Abstract
Eukaryotic metabolism is organised in complex networks of enzyme catalysed reactions which are distributed over different organelles. To quantify the compartmentalised reactions, quantitative measurements of relevant physiological variables in different compartments are needed, especially of cofactors. NADP(H) are critical components in cellular redox metabolism. Currently, available metabolite measurement methods allow whole cell measurements. Here a metabolite sensor based on a fast equilibrium reaction is introduced to monitor the cytosolic NADPH/NADP ratio in Saccharomyces cerevisiae: 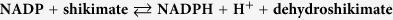 . The cytosolic NADPH/NADP ratio was determined by measuring the shikimate and dehydroshikimate concentrations (by GC-MS/MS). The cytosolic NADPH/NADP ratio was determined under batch and chemostat (aerobic, glucose-limited, D = 0.1 h−1) conditions, to be 22.0 ± 2.6 and 15.6 ± 0.6, respectively. These ratios were much higher than the whole cell NADPH/NADP ratio (1.05 ± 0.08). In response to a glucose pulse, the cytosolic NADPH/NADP ratio first increased very rapidly and restored the steady state ratio after 3 minutes. In contrast to this dynamic observation, the whole cell NADPH/NADP ratio remained nearly constant. The novel cytosol NADPH/NADP measurements provide new insights into the thermodynamic driving forces for NADP(H)-dependent reactions, like amino acid synthesis, product pathways like fatty acid production or the mevalonate pathway.
. The cytosolic NADPH/NADP ratio was determined by measuring the shikimate and dehydroshikimate concentrations (by GC-MS/MS). The cytosolic NADPH/NADP ratio was determined under batch and chemostat (aerobic, glucose-limited, D = 0.1 h−1) conditions, to be 22.0 ± 2.6 and 15.6 ± 0.6, respectively. These ratios were much higher than the whole cell NADPH/NADP ratio (1.05 ± 0.08). In response to a glucose pulse, the cytosolic NADPH/NADP ratio first increased very rapidly and restored the steady state ratio after 3 minutes. In contrast to this dynamic observation, the whole cell NADPH/NADP ratio remained nearly constant. The novel cytosol NADPH/NADP measurements provide new insights into the thermodynamic driving forces for NADP(H)-dependent reactions, like amino acid synthesis, product pathways like fatty acid production or the mevalonate pathway.
The ratios of NAD/NADH and NADPH/NADP (nicotinamide adenine dinucleotide (phosphate)) determine the intracellular redox potential which influence the thermodynamic driving force of many reactions in vivo. However, the cofactors are compartmentalised in Saccharomyces cerevisiae, especially NADP(H) itself is not transported between compartments in S. cerevisiae. Furthermore, the majority of NAD(P)H is found to be protein- or enzyme-bound in vivo in erythrocytes (in rat) and plant systems1,2, but only the fraction of free cofactors actually drives the reaction rates3. Currently, most measurement techniques only allow to measure whole cell amounts, which cannot distinguish between different compartments nor between free and protein-bound concentrations. These whole cell measurements are not sufficient for a comprehensive understanding and description of the in vivo thermodynamics of compartmentalised reactions, like for amino acid synthesis and product pathways like farnesene.
In S. cerevisiae, the NADPH producing reactions, e.g. glucose-6-P dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH) of the Pentose Phosphate Pathway (PPP), are reactions with large thermodynamic driving force, which allows to achieve a high NADPH/NADP ratio. In contrast NADPH consuming reactions typically operate close to the equilibrium4, therefore the NADPH/NADP concentration ratio has a significant contribution to the driving force of the synthesis reactions, e.g. fatty acids, amino acids and products from the mevalonic acid pathway.
The fatty acids are important precursors for the production of fuels and chemicals5. Fatty acids biosynthesis is initiated by the reduction of acetyl-CoA and CO2 to malonyl-CoA (in the cytosol), with NADPH as cofactor. A high cytosolic NADPH/NADP ratio (high driving force) is critical to obtain high production rates. A high cytosolic NADPH level is also a critical bottleneck in amino acid production, one of the largest classes of fermentation products whose syntheses are closely correlated with the availability of NADPH, e.g. L-arginine6 and L-lysine7. Additionally, there are many other reduced product reactions which depend on a high reduction potential. One prominent example is reducing pathways with aldehyde intermediates, which are toxic, and the concentration in the cell needs to be kept low. The aldehyde is usually reduced to alcohols (-OH) by NADH- or NADPH- dependent dehydrogenases. One example is the microbial production of 1,3-propanediol (PDO) in Escherichia coli. It was found that by switching the NADH-dependent dehydrogenase DhaT to the NADPH-dependent dehydrogenase YqhD for the conversion of the toxic aldehyde 3-HPA to PDO, a higher PDO titer was achieved and the differences in the cofactor reduced/oxidised ratios contributed to the high PDO titer8,9.
Subcellular quantification has been approached by non-aqueous fractionation in mammalian tissues and plants, a successful approach but very laborious10. Transcriptional sensor was used to monitor the intracellular NADH/NAD redox state11. Alternatively, metabolite sensor reactions have been proposed to monitor compartment specific concentrations (or ratios), especially for cofactor couples like NAD/NADH, NADPH/NADP, and ATP/ADP12,13,14,15. Malic enzyme and isocitrate dehydrogenase have been used to determine the cytosolic free NADPH/NADP ratio in rat liver and mouse pancreatic islets13,16. Especially, the cytosolic free NADPH/NADP ratio was found to correlate with increasing extracellular glucose concentrations. Ratios up to 57.8 were found which was 1 to 2 orders of magnitude different from the whole-cell NADPH/NADP ratio (approx. 1.2)16. Unfortunately, both reactions (malic enzyme and isocitrate dehydrogenase) and all previously described sensor reactions can’t be applied as sensor reaction in S. cerevisiae because of their localization in the cytosol and/or mitochondria, and reported unspecific cofactor binding (NAD or NADP)17,18.
Here we focus on the cytosolic free NADPH/NADP ratio by using a heterologous metabolite sensor reaction. For the proper function as a sensor reaction, several criteria have to be fulfilled15,19:(1) the enzyme is specific for its coenzyme; (2) the enzyme activity has to be high enough to establish equilibrium between the reactants; (3) the equilibrium constant (Keq) has to be known; (4) the reaction and the measured reactants (except the ones to be determined) are only present in the compartment of interest and can be measured. The reaction can be a dead-end or within a pathway, as long as the enzyme capacity is sufficient to reach pseudo equilibrium.
A heterologous candidate reaction is shikimate dehydrogenase (EC 1.1.1.25), which is NADP specific and only in the cytosol in E. coli20,21. The specific activity of shikimate dehydrogenase (a multifunctional arom enzyme) in S. cerevisiae was very low22,23, which is also only in the cytosol24. Therefore, the shikimate dehydrogenase from E. coli, which has a high activity (19.9 units/mg)25 was overexpressed in yeast CEN.PK 113-5D, and we assume the expressed shikimate dehydrogenase activity is only cytosolic and the SA/DHS accumulates only in the cytosol. With this reaction in place, the cytosolic NADPH/NADP ratio under different batch, chemostat and dynamic perturbation conditions was studied.
Results and Discussion
NADPH/NADP Metabolite sensor reaction
The genetically introduced NADPH/NADP sensor reaction is catalysed by shikimate dehydrogenase (EC 1.1.1.25) (aroE):
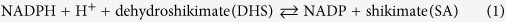 |
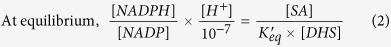 |
Both SA and DHS can be measured using metabolomics approach (GS-MS/MS, Supplementary figure S1). The apparent equilibrium constant Keq’ is 0.26 at specified reference conditions of pH = 7.0, and ionic strength I = 0.25 M26. The reported NADPH/NADP ratios here were calculated assuming intracellular pH = 7.0.
In the cellular system NADPH is a conserved moiety. In case of a reaction a proton is released, while a proton is produced when NADPH is oxidised.
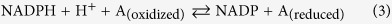 |
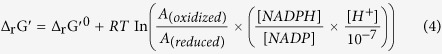 |
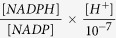 can be replaced by the measured SA/DHS ratio, the Gibbs energy of a reduction reaction became:
can be replaced by the measured SA/DHS ratio, the Gibbs energy of a reduction reaction became:
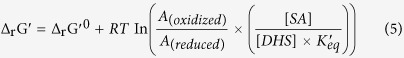 |
Please note that this substitution shows that for the thermodynamic analysis of any reaction involving NADPH/NADP, no assumption on the pH is required (cancels out). For the ratio itself an assumption is needed, but this does not bias the thermodynamic interpretations. Consequently, even if the pH is estimated incorrect, the ΔrG′ difference of the NADP(H) independent reaction is calculated correctly. Clearly, this also holds true under dynamic conditions, like the cytosolic pH would rapidly drop after the glucose pulse27,28,29,30,31, and the effect of changes of the cytosolic pH on the calculated thermodynamic driving force are circumvented. This is another advantage to use the sensor cytosolic NADPH/NADP ratio instead of whole cell ratio measurements.
Influence of the sensor reaction on cell physiology
The successful expression of shikimate dehydrogenase was confirmed by enzymatic activity measurement of cell extracts. The CEN.PK-aroE strain displayed a high shikimate dehydrogenase activity (9.68 U/mgprotein, batch condition), while the reference strain only reached 0.048 U/mgprotein. The activity of G6PDH was comparable in both the CEN.PK-aroE and reference strain (0.70 vs. 0.57 U/mgprotein).
In S. cerevisiae, the oxidative Pentose Phosphate Pathway (oxPPP) is the major source of NADPH in the cytoplasm, and it has been observed that 2% to 60% of the consumed glucose is oxidised in the oxPPP under aerobic conditions32,33. Assuming the maximum rate of oxPPP is 60% of the maximum glucose uptake rate (2.9 μmol/(gDW∙s)34, a NADPH production rate of 1.74 μmol/(gDW∙s) is obtained. Assuming 1 gDW contains 330 mg soluble protein35, the SDH activity of 9.68 U/mg protein (at 25 °C) corresponds to a maximal activity of 53.24 μmol/(gDW∙s), about 15 times higher (maybe even higher at 30 °C) than the estimated maximal NADPH flux, suggesting that the SA dehydrogenase enzyme activity is high enough to keep the sensor reaction close to equilibrium.
To analyse whether the sensor reaction influences the metabolism, the batch growth rate and metabolite levels during steady state were compared to wildtype values. The batch growth rate of CEN.PK-aroE was 0.32 h−1, similar to the reference strain (CEN.PK113-7D) rate (0.33 h−1). There was no significant difference between intracellular concentrations of most intermediates in central metabolism of the two strains under glucose-limited steady state conditions (D = 0.1 h−1) (Supplementary Table S1). This indicates that the expression of shikimate dehydrogenase had no considerable impact on the central metabolism under these growth conditions.
External supplementation of SA for the sensor reaction
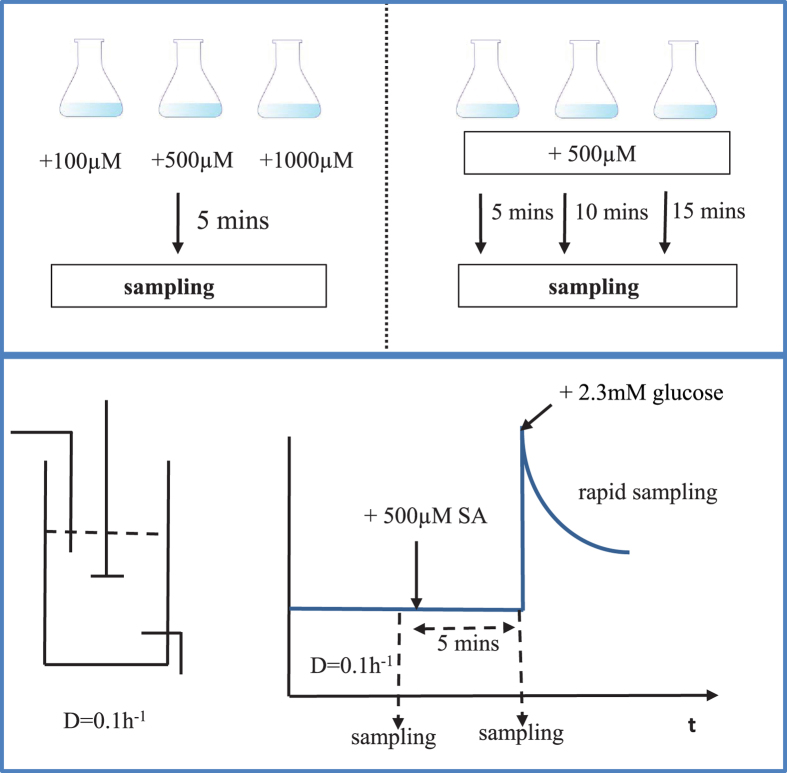
Different experiments were performed (Fig. 1) to evaluate the influence of:
SA addition on intracellular metabolites;
SA incubation time on intracellular metabolites;
SA uptake rate and conversion on NADPH production.
Figure 1. Experiments setup to evaluate the influence.
(1) SA addition on intracellular metabolites; (2) SA incubation time on intracellular metabolites; (3) SA uptake and conversion on NADPH production.
The intracellular SA and DHS concentrations were very low in the sensor strain (0.0133 μmol/gDW and below the detection limit, respectively). To increase the intracellular concentrations, SA was added to the culture broth. The addition in batch culture resulted in a 10 fold increased intracellular SA (0.165 μmol/gDW) and measurable DHS (0.0025 μmol/gDW) amounts.
For batch experiments with different amounts of SA addition, the calculated cytosolic free NADPH/NADP ratio was 35.3 ± 2.5, and there was no clear trend for different amounts of SA supplementation in batch condition (Fig. 2A). The cytosolic free NADPH/NADP ratio was very similar although the SA and DHS concentrations were very different. With 100 μM, the intracellular concentrations were still low, thus a supplementation with 500 μM SA was selected for the following experiments.
Figure 2. Cytosolic NADPH/NADP ratio in batch experiments.
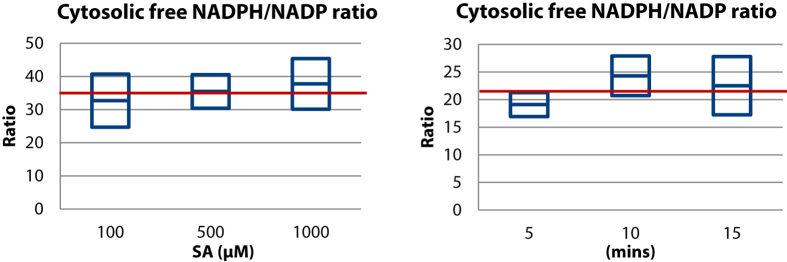
(A) Cytosolic NADPH/NADP ratio (based on sensor reaction) during batch growth with different amounts of supplementation, 100, 500 and 1000 μM SA. (B) 5, 10 and 15 minutes after addition of 500 μM SA. The error bar represents the standard deviations of the mean of three independent cultivations.
With different SA incubation time, the average cytosolic free NADPH/NADP ratio was 22.0 ± 2.6 (Fig. 2B). After 10 minutes, the ratio increased slightly and it decreased at 15 minutes. There was no notable difference for the average ratio of 5, 10, 15 minutes, therefore we choose 5 minutes, also in view of the time monitored after the pulse. The ratio was lower compared to the previous batch cultivation, but it has to be noted that a different medium (urea as nitrogen source) was used. Within one medium condition, the results were consistent.
Based on these experiments, the supplementation of 500 μM SA and sampling 5 minutes after the addition were chosen for the chemostat experiment. The extracellular addition of SA resulted in a SA uptake rate of 0.005 μmol/(gDW∙s) (Fig. 3), which was calculated as the average rate of 3600 seconds. Assuming that all SA taken up by the cell is dehydrogenated to DHS, an extra NADPH production rate of 0.005 μmol/(gDW∙s) is obtained (~0.29 % of the maximum oxPPP production). It is assumed that this very small putative production has no impact on the metabolic flux distribution, nor the NADPH/NADP ratio. There was no considerable difference of intracellular concentrations of glycolytic and PPP pathway intermediates between the steady state condition and 5 minutes after the addition of SA, except the SA and DSA increased substantially as expected (Supplementary Table S1). Several intermediates in central metabolism were slightly increased, e.g. G6P, F6P, but the difference was not substantial. The results indicate that the intracellular concentration of the sensor reaction metabolites can be boosted by external supplementation without disturbing the metabolic flux distribution.
Figure 3. Extracellular glucose, SA and DHS concentrations during the chemostat glucose pulse experiment.
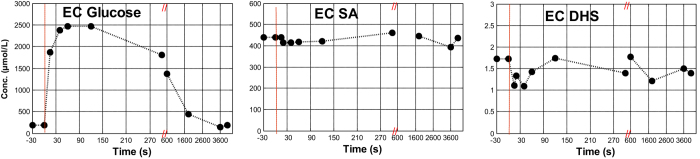
Data points before t = 0 represent the state 5 mins after the SA addition, just before the glucose pulse.
Shikimate transport
For E. coli, ShiA is identified as high affinity transporter for shikimate which is assumed to function as a proton/shikimate symport36. However, the SA transport mechanism in S. cerevisiae has not yet been identified. Using the intracellular and extracellular measurement, we could analyse the thermodynamic feasibilities of different putative uptake mechanisms. The equilibrium ratio of SAin/SAout is calculated for a range of pHin (6.5–7.0) and pmf (0.15–0.2) for uniport (n = 0), symport (n = 1), and antiport (n = −1) for SA- species and pHout = 5.0. pKa of SA is 4.48.
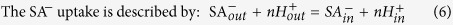 |
The total acid ratio of SAin/SAout at thermodynamic equilibrium can be calculated by:
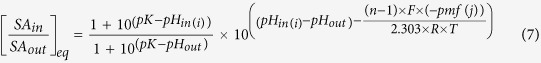 |
Assuming uniport, an equilibrium ratio of SAin/SAout between 0.012 to 0.25 is expected (Supplementary Figure S2). In case of symport, the ratio would be between 24.4 to 78.0. Assuming antiport, the ratio would be between 1 × 10−5 to 5 × 10−4. In our experiments, an average SAin/SAout ratio of 0.3 was found (Supplementary Figure S2). Therefore the transport cannot be antiport, it could be uniport which operates of pseudo equilibrium or symport far from equilibrium. Given the biochemical exchanges and the slow SA uptake rate of 0.005 μmol/(gDW∙s), it is concluded that SA transport occurs as symport, far from equilibrium.
Steady state cytosolic free and whole cell NADPH/NADP ratio
The NADP biomass specific amount found in different studies are comparable (0.2 ~ 0.5 μmol/gDW), but the discrepancy for NADPH is large, ranging from 0.056 to 1.80 μmol/gDW (Table 1). The whole cell biomass specific amount of NADPH + NADP (0.948 μmol/gDW) was comparable to values reported earlier (Table 1). The steady state whole cell NADPH/NADP ratio was 1.05 ± 0.08, which is similar to the ratio previously reported for S. cerevisiae CBS 7336 using enzymatic based measurements37; but different from observations in CEN.PK113-7D by enzymatic method (0.29)17; and a ratio of 4.8 by LC-MS/MS method34. The redox couple NADP(H) is generally considered a challenging target for quantitative analysis38. The chemical instability of NADPH in a variety of conditions (especially the sample extraction and process conditions) hinders the accurate determination, while the oxidised form NADP is more stable, even at low pH39. The different stabilities are most likely the explanation of the very different reported values of NADPH, while NADP concentrations are comparable.
Table 1. Comparison of NADP(H) results.
| Strain | Vaseghi, et al.37 | Moreira dos Santos, et al.17 | Suarez-Mendez, et al.34 | This work |
|---|---|---|---|---|
| CBS 7336 | CEN.PK113-7D** | CEN.PK113-7D | CEN.PK-aroE | |
| Cultivation condition | Glucose-limited, aerobic, D = 0.1 h−1 | |||
| Extraction method | alkaline and acidic solvent | alkaline and acidic solvent | boiling ethanol | boiling ethanol |
| Analysis | enzymatic assay | enzymatic assay | LC-MS/MS | LC-MS/MS |
| NADPH (μmol/gDW) | ~0.289* | 0.056 | 1.80 | 0.486 |
| NADP (μmol/gDW) | ~0.281* | 0.19 | 0.37 | 0.462 |
| NADPH + NADP (μmol/gDW) | 0.55 | 0.246 | 2.17 | 0.948 |
| NADPH/NADP ratio | 1.03 | 0.29 | 4.86 | 1.05 |
*Amounts were converted to μmol/gDW by using a cellular volume of 1.7 mL/gDW.
**The cultivation presented persistent metabolic oscillations.
Based on the metabolite sensor, the glucose-limited steady state cytosolic free NADPH/NADP ratio under aerobic condition (D = 0.1 h−1) was 15.6 ± 0.60, which is much higher than the whole cell NADPH/NADP ratio (1.05 ± 0.08). This large difference of NADPH/NADP ratio was also observed in mouse pancreatic islets (26.8 (cytosolic free) vs. 1.2 (whole-cell ratio))16. The higher cytosolic free NADPH/NADP ratio is assumed to be due to a large fraction of cytosolic NADP being protein-bound13. Recently using another sensor reaction, the in vivo free cytosolic NAD/NADH ratio in S. cerevisiae was estimated to be higher than 100 under aerobic carbon-limited condition favouring oxidative processes, which was again very different from the whole cell average NAD/NADH ratio of 7.515. We should note that the ratio of redox cytosolic ratios (NADPH/NADP)/(NAD/NADH) is estimated at (15.6/0.01 ≈ 1560), showing a redox Gibbs energy advantage for NADPH of R T ln(1560) = 18.2 kJ/mol. Therefore, these effects cannot be observed by direct measurement of total concentration of NADP(H).
Cytosolic free and whole cell NADPH/NADP ratio in response to a glucose pulse
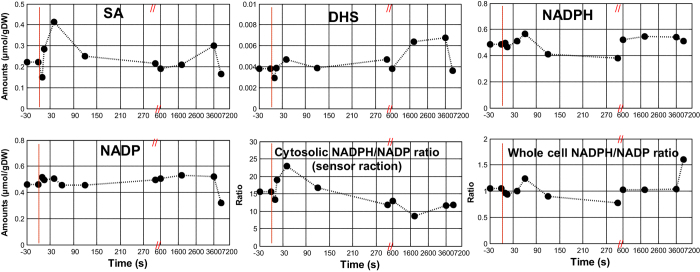
The extracellular responses of glucose, SA and DHS are shown in Fig. 3. The average uptake rate of glucose in the first 10 minutes was 0.54 μmol/(gDW∙s), and comparable to previous pulse experiments40, which is about 1.6 times higher than the steady state (0.33 μmol/(gDW∙s)). There was no considerable changes of extracellular SA in response to the glucose pulse, and the extracellular DHS decreased from 1.72 to 1.10 during the first seconds of the glucose pulse, and increased back to pre-pulse value in two minutes. The intracellular concentration of SA changed about 2 fold (Fig. 4, from 0.22 μmol/gDW to 0.41 μmol/gDW). This shows that the SA transport is not affected by changing intracellular SA level, confirming that SA transport is the irreversible symport mechanism.
Figure 4. Intracellular SA, DHS, NADPH, NADP amounts, cytosolic and whole cell NADPH/NADP ratio during the pulse experiments.
Data points before t = 0 represent the state 5 mins after the SA addition, just before the glucose perturbation.
After the glucose pulse, the whole cell NADPH increased slightly and then decreased back to the steady state value in 120 seconds. The whole cell NADP remained nearly constant (Fig. 4). The intracellular biomass specific amount of SA increased substantially after the glucose pulse, while there was no substantial change of the DHS level (Fig. 4).
The steady state sensor reaction based cytosolic free NADPH/NADP ratio was 15.6 ± 0.60. During the glucose pulse, the cytosolic ratio decreased first and then increased immediately to 23 (that was found in batch condition) in 1 minute and then decreased back to the pre-pulse level within 3 mins (Fig. 4). The first decrease could due to the delay in oxPPP, because it needs to adjust to the high metabolic flux and the glycolysis is faster. The increase suggests a rapid increase of the flux towards G6PDH and 6PGDH, which could cause the rapid production of NADPH and decrease of free NADP, while NADPH was consumed by the anabolic reactions41. Therefore, the cytosolic free NADPH/NADP ratio responded differently to the whole cell NADPH/NADP ratio to a glucose pulse.
Cytosolic NADPH/NADP related thermodynamics and kinetics
The cytosolic NADPH/NADP ratio is important to elucidate in vivo cytosolic pathway kinetics. For G6PDH and 6PGDH of the oxPPP, the overall reaction is:
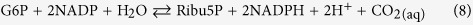 |
The Gibbs free energy (ΔrG′, kJ/mol) was calculated:
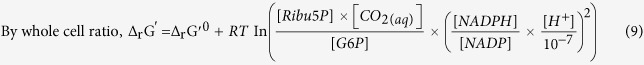 |
Using the sensor reaction based cytosolic NADPH/NADP ratios:
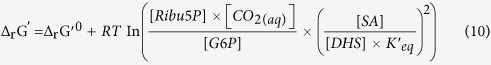 |
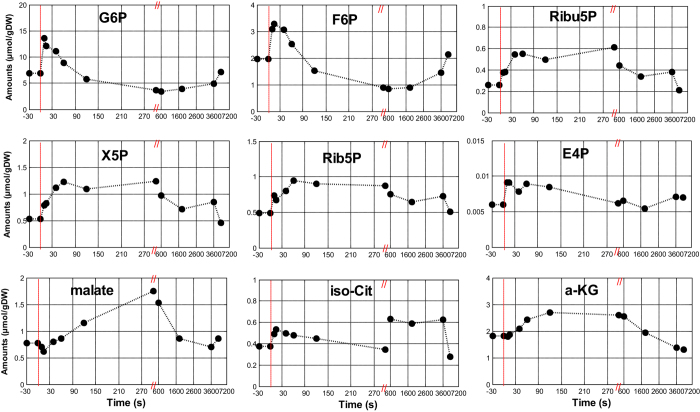
Here, the G6P and Ribu5P were measured (Fig. 5), and note that the Gibbs energy of reaction is not influenced by cytosolic pH because we use the sensor reaction. When S. cerevisiae was grown at pH = 5, the concentration of CO2 in equilibrium with the concentration of dissolved CO2 is in the order of 5% of the dissolved CO2 concentration, and therefore the exchange between these two pools was neglected in the calculations42,43. Assuming that the concentration change of CO2(aq) (estimated of 1 mmol/L using measured off-gas CO2 levels) during the pulse is small, the ΔrG′ can be estimated by sensor reaction or whole cell NADPH/NADP ratios and metabolite measurements (Fig. 6).
Figure 5. Intracellular metabolite amounts after a glucose pulse.
Data points before t = 0 represent the state 5 mins after the SA addition and before the glucose perturbation.
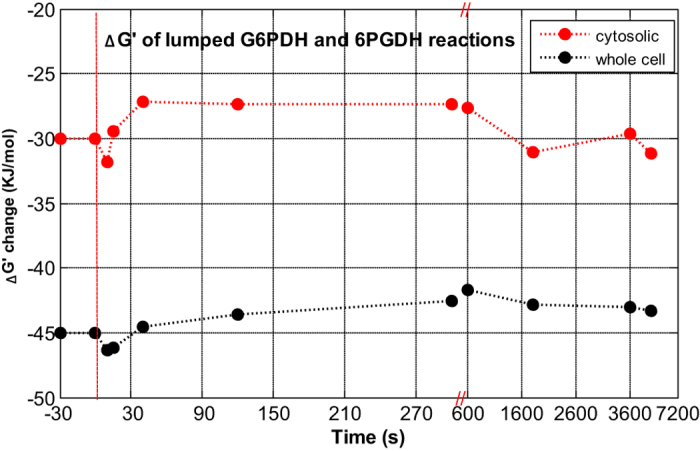
Figure 6. The profile of Gibbs free energy (ΔrG′, kJ/mol) for the lumped G6PDH and 6PGDH reactions.
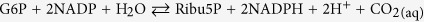 in the oxPPP by using cytosolic and whole cell NADPH/NADP ratio.
in the oxPPP by using cytosolic and whole cell NADPH/NADP ratio.
The sensor based estimation of steady state ΔrG′ for the oxPPP is −29 kJ/mol, while it is −42 kJ/mol using whole cell ratios. After the pulse, the ΔrG′ (by using cytosolic NADPH/NADP ratio) decreased immediately in 10 seconds (about 2 kJ/mol), indicating a higher thermodynamic driving force to allow for a higher flux through the PPP after the pulse. After 10 seconds, ΔrG′ increased above the steady state levels, and the thermodynamic driving force is reduced. This is remarkable because it is expected that the PPP flux increases, and this occurrence of reduced driving force suggest that allosteric mechanisms influence the flux. Using whole cell NADPH/NADP ratio measurement, the thermodynamic driving force is mostly due to the concentration changes of Ribu5P and G6P. The response was slower and the change was smaller than using cytosolic NADPH/NADP ratio (Fig. 6). The dynamic changes of other interested metabolites are shown in Fig. 5.
The NADPH producing oxPPP has a very large ΔrG′ and is far from the equilibrium, while NADPH consuming reactions are typically close to equilibrium reactions. However, a high driving force is important for the production rate. As mentioned before, one strategy for the microbial production of PDO was to switch NADH-dependent dehydrogenase DhaT to NADPH-dependent dehydrogenase YqhD. The cytosolic free NADH/NAD ratio was estimated to be 0.01 (assuming cytosolic pH 7.0)15, while the cytosolic free NADPH/NADP ratio was found to be 15.6 by sensor reaction. The driving force (ΔrG′) is then 18.2 kJ/mol larger using NADPH as a cofactor than NADH as a cofactor. This much higher driving force has probably led to a much lower level of the toxic aldehyde 3-HPA, allowing a higher PDO concentrations.
Model based estimation of fluxes
To obtain an estimation of the dynamic response of the PPP flux, the model of Vaseghi, et al.37 was used. The steady state and dynamic fluxes into PPP and the flux split ratio into glycolysis and PPP are shown in Fig. 7. The detailed calculation is shown in S1 Appendix.
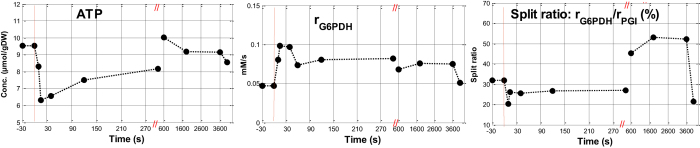
Figure 7. Model based estimation of the intracellular flux of G6PDH and the split ratio of flux into PPP and glycolysis during the pulse experiments.
Data points before t = 0 represent the state 5 mins after the SA addition, just before the glucose perturbation.
The steady state glucose uptake rate was 0.194 mM/s. The steady state flux in the PPP was 0.047 mM/s, which was 32% of the flux through the glycolysis (0.194–0.047 = 0.147 mM/s), which is lower than previous reported37, but well in the range of 2%–60%32,44. After the glucose pulse, it increased immediately and achieved a maximum flux of 0.098 mM/s in 15 seconds, and decreased slightly back to the steady state level (Fig. 7).
The glucose uptake rate was high directly after the pulse and decreased in time. Three different phases were distinguished based on the observed NADPH/NADP ratio profile and extracellular profile: 0–60 seconds, 60–600 seconds, 600–1800 seconds. The split ratio into PPP and glycolysis was calculated for the different phases. Both PPP and glycolytic flux increased after the glucose pulse, and the split ratio into PPP decreased from 32% to 20% of the glycolytic flux in 10 seconds and then increased back to the steady-state ratio (Fig. 7). This result is similar but more moderate compared to the observation of immediate decreasing of the split ratio from 56% to 10% after the glucose pulse observed by Vaseghi et al.37. A similar trend was observed by Frick and Wittmann45, here the contribution of the PPP decreased with increasing glucose uptake rate.
The enzyme activity of G6PDH was reported to be inhibited by NADPH/NADP ratio and the ATP level37. After the glucose pulse, the ATP concentration decreased (Fig. 7), and the oxPPP reaction rate increased. After 3 minutes, the cytosolic NADPH/NADP ratio dropped below the steady state ratio, which could be due to the decreased G6PDH activity or significantly increased NADPH consumption in biosynthetic reactions.
Conclusions
The SA-based cytosolic NADPH/NADP sensor reaction was used to determine the cytosolic free NADPH/NADP ratio under different conditions. The sensor reaction reacted rapidly to metabolic perturbations. The cytosolic free NADPH/NADP ratio was 15.6 ± 0.60 in glucose-limited chemostat (D = 0.1 h−1) condition, while the measured whole cell NADPH/NADP ratio was more than 10 times lower (1.05 ± 0.08). In response to a glucose pulse, the cytosolic NADPH/NADP ratio increased very rapidly and reached the steady state ratio after about 3 mins, while the whole cell NADPH/NADP ratio didn’t change substantially. The novel cytosolic NADPH/NADP ratio provides new insight on the thermodynamic driving force of NADPH dependent reactions, and will be useful in production strains of reduced products (alcohols, fuels) to identify putative bottlenecks on NADPH supply and problems of thermodynamic driving force in product pathways.
Materials and Methods
Strain and plasmid construction
First, a multicopy plasmid for yeast containing the aroE gene (encoding shikimate dehydrogenase (SDH)) under expression of the strong promoter TDH3 was constructed. The aroE gene was amplified from E. coli DH5α (18258-012, Invitrogen) genomic DNA with PCR using primer pairs SDH forward pTDH3 overlap (5′ AGTTTCGACGGATTCTAGAACTAGTATGGAAACCTATGCTGTTTTTGGTAATC 3′), SDH reverse tCYC1 overlap (5′ TAACTAATTACATGACTCGAGTCACGCGGACAATTCCTCCTG 3′) (Sigma-Aldrich) and Phusion® Hot Start II High Fidelity Polymerase (Thermo Scientific, Waltham, MA). The PCR product was purified from 1% agarose (w/v) gel using the Zymoclean™ kit (Zymo Research, Irvine, CA). The plasmid pAG426GPD-ccdB (Addgene, Cambridge, MA, USA) was restricted with SpeI and XhoI (Thermo Scientific), following gel purification of its backbone. The aroE cassette was assembled into this backbone using In-Fusion cloning (Clontech, Mountain View, CA, USA). The resulting plasmid pAG426-GPD-SDH was transformed into E. coli and confirmed using restriction analysis with SnaBI, SpeI and XhoI (Thermo Scientific). The plasmid was then transformed to S. cerevisiae strain CEN.PK113-5D using the lithium acetate protocol46, yielding strain CEN.PK-aroE.
Enzyme activity measurement
The cell extract from the culture samples were prepared by sonication method as described47. Protein concentrations in cell extracts were measured with the Lowry method48, and bovine serum albumin (BSA) (Sigma-Aldrich, The Netherland) was used as a standard. The activity of shikimate dehydrogenase was assayed in the direction of NADPH production. The assay was conducted in a final volume of 500 μL that contained: 100 mM Tris-HCl, pH 9.0, 4 mM shikimate (Sigma-Aldrich, The Netherland), 2 mM NADP and 5 μL cell extract. The enzyme activity was assayed by continuously monitoring the increase in NADPH absorbance at 340 nm for 30 mins at 25 °C49. Additionally, the enzymatic activity of G6PDH was measured according to the protocol of Postma, et al.35.
External SA supplementation for the sensor reaction
The intracellular SA and DHS concentrations were too low for accurate measurement in the CEN.PK-aroE strain. Therefore, SA was added to the extracellular space (Fig. 1) . To analyse if there was an influence of SA supplementation on the NADP(H) level, different amounts of SA were added to batch shake flask cultures (NH4+ as nitrogen source) (100 μM, 500 μM and 1000 μM residual SA). After 5 minutes, the intracellular SA and DHS were measured. To measure if there was an influence of SA incubation time, the supplementation (500 μM) was performed in batch (urea as nitrogen source) and samples were taken after 5, 10 and 15 minutes, respectively. The membrane transport rate and equilibrium was evaluated by a pulse experiment, where a concentrated SA solution (5 mL of 100 mM) was rapidly added to a chemostat culture (1L) using a syringe, resulting in a 500 μM extracellular SA concentration. After 5 minutes, a glucose pulse was added, resulting in a 2.3 mM increase of the residual glucose concentration.
Media and cultivation
For the working stock, S. cerevisiae CEN.PK-aroE strain was cultured aerobically in 500 mL shake flask at 30 °C using 100 mL synthetic medium with 20 g/L glucose as carbon source50. After overnight growth, glycerol was added (final concentration 30% (v/v)) and 1 mL aliquots were stored in sterile vials at −80 °C.
Shake flask cultures were performed with synthetic media containing 20 g/L glucose50. NH4+ was used as nitrogen source for the set of batch experiments with/without SA supplementation. Urea was used as nitrogen source for batch experiments with different SA incubation times to avoid decreasing of the extracellular pH.
The glucose-limited chemostat cultivation was performed using a low-salt Verduyn minimal medium51 with 7.5 g/L glucose in a 2 L bioreactor (Applikon, The Netherlands) with 1 L working volume. The dilution rate was set to 0.1 h−1; pH was kept constant at 5.0 by automatic addition of 2 M KOH as described previously52. The temperature was controlled at 30 °C and the head space overpressure was kept at 0.3 bars. The aeration rate was 0.5 vvm and the stirrer speed was 600 rpm. Cultures were assumed to be in steady state after 5 volume changes, as confirmed by constant CO2, O2 and DO levels. The biomass dry weight concentration was measured as described earlier34.
Glucose pulse experiments in continuous cultivation
To perform the glucose pulse experiment, 5 minutes after the addition of 500 μM SA, a concentrated glucose solution (30 mL of 79 mM glucose) was rapidly added to a chemostat culture using a syringe in glucose-limited chemostat, resulting in a 2.3 mM increase of the residual glucose concentration, which was 0.18 mM. For the continous culture, the steady samples were measured in triplicate by taking three independent samples from the same cultivation. During the dynamic glucose pulse experiment, only single samples could be taken and this experiment has only been performed once (very labor intensive). Please take into account that timeseries measurements contain a certain level of redundancy because of the dense sampling in time.
Intracellular metabolites
The sampling, quenching, extraction and analysis of the intracellular metabolites was similar to the methods described earlier in Wahl, et al.53 using a rapid sampling device. Briefly, 1.2 mL broth was withdrawn from the bioreactor and immediately quenched in 6 mL cold (−40 °C) 100% methanol, followed by vortexing and weighing. For shake flask cultures, samples were taken directly from the flask by pipetting and immediate quenching in 6 mL cold (−40 °C) 100% methanol. Clearly, this latter sampling was slower compared to the rapid sampling device, nevertheless batch and chemostat are different conditions and we expect representative results with this approach because the samples contain excess substrate and oxygen limitation is not reached during the transfer of about 3 seconds. 120 μL of 13C cell extract were added to the tube as an internal standard54. The intracellular metabolites were extracted and analysed by GC-MS55 or LC-MS/MS56,57, including whole cell NADPH and NADP concentrations58. The SA and DHS were analysed by GC-MS/MS, and the collision energy (CE) for SA is 35 and for DHS is 5 eV. The MS/MS fragments for SA is 372.1/73.1(12C) and 379.1/73.1(13C, internal standard), and the MS/MS fragments for DHA is 417.1/386.1 (12C) and 424.1/393.1(13C). Amounts were converted to intracellular concentrations by using a cellular volume of 1.7 mL/gDW59. Please note that in this work, we focus on the NADP/NADPH ratio. Therefore only the concentration ratio is the determining factor. Even if the volume assumption was severely violated, the amount of SA/DHS will be correct as all cytosolic concentrations are affected by the conversion (from the raw data μmol/gDW to cytosolic concentration).
Extracellular metabolites
Samples for extracellular metabolite analysis were taken using the cold steel beads method as described earlier60. The concentration of glucose was determined by HPLC, GC-MS or enzymatically as described elsewhere34. The extracellular concentrations of SA and DHS were determined by GC-MS/MS. Here, an aliquot of 100 μL sample was mixed with 20 μL of13C cell extract and further processed following the method described in Wahl, et al.53.
Additional Information
How to cite this article: Zhang, J. et al. Determination of the Cytosolic NADPH/NADP Ratio in Saccharomyces cerevisiae using Shikimate Dehydrogenase as Sensor Reaction. Sci. Rep. 5, 12846; doi: 10.1038/srep12846 (2015).
Supplementary Material
Acknowledgments
This project was funded by the Kluyver Centre for Genomics of Industrial Fermentation ((KCII 1.1.03), www.kluyvercentre.nl) to J.Z, A.W, J.H, which is part of the Netherlands. Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NWO).
Footnotes
Author Contributions Conceived and designed the experiments: J.Z., J.H. and S.W. Performed the experiments: J.Z. and H.R. Analytical work: J.Z., A.P., R.M.S., C.R. and J.D. Wrote the paper: J.Z. and S.W. All authors reviewed the manuscript.
References
- Canepa L., Ferraris A. M., Miglino M. & Gaetani G. F. Bound and unbound pyridine dinucleotides in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Biochimica et Biophysica Acta (BBA)-General Subjects 1074, 101–104 (1991). [DOI] [PubMed] [Google Scholar]
- Latouche G., Cerovic Z. G., Montagnini F. & Moya I. Light-induced changes of NADPH fluorescence in isolated chloroplasts: a spectral and fluorescence lifetime study. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1460, 311–329 (2000). [DOI] [PubMed] [Google Scholar]
- Pollak N., Dolle C. & Ziegler M. The power to reduce: pyridine nucleotides-small molecules with a multitude of functions. Biochem. J 402, 205–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruice P. Y. Organic Chemistry 7th Edition. (Prentice Hall, 2013). [Google Scholar]
- Runguphan W. & Keasling J. D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21, 103–113 (2014). [DOI] [PubMed] [Google Scholar]
- Park S. H. et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nat Commun 5, 4618, 10.1038/ncomms5618 (2014). [DOI] [PubMed] [Google Scholar]
- Bommareddy R. R., Chen Z., Rappert S. & Zeng A.-P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab. Eng. 25, 30–37 (2014). [DOI] [PubMed] [Google Scholar]
- Emptage M., Haynie S. L., Laffend L. A., Pucci J. P. & Whited G. inventors; E. I. Du Pont De Nemours And Company, Assignee. Process for the biological production of 1, 3-propanediol with high titer. United States patent US6.514,733 B1. 2003 Feb 04.
- Nakamura C. E. & Whited G. M. Metabolic engineering for the microbial production of 1, 3-propanediol. Curr. Opin. Biotechnol. 14, 454–459 (2003). [DOI] [PubMed] [Google Scholar]
- Soboll S., Scholz R. & Heldt H. W. Subcellular metabolite concentrations. Dependence of mitochondrial and cytosolic ATP systems on the metabolic state of perfused rat liver. Eur. J. Biochem. 87, 377–390 (1978). [DOI] [PubMed] [Google Scholar]
- Wang E. et al. Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD+ binding in solution. J. Mol. Biol. 408, 670–683 (2011). [DOI] [PubMed] [Google Scholar]
- Williamson D. H., Lund P. & Krebs H. A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103, 514–527 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech R. L., Eggleston L. V. & Krebs H. A. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 115, 609–619 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech R., Raijman L. & Krebs H. Equilibrium relations between the cytoplasmic adenine nucleotide system and nicotinamide-adenine nucleotide system in rat liver. Biochem. J 117, 499–503 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canelas A. B., van Gulik W. M. & Heijnen J. J. Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol. Bioeng. 100, 734–743, 10.1002/bit.21813 (2008). [DOI] [PubMed] [Google Scholar]
- Hedeskov C., Capito K. & Thams P. Cytosolic ratios of free [NADPH]/[NADP+] and [NADH]/[NAD+] in mouse pancreatic islets, and nutrient-induced insulin secretion. Biochem. J 241, 161–167 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira dos Santos M., Raghevendran V., Kötter P., Olsson L. & Nielsen J. Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab. Eng. 6, 352–363 (2004). [DOI] [PubMed] [Google Scholar]
- Lu Q. & McAlister-Henn L. Peroxisomal localization and function of NADP+-specific isocitrate dehydrogenases in yeast. Arch. Biochem. Biophys. 493, 125–134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D., Lund P. & Krebs H. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103, 514–527 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv H. & Gilvarg C. Aromatic biosynthesis XIV. 5-Dehydroshikimic reductase. J. Biol. Chem. 213, 787–795 (1955). [PubMed] [Google Scholar]
- Ishihama Y. et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L. D., Gillies F. M. & Coggins J. R. Over-expression of the yeast multifunctional arom protein. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1216, 417–424 (1993). [DOI] [PubMed] [Google Scholar]
- Duncan K., Edwards R. & Coggins J. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem. J 246, 375–386 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- Anton I. A. & Coggins J. R. Sequencing and overexpression of the Escherichia coli aroE gene encoding shikimate dehydrogenase. Biochem. J 249, 319–326 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A., Noor E., Bar-Even A. & Milo R. eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–775 (2011). 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hollander J., Ugurbil K., Brown T. & Shulman R. Phosphorus-31 nuclear magnetic resonance studies of the effect of oxygen upon glycolysis in yeast. Biochemistry 20, 5871–5880 (1981). [DOI] [PubMed] [Google Scholar]
- Caspani G., Tortora P., Hanozet G. M. & Guerritore A. Glucose-stimulated cAMP increase may be mediated by intracellular acidification in Saccharomyces cerevisiae. FEBS letters 186, 75–79 (1985). [Google Scholar]
- Thevelein J. M. et al. Regulation of the cAMP level in the yeast Saccharomyces cerevisiae: the glucose-induced cAMP signal is not mediated by a transient drop in the intracellular pH. J Gen Microbiol 133, 2197–2205 (1987). [DOI] [PubMed] [Google Scholar]
- Orij R., Postmus J., Ter Beek A., Brul S. & Smits G. J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155, 268–278 (2009). [DOI] [PubMed] [Google Scholar]
- Kresnowati M., Suarez Mendez C., Groothuizen M., Van Winden W. & Heijnen J. Measurement of fast dynamic intracellular pH in Saccharomyces cerevisiae using benzoic acid pulse. Biotechnol. Bioeng. 97, 86–98 (2007). [DOI] [PubMed] [Google Scholar]
- Bruinenberg P. M., Van Dijken J. P. & Scheffers W. A. A theoretical analysis of NADPH production and consumption in yeasts. J Gen Microbiol 129, 953–964 (1983). [DOI] [PubMed] [Google Scholar]
- Winden W. A. et al. Metabolic‐flux analysis of Saccharomyces cerevisiae CEN. PK113‐7D based on mass isotopomer measurements of 13C‐labeled primary metabolites. FEMS Yeast Res. 5, 559–568 (2005). [DOI] [PubMed] [Google Scholar]
- Suarez-Mendez C. A., Sousa A., Heijnen J. J. & Wahl A. Fast “Feast/Famine” cycles for studying microbial physiology under dynamic conditions: a case study with Saccharomyces cerevisiae. Metabolites 4, 347–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma E., Verduyn C., Scheffers W. A. & Van Dijken J. P. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55, 468–477 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipp M., Camakaris H. & Pittard A. Cloning and analysis of the shiA gene, which encodes the shikimate transport system of Escherichia coli K-12. Gene 209, 185–192 (1998). [DOI] [PubMed] [Google Scholar]
- Vaseghi S., Baumeister A., Rizzi M. & Reuss M. In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab. Eng. 1, 128–140 (1999). [DOI] [PubMed] [Google Scholar]
- Ortmayr K. et al. Sample preparation workflow for the liquid chromatography tandem mass spectrometry based analysis of nicotinamide adenine dinucleotide phosphate cofactors in yeast. J Sep Sci 37, 2185–2191, 10.1002/jssc.201400290 (2014). [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Passonneau J. V. & Rock M. K. The stability of pyridine nucleotides. J. Biol. Chem. 236, 2756–2759 (1961). [PubMed] [Google Scholar]
- Wu L. et al. Short-term metabolome dynamics and carbon, electron, and ATP balances in chemostat-grown Saccharomyces cerevisiae CEN. PK 113-7D following a glucose pulse. Appl. Environ. Microbiol. 72, 3566–3577 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302, 10.1038/nature13236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Lange H., Van Gulik W. & Heijnen J. Determination of in vivo oxygen uptake and carbon dioxide evolution rates from off‐gas measurements under highly dynamic conditions. Biotechnol. Bioeng. 81, 448–458 (2003). [DOI] [PubMed] [Google Scholar]
- de Jonge L., Heijnen J. & van Gulik W. Reconstruction of the oxygen uptake and carbon dioxide evolution rates of microbial cultures at near-neutral pH during highly dynamic conditions. Biochem. Eng. J. 83, 42–54 (2014). [Google Scholar]
- van Winden W. et al. Metabolic-flux analysis of Saccharomyces cerevisiae CEN. PK113-7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEMS Yeast Res. 5, 559 (2005). [DOI] [PubMed] [Google Scholar]
- Frick O. & Wittmann C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microbial cell factories 4, 30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D. & Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 (2002). [DOI] [PubMed] [Google Scholar]
- de Kok S. et al. Increasing free-energy (ATP) conservation in maltose-grown Saccharomyces cerevisiae by expression of a heterologous maltose phosphorylase. Metab. Eng. 13, 518–526, 10.1016/j.ymben.2011.06.001 (2011). [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L. & Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951). [PubMed] [Google Scholar]
- Chaudhuri S., Anton I. A. & Coggins J. R. Shikimate dehydrogenase from Escherichia coli. Methods Enzymol. 142, 315–320 (1987). [DOI] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W. A. & van Dijken J. P. Physiology of Saccharomyces Cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol 136, 395–403 (1990). [DOI] [PubMed] [Google Scholar]
- Canelas A. B. et al. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 81, 7379–7389 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. A fast sensor for in vivo quantification of cytosolic phosphate in Saccharomyces cerevisiae. Biotechnol. Bioeng. 10.1002/bit.25516 (2014). [DOI] [PubMed] [Google Scholar]
- Wahl S. A. et al. in Methods Mol Biol Metabolic Flux Analysis: Methods and Protocols (eds Krömer Jens O., Nielsen Lars K. & Blank Lars M.) Ch. Quantitative Metabolomics Using ID-MS, 91–105 (Springer, 2014). [Google Scholar]
- Wu L. et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 336, 164–171 (2005). [DOI] [PubMed] [Google Scholar]
- Cipollina C. et al. A comprehensive method for the quantification of the non-oxidative pentose phosphate pathway intermediates in Saccharomyces cerevisiae by GC–IDMS. J. Chromatogr. 877, 3231–3236 (2009). [DOI] [PubMed] [Google Scholar]
- Seifar R. M. et al. Simultaneous quantification of free nucleotides in complex biological samples using ion pair reversed phase liquid chromatography isotope dilution tandem mass spectrometry. Anal. Biochem. 388, 213–219 (2009). [DOI] [PubMed] [Google Scholar]
- van Dam J. C. et al. Analysis of glycolytic intermediates in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionization with tandem mass spectrometric detection. Anal. Chim. Acta 460, 209–218 (2002). [Google Scholar]
- Seifar R. M. et al. Quantitative analysis of intracellular coenzymes in Saccharomyces cerevisiae using ion pair reversed phase ultra high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. 1311, 115–120 (2013). [DOI] [PubMed] [Google Scholar]
- Canelas A. B., Ras C., ten Pierick A., van Gulik W. M. & Heijnen J. J. An in vivo data-driven framework for classification and quantification of enzyme kinetics and determination of apparent thermodynamic data. Metab. Eng. 13, 294–306 (2011). [DOI] [PubMed] [Google Scholar]
- Mashego M. R., van Gulik W. M., Vinke J. L., Visser D. & Heijnen J. J. In vivo kinetics with rapid perturbation experiments in Saccharomyces cerevisiae using a second-generation BioScope. Metab. Eng. 8, 370–383 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.