 T2DM specific ATs in the signature, we only found up to 10 simultaneously in a single patient’s record. The precision is significantly better than by chance and increases above 80% with when at least 4 ATs are matched. At 6 or more distinct ATs the recall falls to below 5% (Figure 1).
Figure 1.
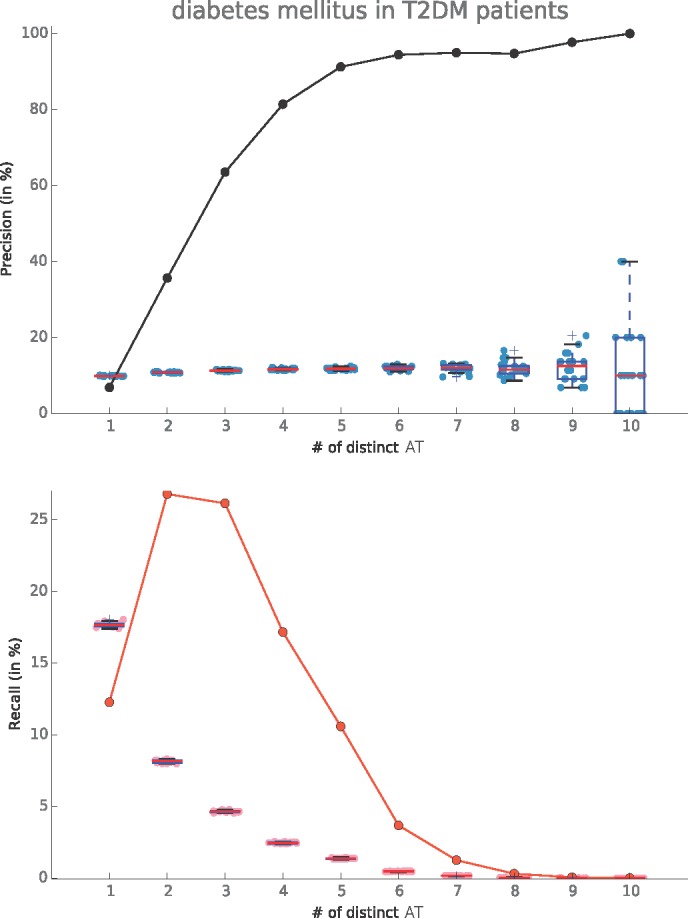
(left) Precision and Recall curves for Diabetes Mellitus signatures tested on T2DM patients
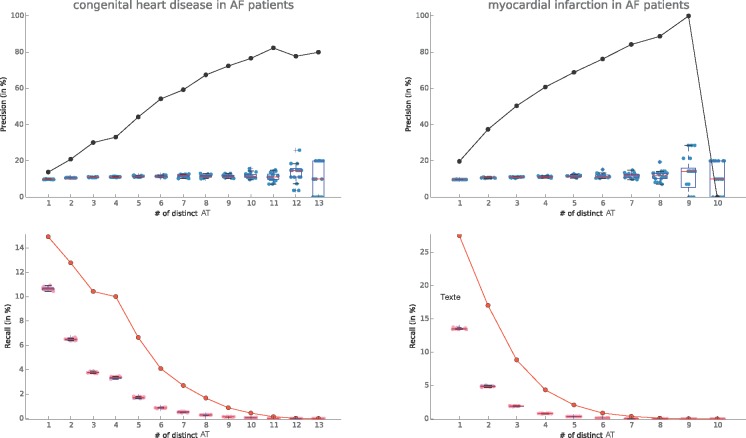
We also explored the cohorts of 80,163 patients with Atrial Fibrillation and evaluated the signatures of two of AF’s known comorbidities: myocardial infarction19, and congenital heart disease20. We observed an interesting precision for Congenital Heart Disease (Figure 2.a.) reaching a plateau around 80% from 10 distinct ATs. Myocardial Infarction (Figure 2.b.) presented a better precision, needing only 6 distinct ATs to reach 80%. However, despite a better initial recall, we witnessed a faster drop in sensitivity for the myocardial infarction signature than the congenital heart disease one. Finally, we observed that for 10 distinct ATs the predicted set of patients was so small that the precision fell to zero.
Figure 2.
(a) Congenital heart disease and (b) Myocardial infarction signatures in Atrial Fibrillation patients
Discussions
In this paper we present a novel automated EHR phenotyping algorithm by defining signatures of abnormal laboratory tests and scanning for matches in a patient’s longitudinal medical record. These signatures are knowledge-driven and rely on only one type of clinical data helping to minimize biases and improve interoperability. Since the signatures are knowledge-based they are not directly exposed to any clinical data before they are used for phenotyping. In total we generated 858 disease signatures. We validated two (atrial fibrillation and type 2 diabetes mellitus) of these signatures against a reference cohort of patients identified using eMERGE algorithms available at PheKB.org. We did not revalidate the PheKB algorithms in the CUMC database, however, previous implementations showed a 98% Positive Predictive Value for AF, and between 98 and 100% for T2DM.
In future studies, we would like to consider co-occurrences of those signatures across time. We might consider restricting the time windows from 1 to 12 months in patients’ records and look for the phenotype signatures, keeping only the maximum number of distinct simultaneous ATs in these windows. It might improve the precision of our predictions since some patients present sparse clinical pathology reports. Dynamical phenotyping using those reports has shown promising opportunities21. We would also like to investigate the potential of combining different phenotypes signatures. We also envision a possible approach for robustness assessment, which would consist of mapping ontological terms, in this example a DOID term, to ICD-9-CM diagnoses codes. This would allow us to evaluate performance of our all or most of our generated phenotype signatures systematically.
The EHR systems are in constant evolution, and many efforts are focused on designed new models learning from data and mitigate complex, inaccurate and frequently missing clinical values4. Indeed, the need for normalization in the information models that are use and the use of standardized vocabularies would ensure a better end-to-end connectivity over platforms allowing more reliable high-throughput phenotyping6. Meanwhile, as clinical notes still remain a critical source of information for phenotypic characteristics, phenotyping techniques using natural language processing (NLP) has been widely used and are gaining popularity22. The term of “Verotype” as a matching of genotype, phenotype and disease subtype has also been described23 to make a step forward to personalized medicine. The systematic inclusion of genotype and phenotype data in future EHR would be critical for this purpose24.
Conclusion
We presented Ontology-driven Reports-based Phenotyping with Unique Signatures (ORPheUS), a knowledge-based automated method for EHR-phenotyping, using only clinical pathology reports. We evaluated the performances of our phenotype signatures for T2DM and AF and demonstrated the potential use of this method for phenotyping. Our ontology-driven approach could allow us in future work to use other medical semantic fields and study for example adverse events signatures.
References
- 1.Li Q, Melton K, Lingren T, Kirkendall ES, Hall E, Zhai H, et al. Phenotyping for patient safety: algorithm development for electronic health record based automated adverse event and medical error detection in neonatal intensive care. Journal of the American Medical Informatics Association. 2014 Sep;21(5):776–84. doi: 10.1136/amiajnl-2013-001914. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorberbaum T, Nasir M, Keiser MJ, Vilar S, Hripcsak G, Tatonetti NP. Systems pharmacology augments drug safety surveillance. Clin Pharmacol Ther. 2014 Nov 1; doi: 10.1002/cpt.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro VM, Mahamaneerat W, Gainer VS, Ananthakrishnan AN, Porter AJ, Wang TD, et al. Evaluation of matched control algorithms in EHR-based phenotyping studies: A case study of inflammatory bowel disease comorbidities. J Biomed Inform. 2014 Sep 6; doi: 10.1016/j.jbi.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2013 Jan 1;20(1):117–21. doi: 10.1136/amiajnl-2012-001145. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Science Translational Medicine. American Association for the Advancement of Science. 2011 Apr 20;3(79):79re1–79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pathak J, Bailey KR, Beebe CE, Bethard S, Carrell DC, Chen PJ, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. Journal of the American Medical Informatics Association. 2013 Dec;20(e2):e341–8. doi: 10.1136/amiajnl-2013-001939. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overby CL, Weng C, Haerian K, Perotte A, Friedman C, Hripcsak G. Evaluation considerations for EHR-based phenotyping algorithms: A case study for drug-induced liver injury. AMIA Jt Summits Transl Sci Proc. 2013;2013:130–4. [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Simon G, Chute CG, Pathak J. Using association rule mining for phenotype extraction from electronic health records. AMIA Jt Summits Transl Sci Proc. 2013;2013:142–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Ho JC, Ghosh J, Steinhubl S, Stewart W, Denny JC, Malin BA, et al. Limestone: High-throughput candidate phenotype generation via tensor factorization. J Biomed Inform. 2014 Jul 16; doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peissig PL, Santos Costa V, Caldwell MD, Rottscheit C, Berg RL, Mendonca EA, et al. Relational machine learning for electronic health record-driven phenotyping. J Biomed Inform. 2014 Jul 15; doi: 10.1016/j.jbi.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith SD, Thompson NR, Rathore JS, Jehi LE, Tesar GE, Katzan IL. Incorporating patient-reported outcome measures into the electronic health record for research: application using the Patient Health Questionnaire (PHQ-9) Qual Life Res. 2014 Aug 7;:1–9. doi: 10.1007/s11136-014-0764-y. Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- 12.Hripcsak G, Knirsch C, Zhou L, Wilcox A, Melton G. Bias associated with mining electronic health records. J Biomed Discov Collab. 2011;6(0):48–52. doi: 10.5210/disco.v6i0.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucl Acids Res. 2009 Jul;37(Web Server issue):W170–3. doi: 10.1093/nar/gkp440. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust Replication of Genotype-Phenotype Associations across Multiple Diseases in an Electronic Medical Record. The American Journal of Human Genetics. 2010 Apr;86(4):560–72. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association. 2012 Mar;19(2):212–8. doi: 10.1136/amiajnl-2011-000439. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W-Q, Leibson CL, Ransom JE, Kho AN, Caraballo PJ, Chai HS, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association. 2012 Mar;19(2):219–24. doi: 10.1136/amiajnl-2011-000597. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clinical Chemistry. 2003 Apr;49(4):624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 18.Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, et al. Disease Ontology: a backbone for disease semantic integration. Nucl Acids Res. 2012 Jan;40(Database issue):D940–6. doi: 10.1093/nar/gkr972. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soliman EZ, Safford MM, Muntner P, Khodneva Y, Dawood FZ, Zakai NA, et al. Atrial Fibrillation and the Risk of Myocardial Infarction. JAMA Intern Med. 2014 Jan 1;174(1):107–14. doi: 10.1001/jamainternmed.2013.11912. American Medical Association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrade J, Khairy P, Dobrev D, Nattel S. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circulation Research. 2014 Apr 25;114(9):1453–68. doi: 10.1161/CIRCRESAHA.114.303211. Lippincott Williams & Wilkins. [DOI] [PubMed] [Google Scholar]
- 21.Albers DJ, Elhadad N, Tabak E, Perotte A, Hripcsak G. Dynamical phenotyping: using temporal analysis of clinically collected physiologic data to stratify populations. Garcia-Ojalvo J, editor. PLoS ONE. 2014;9(6):e96443. doi: 10.1371/journal.pone.0096443. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014 Mar;21(2):221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boland MR, Hripcsak G, Shen Y, Chung WK, Weng C. Defining a comprehensive verotype using electronic health records for personalized medicine. Journal of the American Medical Informatics Association. 2013 Dec;20(e2):e232–8. doi: 10.1136/amiajnl-2013-001932. BMJ Publishing Group Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey LJ, Lenert L, Lopez-Campos G. EHR Big Data Deep Phenotyping. Contribution of the IMIA Genomic Medicine Working Group. Yearb Med Inform. 2014;9(1):206–11. doi: 10.15265/IY-2014-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]