Abstract
It is unclear the extent to which best practices for phenotyping disease states from electronic medical records (EMRs) translate to phenotyping adverse drug events. Here we use statin-induced myotoxicity as a case study to identify best practices in this area. We compared multiple phenotyping algorithms using administrative codes, laboratory measurements, and full-text keyword matching to identify statin-related myopathy from EMRs. Manual review of 300 deidentified EMRs with exposure to at least one statin, created a gold standard set of 124 cases and 176 controls. We tested algorithms using ICD-9 billing codes, laboratory measurements of creatine kinase (CK) and keyword searches of clinical notes and allergy lists. The combined keyword algorithms produced were the most accurate (PPV=86%, NPV=91%). Unlike in most disease phenotyping algorithms, addition of ICD9 codes or laboratory data did not appreciably increase algorithm accuracy. We conclude that phenotype algorithms for adverse drug events should consider text based approaches.
Introduction
Increasingly electronic medical records are leveraged for secondary use in research or quality improvement. These efforts require precise approaches to identify both disease status and drug outcomes (efficacy and adverse reactions). Much work has been done on phenotyping disease status and raw clinical measurements (laboratory values, etc).1 Most algorithms use ICD-9 codes alone or in concert with laboratory values, medications, and/or keywords and often require repeated measurements (i.e., multiple instances of the same ICD-9 code). This works well for disease identification, but it is unclear how well these approaches translate to identifying adverse drug reactions. For example, while repeated measurements help ensure accuracy of disease diagnosis, these should only be a single event for drug adverse reactions (ADR).
This study compares multiple phenotyping approaches for identifying statin-related myotoxicity to highlight potential best practices for identifying adverse drug events. Statins are widely used drugs that decrease risk for cardiovascular disease.2 Muscle toxicity is the most common side effect (1–5% in randomized controlled trials, 9–20% in observational studies) and reason for statin cessation.3 Statin-induced myotoxicity is an excellent case study for the challenges involved in phenotyping adverse drug events as it falls along a spectrum of reactions from simple muscle pain to severe muscle break down. Although efforts define different categories along this spectrum will help4, the best approach extract this phenotype is unclear.
There has been much work done on phenotyping this reaction in EMRs for pharmacovigilance5–8, identification of drug interactions9, investigation of allergy documentation behaviors10, and for use in genetic studies11–15. However, they each have used different approaches and even different phenotype definitions. One study used ICD-9 codes9 while many others have used creatine kinase (CK, an indication of muscle breakdown). Many omit CK measurements taken in concert with troponin (indicative of suspected myocardial infarction)15, or require mild or moderate6,11,14 (defined as 3× the upper limit of normal) elevations of the enzyme. Still others require complex temporal sequencing of CK elevations.7 Natural language processing (NLP) has been used with success both alone,8,10 and in combination with the previous approaches.5,12,13 Given these differences, it is unclear which approach is most accurate. Here we use algorithms outlined in these previous studies on a consistent phenotype definition to identify the best approach for identifying this adverse reaction.
Methods
Gold Standard Development
We selected 300 individual for this study from Vanderbilt’s biorepository linked to deidentified electronic medical records: BioVU.16 All individuals to have at least one mention of a statin (see Table 1) as defined using the NLP tool MedEx17 in the problem list or a history and physical note. Of these, we selected 138 records containing one or more of the following stems near the statin mention: cramp, muscle, pain, myo (myopathy/myositis), mya (myalgia), rha (rhabdomyolysis), ache, weak, hold, dc (discontinue). The remaining 162 records were selected randomly from a group of 3,870 individuals selected for statin exposure and genotype data availability. All records were reviewed by two reviewers (JDM, LKW) with discrepancies reconciled by JFP. Statin-intolerant individuals were considered cases unless attributed to another symptom (e.g. elevation of liver enzymes). In the case of statin holds or discontinuations, we only considered individuals as cases if specifically attributed to myotoxic side effects. We used Cohen’s kappa to assess agreement between the reviewers.
Table 1.
Statin Names Used for Searches
| Generic | Trade Name |
|---|---|
| Atorvastatin | Lipitor, Caduet |
| Fluvastatin | Lescol |
| Lovastatin | Mevacor, Altocor, Altoprev |
| Pitavastatin | Livalo, Pitava |
| Pravastatin | Pravachol |
| Rosuvastatin | Crestor |
| Simvastatin | Zocor, Vytorin, Simcor |
Also: statin, statins, & hmg
Phenotyping Algorithms
ICD-9-CM Algorithms: We used two types of ICD-9 codes identified in a previous study9 − those specifying drug toxicity/adverse reaction and those specifying muscle symptoms. Codes specific to drug toxicity (359.4, 972.2, 995.29, E942.2, and E980.4) could occur at any time in their medical record. Codes related to muscle effects (359.8, 359.9, 728.87, 728.88, 728.9, 729., and 791.3) had to occur after the first statin mention in the record.
Creatine-Kinase Algorithms: CK measurements are often used to identify statin myotoxicity.7,12,15 Using these approaches, we tested five algorithms based on CK measurements after the first statin. 1) Moderate CK elevation (> 500 IU/L). 2) Any CK measurement that did not occur on the same day as a troponin measurement. 3) Moderate CK elevation (>500 IU/L) that did not occur on the same day as a troponin measurement. 4) Single CK measurement of any value within a 5 day period. 5) Single moderate CK elevation (>500 IU/L) within a 5 day period. Algorithms 4 and 5 were new algorithms premised on the hypothesis that CK measurements occurring in isolation (only CK measurement within 5 day window) may correlate with outpatient suspected statin myotoxicity.
Allergy Algorithms: Following a similar approach to Skentzos et.al., we investigated using allergy lists as a phenotyping approach. We extracted the allergy and adverse reactions section from the problem list and high value documents (history and physical, clinic notes, inpatient progress notes, etc.) using the tool SecTag.18 We then split the allergy sections in both problem list and high value documents into single allergy listings and extracted all statins from Table 1. From this list we removed any obvious reaction unrelated to myotoxicity (e.g. elevation of liver function tests, nausea, etc.). Given that the most common adverse reactions to statins are related to myotoxicity, we counted any allergy without a specified reaction as a myopathy case. To identify keywords for this filtering, we reviewed allergy listings in 300 independent records (hereafter referred to as the training cohort) in the larger data set from which our review sample was drawn. Original algorithms used keywords derived from this training sample, while corrected algorithms also include keywords identified in the gold standard population. The combined definition classifies cases with a listed allergy in either or both document types.
Keyword Algorithms: Natural language processing has previously been successfully applied to this phenotyping task.10,13 However these algorithms are not freely available. We created algorithms that identify myotoxicity related keywords in clinical communications and high value documents. As before, the original algorithms used inclusion and exclusion keywords derived from the training set and the corrected from both the training and gold standard set. We processed all clinical communications and high value documents with one or more statins in Table 1 and extracted phrases of different length around the statin keywords. We used a phrase length of 30 words (15 words before and after each statin mention) for clinical communications given the number of extraneous identifiers and dates listed, and a phrase length of 16 words (8 words before and after each statin) for high value documents. All code can be found at: https://github.com/laurakwiley/StatinMyopathy.git
Combination Algorithms: We investigated the maximal benefit of combining algorithms together by creating two algorithms to identify 1) the greatest possible sensitivity and 2) the best balance of sensitivity and specificity.
Algorithm Evaluation
First we evaluated each algorithm’s ability to classify patients as cases (have statin myotoxicity) or controls (do not have statin myotoxicity) using four metrics: sensitivity (probability the algorithm classifies patient as a case when they have statin myotoxicity), specificity (probability the algorithm classifies patient as a control when they do not have statin myotoxicity), positive predictive value (PPV, probability the patient has statin myotoxicity when the algorithm classifies them as a case) and negative predictive value (NPV, probability the patient does not have statin myotoxicity when the algorithm classifies them as a control). To compare the different algorithms, we plotted algorithms in ROC (Receiver Operator Characteristic) space (comparison of true positive rate to false positive rate).
Additionally, we assessed the ability of the algorithms to identify specific statins associated with the adverse reaction. We used Cohen’ kappa to measure the accuracy (i.e., identify only statins known to cause the adverse reaction) and the completeness (i.e., identify all of the statins known to cause the adverse reaction). This statistic evaluates agreement between two methods of classification by identifying the percent agreement of the two methods and the probability of random agreement between the methods. To assess accuracy, agreement was defined where the statin from the algorithm is present in the gold standard. Statins in the Gold Standard, but not in the algorithm did not count as disagreement. To assess the completeness, statins from the algorithm that were not in the gold standard did not count as disagreement. We tested statin lists from the allergy and keyword algorithms only.
Results
There were a total of 124 cases and 176 controls identified from manual review. Of the 300 individuals reviewed, 157 (52.3%) were male. The majority (251, 83.7%) were white, 12.7% (38) were African American. The mean patient age was 66.58 (SD=14.0). Table 2 details the statin distribution in this population. The two reviewers achieved high agreement (κ = 0.986). The mean number of statins prescribed and statins associated with myopathy per patient were 2.6 (SD=1.12) and 1.7 (SD=0.99) respectively.
Table 2.
Review Population Statins (n=300)
| Characteristic | n (%) |
|---|---|
| Statin Ever Prescribed1 / Causing Myotoxic Event1,2 | |
| Atorvastatin | 150 (50%) / 64 (51.6%) |
| Fluvastatin | 25 (8.3%) / 5 (4.0%) |
| Lovastatin | 42 (14.0%) / 11 (8.9%) |
| Pitavastatin | 1 (0.3%) / 0 (0%) |
| Pravastatin | 99 (33.0%) / 31 (25%) |
| Rosuvastatin | 60 (20.0%) / 19 (15.3%) |
| Simvastatin | 235 (78.3%) / 74 (59.7%) |
| Not Specified | 0 (0%) / 8 (6.5%) |
Includes overlap (i.e., individuals on multiple statins).
Percentages of 124 myopathy events.
ICD-9-CM and Creatine-Kinase Algorithms: Performance metrics for the ICD9 and creatine kinase algorithms are presented in Table 3. Two ICD-9-CM codes (972.2 and 791.3) did not occur at all in our population and were excluded from further analysis.
Table 3.
Performance of ICD9 and Creatine Kinase Algorithms
| (95%CI) | |||||
|---|---|---|---|---|---|
| (n) | Sensitivity | Specificity | PPV1 | NPV1 | |
| ICD9 Algorithms | |||||
| Drug Related2 | 4 | 0.02 (0.01–0.07) | 0.99 (0.97–1.00) | 0.75 (0.19–0.99) | 0.59 (0.53–0.65) |
| Muscle Related3 | 82 | 0.47 (0.38–0.56) | 0.86 (0.80–0.91) | 0.71 (0.60–0.80) | 0.70 (0.63–0.76) |
| All Codes | 84 | 0.48 (0.39–0.57) | 0.86 (0.80–0.91) | 0.70 (0.59–0.80) | 0.70 (0.63–0.76) |
| Creatine Kinase Algorithms | |||||
| CK > 500 IU/L | 72 | 0.32 (0.24–0.41) | 0.81 (0.75–0.87) | 0.54 (0.42–0.66) | 0.64 (0.57–0.70) |
| CK (any value), no troponin | 224 | 0.89 (0.82–0.94) | 0.35 (0.28–0.43) | 0.49 (0.42–0.55) | 0.83 (0.73–0.91) |
| CK > 500 IU/L, no troponin | 61 | 0.30 (0.22–0.38) | 0.86 (0.80–0.91) | 0.59 (0.46–0.71) | 0.64 (0.58–0.70) |
| CK (any value), single measurement4 | 55 | 0.30 (0.22–0.38) | 0.89 (0.84–0.93) | 0.65 (0.51–0.78) | 0.65 (0.59–0.71) |
| CK > 500 IU/L, single measurement4 | 15 | 0.07 (0.03–0.14) | 0.97 (0.93–0.99) | 0.60 (0.32–0.84) | 0.60 (0.54–0.66) |
PPV (Positive Predictive Value), NPV (Negative Predictive Value).
Any time.
After first statin mention.
Only CK measurement in 5 day period.
Allergy and Keyword Algorithms: We reviewed 27,637 problem list allergy sections and 4,580 high value document allergy sections from our training cohort to create our original allergy-based phenotyping algorithm. We applied that algorithm to 33,971 problem list and 6,652 high value document allergy sections in our gold standard population. Performance metrics for the original and corrected algorithms applied to the problem list, high value document and a combination of both allergy sections are presented in Table 4. To create the original keyword-based algorithms, we reviewed 1,220 clinical communications and 10,617 high value documents from our training set. We applied that algorithm to 1,925 clinical communications and 15,040 high value documents in our gold standard. Performance values for the keyword algorithms are also presented in Table 4.
Table 4.
Performance of Allergy and Keyword Algorithms
| Originial2 (95% CI) Corrected2 (95% CI) |
|||||
|---|---|---|---|---|---|
| (n) | Sensitivity | Specificity | PPV1 | NPV1 | |
| Allergy Algorithms | |||||
|
| |||||
| Problem List | 75 | 0.58 (0.49–0.67) | 0.98 (0.94–0.99) | 0.95 (0.87–0.99) | 0.77 (0.71–0.83) |
| 71 | 0.57 (0.48–0.66) | 1.00 (0.97–1.00) | 1.00 (0.93–1.00) | 0.77 (0.71–0.82) | |
|
| |||||
| High Value Documents | 86 | 0.62 (0.53–0.71) | 0.94 (0.90–0.97) | 0.88 (0.80–0.94) | 0.79 (0.72–0.84) |
| 78 | 0.61 (0.52–0.70) | 0.99 (0.96–1.00) | 0.97 (0.91–1.00) | 0.78 (0.72–0.84) | |
|
| |||||
| Combined | 89 | 0.64 (0.55–0.72) | 0.94 (0.89–0.97) | 0.88 (0.79–0.94) | 0.79 (0.73–0.84) |
| 80 | 0.63 (0.54–0.71) | 0.99 (0.96–1.00) | 0.97 (0.91–1.00) | 0.79 (0.73–0.84) | |
|
| |||||
| Keyword Algorithms | |||||
|
| |||||
| Clinical Comm. | 65 | 0.45 (0.36–0.54) | 0.94 (0.90–0.97) | 0.85 (0.74–0.92) | 0.71 (0.65–0.77) |
| 60 | 0.46 (0.37–0.55) | 0.98 (0.95–1.00) | 0.98 (0.95–1.00) | 0.72 (0.66–0.78) | |
|
| |||||
| High Value Documents | 104 | 0.68 (0.59–0.76) | 0.88 (0.83–0.93) | 0.80 (0.71–0.87) | 0.80 (0.74–0.85) |
| 108 | 0.75 (0.66–0.82) | 0.91 (0.86–0.95) | 0.86 (0.78–0.92) | 0.84 (0.78–0.89) | |
|
| |||||
| Combined | 122 | 0.78 (0.69–0.85) | 0.85 (0.79–0.90) | 0.78 (0.69–0.85) | 0.85 (0.79–0.90) |
| 117 | 0.81 (0.73–0.87) | 0.90 (0.85–0.94) | 0.85 (0.78–0.91) | 0.87 (0.81–0.91) | |
PPV (Positive Predictive Value), NPV (Negative Predictive Value);
Original: algorithm built on training dataset. Corrected: revised algorithm with text extracted from the gold standard set.
Combined Algorithms and Algorithm Comparison: The most sensitive algorithm (sens. = 0.99, spec. = 0.32, PPV = 0.51) included: all corrected allergy, all corrected keyword, the second CK (any value of CK, no troponin measurement) and the ICD9 algorithms and identified all but one case in our data set. The second most sensitive algorithm (sens. = 0.94, spec. = 0.84, PPV = 0.81) contained: all corrected allergy algorithms, all corrected keyword algorithms, and all ICD9 codes. This algorithm identified 116 of our 124 cases. The algorithm with the best balance of sensitivity and specificity (sensitivity = 0.88, specificity = 0.90, PPV = 0.86) included only the corrected allergy algorithms and the corrected keyword algorithms and identified 109 of our cases. A comparison of all algorithms in ROC (receiver operator characteristic) space is presented in Figure 1.
Figure 1.
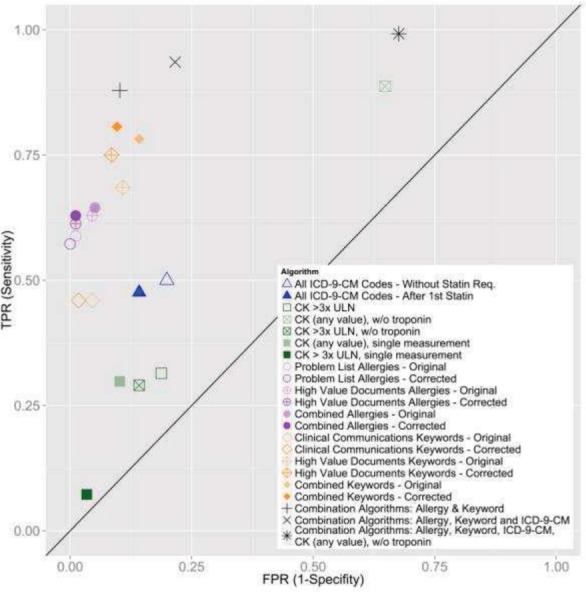
Receiver Operator Characteristic Graph Comparison of Phenotyping Algorithms Comparison of true positive (TPR) vs false positive rate (FPR). Points in upper left are better classifiers.
Statin-Specific Evaluation: There was moderate agreement (κ = 0.433) between the gold standard list of statins causing adverse reactions to the statin list extracted using the combined allergy algorithm. The accuracy (tolerant of false negatives) and completeness (tolerant of false positives) showed substantial agreement (κ =0.625 and κ =0.65 respectively). Broken down by allergy origin, problem list and history and physical derived allergies had almost perfect agreement for accuracy (κ = 0.932 and κ = 0.874 respectively). There was moderate agreement (κ = 0.498) between the gold standard and the combined keyword algorithm statin list. However, none of the accuracy agreement statistics for this algorithm were higher than κ = 0.693 (substantial agreement).
Discussion
Many studies have identified statin induced myotoxicity from EMRs, but the performance of these methods at other institutions has been unclear. In this study, we tested multiple phenotyping approaches to identify best practices. Most phenotyping algorithms are assessed by positive and negative predictive values, but it is important to consider the goal. For some adverse reactions or study designs it is necessary to extract every case (i.e., high sensitivity) even with many false positives (i.e., low specificity). In other instances, the reverse may be desired.
Two of our algorithms used structured data: ICD-9 codes or CK measurements. For the ICD9 algorithms, codes specific for reactions to antilipemic drugs were use so infrequently as to be meaningless for phenotyping. Even with the more common muscle related codes, the sensitivity was ≤50% and should not be used for phenotyping. The more common approach of using CK levels also performed poorly (PPV = 54%, sensitivity = 32%) and was insufficient for identifying the myopathy adverse reaction. Although this test is discriminating in a clinical trial, at a tertiary care facility and Level 1 trauma center, CK are routinely measured following trauma or cardiac events. Although filtering CK measurements based on time (single CK measurement in a 5 day period) helped remove these off target measurements, the improved specificity came at the cost of decreased sensitivity. Similarly, excluding CK drawn in conjunction with troponin (indicating suspected cardiac event) increased specificity, but the sensitivity (30%) remained too low to be feasible for phenotyping. However if it is essential to identify most cases, using any CK measurement (i.e. any CK value) without troponin is very sensitive (89%). Importantly, over half of the cases identified were false positives and the algorithm could only identify 110 of the 124 cases. The sensitivity improves with inclusion of other algorithms but should only be used in cases where there are resources for manual review to exclude the high number of false positives.
Our text based approaches to identify statin allergy and myopathy related keywords both performed better than structured data approaches. Importantly we greatly improved the text algorithms by including phrases from the gold standard population. Thus it is likely important to adjust the algorithms based on documentation practices at each implementation site. At our institution allergies in the problem list were easy to extract and had an excellent PPV (100%), and acceptable sensitivity (57%). Allergy extraction from high value documents required addition computational time while keyword algorithms required computable narrative text from patient records, which may not be available to all investigators. Interestingly, clinical communications (i.e., notes among care providers and patients) provided extremely specific results (94–98%) even in the uncorrected algorithm. While the sensitivity was marginal (46%), these documents were less common which made the analysis more tractable. In summary, if investigators have access to the text of the medical records, these algorithms offered the best compromise of sensitivity and specificity in our study.
It is common to use of triangulation of multiple types of data to identify cases in phenotyping algorithms.1 However, our results suggest that keywords alone are sufficient to identify statin related muscle adverse reactions. In fact, combination of keywords with ICD9 codes had a lower PPV compared to text models alone (81% and 86% respectively) and inclusion of laboratory measurements decreased specificity to intolerably low levels. Importantly, even our most sensitive algorithm could only identify 123 of the 124 cases. Unless it is critical to identify almost every case with this reaction, text-based approaches alone should be sufficient. Unfortunately, while problem list allergies were very accurate at identifying the specific statin causing the reaction, none of the algorithms had perfect assignments. Though this is somewhat expected (multiple statins are mentioned with respect to both the reaction and future treatment plans), it is a limitation that needs to be addressed in future studies.
There are important limitations with our study. The use of text stems to select our gold standard may have artificially improved performance of the text based algorithms. Additionally, the positive predictive values may be inflated due to artificially increased prevalence of statin-induced myopathy in our gold standard. Finally, the strong improvement from corrected algorithms re-emphasizes the need to tailor these algorithms to each study location.
In conclusion, text based approaches to identify statin related myotoxicity provided the best balance of sensitivity and specificity. Unlike in traditional disease state phenotyping algorithms, using multiple data types such as ICD9 and laboratory measurements did not appreciably increase algorithm performance. When considering developing adverse reaction phenotyping algorithms, text based approaches should be strongly considered.
Acknowledgments
We would like to thank Lisa Bastarache, Joshua Smith, Pedro Texiera, and Robert Carroll for their assistance. Dataset(s) used were obtained from Vanderbilt’s BioVU, supported by institutional funding and the Vanderbilt CTSA grant ULTR000445 from NCATS/NIH. Additional support came from VGER, the Vanderbilt Genome-Electronic Records Project grant from NHGRI – U01HG04603 and the NLM -5T15LM007450.
References
- 1.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc United States. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro RA, Ziegelmann PK, Duncan BB, et al. Impact of statin dose on major cardiovascular events: A mixed treatment comparison meta-analysis involving more than 175,000 patients. Int J Cardiol. 2011 doi: 10.1016/j.ijcard.2011.10.128. [DOI] [PubMed] [Google Scholar]
- 3.Thompson PD, Clarkson PM, Rosenson RS, National Lipid Association Statin Safety Task Force Muscle Safety Expert Panel An assessment of statin safety by muscle experts. Am J Cardiol. United States. 2006 Apr 17;97(8A):69C–76C. doi: 10.1016/j.amjcard.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Alfirevic A, Neely D, Armitage J, et al. Phenotype standardisation for statin-induced myotoxicity. Clin Pharmacol Ther. 2014 doi: 10.1038/clpt.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floyd JS, Heckbert SR, Weiss NS, Carrell DS, Psaty BM. Use of administrative data to estimate the incidence of statin-related rhabdomyolysis. J Am Med Assoc. 2012;307(15):1580–1582. doi: 10.1001/jama.2012.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mareedu RK, Modhia FM, Kanin EI, et al. Use of an Electronic Medical Record to Characterize Cases of Intermediate Statin-Induced Muscle Toxicity. Preventive cardiology. Wiley Online Library. 2009;12(2):88–94. doi: 10.1111/j.1751-7141.2009.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sai K, Hanatani T, Azuma Y, et al. Wiley Online Library. 2013. Development of a detection algorithm for statin-induced myopathy using electronic medical records. Journal of Clinical Pharmacy and Therapeutics. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Plutzky J, Skentzos S, et al. Discontinuation of statins in routine care settings: a cohort study. Ann Intern Med. 2013 doi: 10.7326/0003-4819-158-7-201304020-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duke JD, Han X, Wang Z, et al. Literature Based Drug Interaction Prediction with Clinical Assessment Using Electronic Medical Records: Novel Myopathy Associated Drug Interactions. PLoS Computational Biology. 2012;8(8):e1002614. doi: 10.1371/journal.pcbi.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skentzos S, Shubina M, Plutzky J, Turchin A. Structured vs. Unstructured: Factors Affecting Adverse Drug Reaction Documentation in an EMR Repository. AMIA Annual Symposium Proceedings. 2011:1270. [PMC free article] [PubMed] [Google Scholar]
- 11.Carr DF, OMeara H, Jorgensen AL, et al. SLCO1B1 Genetic Variant Associated With Statin-Induced Myopathy: A Proof-of-Concept Study Using the Clinical Practice Research Datalink. Clinical Pharmacology & Therapeutics. 2013 doi: 10.1038/clpt.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan AA, Ding K, Khader S, Kullo IJ. Association of a Polymorphism in SLCO1B1 with Statin-Induced Myalgias, Myositis and Myopathy: An Electronic Medical Record Based Pharmacogenetic Study. J. Am Coll Cardio. 2011 [Google Scholar]
- 13.Khan AA, Kullo IJ. Genome Wide Association Study of Statin-Induced Myalgias using Natural Language. Circulation. 2012. Retrieved from: http://circ.ahajournals.org/cgi/content/meeting_abstract/126/21_MeetingAbstracts/A17484.
- 14.O’Meara H, Carr DF, Evely J, et al. Electronic Health Records For Biological Sample Collection: Feasibility Study Of Statin-Induced Myopathy Using The Clinical Practice Research Datalink. Br J Clin Pharmacol. 2013 doi: 10.1111/bcp.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dube MP, Zetler R, Feroz Zada Y, et al. Genome-wide analysis of creatine kinase levels in statin-users; Presented at the 63rd Annual Meeting of The American Society of Human Genetics. [Google Scholar]
- 16.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc United States. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny JC, Spickard A, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. United States. 2009;16(6):806–15. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]


