Abstract
The ∼230-residue C-terminal tail of the epidermal growth factor receptor (EGFR) is phosphorylated upon activation. We examined whether this phosphorylation is affected by deletions within the tail and whether the two tails in the asymmetric active EGFR dimer are phosphorylated differently. We monitored autophosphorylation in cells using flow cytometry and found that the first ∼80 residues of the tail are inhibitory, as demonstrated previously. The entire ∼80-residue span is important for autoinhibition and needs to be released from both kinases that form the dimer. These results are interpreted in terms of crystal structures of the inactive kinase domain, including two new ones presented here. Deletions in the remaining portion of the tail do not affect autophosphorylation, except for a six-residue segment spanning Tyr 1086 that is critical for activation loop phosphorylation. Phosphorylation of the two tails in the dimer is asymmetric, with the activator tail being phosphorylated somewhat more strongly. Unexpectedly, we found that reconstitution of the transmembrane and cytoplasmic domains of EGFR in vesicles leads to a peculiar phenomenon in which kinase domains appear to be trapped between stacks of lipid bilayers. This artifactual trapping of kinases between membranes enhances an intrinsic functional asymmetry in the two tails in a dimer.
INTRODUCTION
The epidermal growth factor receptor (EGFR) is a tyrosine kinase that is critical for proper cell growth and differentiation and is frequently mutated in many cancers (1, 2). Human EGFR (HER1, also known as ErbB1, after the erythroblastoma virus oncogene) is a member of a family of four receptors, that includes HER2 (ErbB2), HER3 (ErbB3). and HER4 (ErbB4) (3). These receptors consist of a ligand-binding extracellular module and an intracellular kinase domain linked by a single transmembrane helix (2, 4, 5) (Fig. 1A). EGFR and HER4 are fully functional kinases, and both their extracellular domains bind ligands, allowing them to function as homodimers or heterodimers. HER2 does not have a ligand, and HER3 has impaired kinase activity (3), and these two receptors signal principally through heterodimerization. The HER2/HER3 heterodimer generates one of the strongest proliferative signals in cancer (1).
FIG 1.
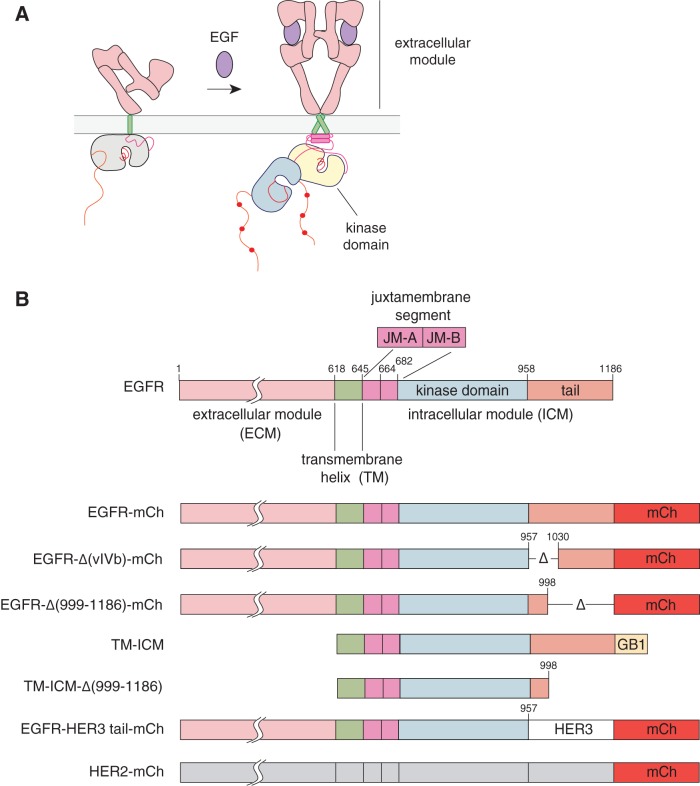
Model for activation of epidermal growth factor receptor (EGFR) and constructs used in this study. (A) Ligand binding to the extracellular domain of the epidermal growth factor receptor induces a conformational change that results in receptor-mediated dimerization and activation. Activation of the intracellular kinase domains is promoted by the formation of an asymmetric dimer, in which one kinase domain (the activator [yellow]) activates the other kinase domain (the receiver [blue]). (B) Domain architecture of human EGFR with domain boundaries highlighted. The domain composition of the EGFR family constructs used in this study is also presented, including EGFR deletion constructs, the EGFR-HER3 tail chimera, and HER2 (Δ, deletion; mCh, mCherry fluorescent protein fusion).
The very C-terminal portion of these receptors, following the kinase domain, is a long tail segment with more than 200 residues. The tails contain several tyrosine residues that undergo autophosphorylation upon activation of the receptor and serve as docking sites for effector proteins that transmit the signal further downstream (6). All but one of the tyrosine residues that are critical for signaling by these receptors are in the tail segment. The one exception is a tyrosine residue in the activation loop of the kinase domain, phosphorylation of which is expected to stabilize the active conformation of EGFR, HER2, and HER4 (7).
The mechanism by which EGFR family members are activated has several distinctive features (2, 4, 5, 8). Binding of EGF results in a conformational change in the extracellular domain, converting an unliganded “tethered” conformation (9) to an extended conformation that forms “back-to-back” dimers in which the ligand does not bridge the two subunits (8, 10, 11) (Fig. 1A). A key step in switching on the receptor is the formation of an asymmetric dimer of the kinase domains, in which one kinase, termed the activator, allosterically activates the other one, termed the receiver (12–17). The transmembrane helices of the receptors also dimerize (18–20).
The tail in human EGFR spans 229 residues, from Gln 958 to Ala 1186 (we use a residue numbering system in which the 24 residues of the signal sequence are not counted). There are seven tyrosine residues within this span that are conserved in EGFR in the jawed vertebrates (Gnathostomes) in which the EGFR family has expanded to four members (see Fig. S1A in the supplemental material). The sequence of the tail is not consistent with regular secondary structure, suggesting that it is flexible. It is therefore surprising that the entire tail segment, and not just the regions immediately flanking the phosphorylation sites, is highly conserved in EGFR sequences across these species (see Fig. S1A). This conservation breaks down when comparing the human EGFR sequence with those of invertebrates, which typically have only one EGFR (see Fig. S1B). Within the human EGFR family members, the sequences of the kinase-active HER2 and HER4 tails resemble that of EGFR, while that of HER3 is quite different.
The proximal segment of the EGFR tail, spanning residues 958 to 1043, is important for autoinhibition of the receptor. This is highlighted by two cancer mutations that produce variants of EGFR, known as vIVa and vIVb, in which this proximal segment is deleted (21). Pines and coworkers have shown that these variants are characterized by constitutive receptor dimerization, elevated levels of kinase activity without EGF stimulation, and the inappropriate triggering of downstream pathways (22). A part of the proximal tail segment has been visualized in a structure of an EGFR kinase domain variant in which a mutation (V924R) prevents formation of the asymmetric dimer (15). The structure shows three interactions that are consistent with autoinhibition. The first portion of the tail, residues 973 to 977, forms an α-helix, referred to as the AP2 helix because this segment also interacts with the clathrin adapter AP2 (23). The AP2 helix is responsible for holding two kinase domains together in an inactive dimer in the crystal by interacting with the N-lobe of the other kinase in the dimer, and switching between alternative kinase dimers has been invoked as an important function of the tail (24). The region immediately following the AP2 helix, spanning residues 979 to 990, is termed the “electrostatic hook,” and it contains several acidic residues that interact with the hinge region of the kinase domain. Residues 991 to 998 form a β-strand that runs along the surface of the kinase domain in a manner that resembles the latch formed by the juxtamembrane segment, and it thereby prevents the formation of the juxtamembrane latch that is necessary for activation.
In this paper, we address two questions. First, what is the effect of deletions within different regions of the tail on EGFR phosphorylation? Second, is the asymmetry in the kinase domains that form the active dimer reflected in an unequal level of phosphorylation of the activator and receiver tails? To answer these questions, we made overlapping deletions in the tail and also introduced mutations that isolate the effects on the activator and receiver tails and measured the autophosphorylation of EGFR in cells. As expected based on the work of Pines and colleagues (22), the proximal region of the tail inhibits the activity of the receptor. We map the inhibitory segments in finer detail and extend a structural model for the proximal segment of the tail based on two new crystal structures. We show that a localized region of the distal portion of the tail, spanning Tyr 1086, is required for activation loop phosphorylation. We find that tail phosphorylation is asymmetric, with a small, but significant, preference for phosphorylation of the activator tail, rather than the receiver tail. In an attempt to study the autophosphorylation mechanism in vitro, we made the unexpected discovery that membrane incorporation of an EGFR construct consisting of the transmembrane and the intracellular modules causes lipid bilayers to stack on top of one another, with the kinases presumably trapped between membrane layers. This peculiar artifact of in vitro membrane reconstitution is dependent on asymmetric dimer formation by the kinases and results in biphasic kinetics for phosphorylation of tyrosine residues in the tail.
MATERIALS AND METHODS
Flow cytometry.
Human EGFR (UniProt accession no. P00533) constructs were cloned into vectors based on the pEGFP-N1 plasmid (Clontech) using XhoI and SacII restriction sites. Enhanced green fluorescent protein (EGFP) was replaced by either monomeric variants of either mCherry (Clontech) or Cerulean (25) fluorescent proteins using BamHI and NotI sites. The EGFR-HER3 tail chimera was generated by restriction-free cloning to replace the EGFR tail (residues 958 to 1186) with the equivalent tail region of human HER3. Full-length human HER2 was cloned in a manner similar to EGFR.
Cos-7 or HEK-293T cells grown on 6-well plates were transfected with FuGENE (Promega) and serum starved for 16 h. These two cell lines were chosen because of their low endogenous EGFR levels and high transfection efficiency. Cells were dissociated by enzyme-free cell dissociation buffer (Gibco), transferred to 96-well plates, and spun down at 1,800 rpm for 3 min. Samples were treated with 100 ng/ml EGF (Sigma-Aldrich) for 3 min at room temperature. After stimulation, cells were immediately fixed in 2% formaldehyde for 10 min. Samples were spun down and permeabilized in ice-cold methanol for 30 min on ice. After rehydration in staining buffer (phosphate-buffered saline [PBS] supplemented with 0.2% bovine serum albumin [BSA] and 1 mM EDTA), cells were stained with 1:100 dilution of primary antibody. Anti-EGFR-pY845 (antibody to phosphorylated EGFR [phosphorylated tyrosine at position 845]) (catalog no. 2231), anti-EGFR-pY974 (catalog no. 2641), anti-EGFR-pY992 (catalog no. 2235), anti-EGFR pY1068 (catalog no. 2234), anti-HER3-pY1289 (catalog no. 4791), anti-HER2-pY1221 (catalog no. 2243), and anti-pErk1/2-pT202/pY204 (catalog no. 4370) antibodies were purchased from Cell Signaling Technology. Anti-EGFR pY1173 (catalog no. sc-12351) was purchased from Santa Cruz Biotechnology. Samples were then stained with 1:100 dilution of anti-rabbit fluorescein-labeled (fluorescein isothiocyanate [FITC]) antibody (Sigma-Aldrich). Antibody specificity toward EGFR phosphorylated tyrosines was evaluated by Western blotting of EGFR variants in which the corresponding tyrosine residues were mutated to alanine (see Fig. S2 in the supplemental material). The anti-EGFR-pY974 and anti-EGFR-pY1068 antibodies showed some unspecific binding to their corresponding mutant EGFR variants (Y974A and Y1068F, respectively). This was taken into account in the interpretation of the results presented here.
Flow cytometry analysis was done using a BD Bioscience LSR Fortessa cell analyzer (San Jose, CA). A 50-mW, 561-nm, Coherent Sapphire laser (San Jose, CA) was used to excite mCherry, and fluorescence was detected using a 610/20-nm band-pass filter. Cerulean was excited by a 50-mW, 440-nm, Coherent Sapphire laser (San Jose, CA). FITC was excited by a 50-mW, 488-nm, Coherent Sapphire laser (San Jose, CA), and fluorescence was detected using a 525/50-nm band-pass filter.
In experiments where only one type of receptor was transfected, cells were binned based on their EGFR-mCherry expression levels, and the mean and standard error values were calculated for the green channel to obtain phosphorylation density dependence graphs. In cotransfection experiments, two-dimensional histograms were generated using Gnuplot (http://www.gnuplot.info). Here, cells were binned based on the expression levels of two different EGFR constructs, one labeled with mCherry, the other with Cerulean. Mean and standard error values for the green fluorescence intensities, corresponding to phosphorylation levels, were calculated in each bin. Colors were used to represent phosphorylation levels, based on the calculated mean intensities of the green channel, blue meaning less phosphorylation and red meaning more. The color scale is linear, and the scale was set so that it covers the full range of data. The same scale was used for each sample analyzed within the same set of experiments, resulting in comparable graphs for different samples measured on the same day.
Crystal structure determination.
DNA encoding residues 658 to 998 (658–998) or 672 to 1018 (672–1018) of human EGFR was cloned into pFAST BAC HT (Invitrogen). The constructs have an N-terminal six-histidine (6×His) tag and a cleavage site for the tobacco etch virus (TEV) protease (MSYYHHHHHHDYDIPTTENLYFQGAM) that was cleaved prior to the last step of purification. The cleavage leaves the heterologous GAM peptide motif at the N terminus of the EGFR constructs. The mutations were introduced using the QuikChange kit (Stratagene) and confirmed by DNA sequencing. The proteins were expressed in Sf9 cells using the Bac-to-Bac expression system (Gibco BRL) and purified according to the previously described protocol for the EGFR kinase domain (residues 672 to 998) (12).
Crystals of the adenyl-5′-yl imidodiphosphate (AMP-PNP)-bound V924R 658–998 EGFR kinase domain construct were grown using the hanging drop vapor diffusion method. Equal volumes (0.7 μl) of protein [4 mg/ml in 10 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 5 mM AMP-PNP, 10% glycerol, 50 mM NaCl, 2 mM dithiothreitol (DTT), and 2 mM Tris(2-carboxyethyl)phosphine (TCEP)] and crystallization solution (0.2 M ammonium nitrate, 16% polyethylene glycol 3350 [PEG 3350]) were mixed and then equilibrated with a 500-μl reservoir of the same crystallization solution at 20°C. Crystals for the AMP-PNP-bound I682Q 672–1018 kinase domain construct were grown using the hanging drop vapor diffusion method. Equal volumes (0.7 μl) of protein (4 mg/ml in 10 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 5 mM AMP-PNP, 10% glycerol, 50 mM NaCl, 2 mM DTT, and 2 mM TCEP) and a different crystallization solution (16% PEG 6000, 0.1 M morpholineethanesulfonic acid [MES] [pH 5.5], 0.1 M ammonium chloride) were mixed and then equilibrated with a 500-μl reservoir of the same crystallization solution at 20°C. These crystals were further optimized by seeding into the crystallization condition containing 11% PEG 6000, 0.1 M MES (pH 5.5), and 0.15 M ammonium chloride at a 3-mg/ml protein concentration. All crystals were frozen in the crystallization solution containing 20% glycerol.
Diffraction data to 1.55-Å spacings for V924R 658–998 EGFR crystals and to 1.90-Å spacings for the I682Q 672–1018 EGFR crystals were collected at 100 K using synchrotron radiation at beam line 8.2.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory (see Table S1 in the supplemental material). Reflections were integrated and scaled using HKL2000 (26). The structure was determined using molecular replacement with the program Phaser (27) with the inactive EGFR kinase domain (PDB accession no. 3GT8) as a search model. The models were built using Coot (28) and refined using PHENIX (29). MolProbity (30) was used to monitor model quality.
In the V924R crystal structure, electron density maps reveal a peculiar feature, which is that a peptide fragment corresponding to residues Ala 989 to Gln 997 is sandwiched between the N-lobes of the two kinases in the asymmetric unit, without being connected to any other part of the structure (see Fig. S5A and B in the supplemental material). This peptide may have been generated by proteolysis in the crystallization drop and appears to have cocrystallized with the intact kinase molecules. Such electron density of a peptide fragment sandwiched between the N-lobes of the two kinases was not observed in the I682Q crystal structure.
The higher resolution of the present analysis of the EGFR V924V variant allowed us to correct a sequence register error in the previously published model for residue Met 983 to the end of the tail. In the original model, Met 983 bulged out from the turn that connects the electrostatic hook to the strand that blocks the juxtamembrane latch (see Fig. S5C and D in the supplemental material). We now find that Met 983 is incorporated into the strand and that all subsequent residues are moved down by one position in register. This adjustment does not change the backbone trace of the residues, except for the displacement of Met 983, and so the conclusion that the 983–991 segment of the tail blocks the juxtamembrane latch is unaffected. A corrected version of the PDB entry 3GT8 is being uploaded to the Protein Data Bank.
Molecular dynamics.
All molecular dynamics simulations were performed with Amber 14 (31) using the ffSB-99ILDN force field (32) and periodic boundary conditions. Simulations were performed with pmemd module on Nvidia Tesla M2075 or GeForce GTX Titan graphics cards. The starting model was generated as follows. Chain B from the crystallographic model of the I682Q mutant reported in this study, from a late stage of refinement, was stripped of all water molecules, ions, and nucleotides. These coordinates were aligned to those of chain A from PDB entry 2RFE, the crystal structure of human EGFR bound to Mig6 (33). Backbone and Cβ atoms for EGFR residues 1012 to 1014 were placed at the positions of Mig6 residues 349 to 352 (chain E of PDB entry 2RFE). Using Coot (28), residues 991 to 1010 of EGFR were added to connect Asp 990, the last residue in the crystallographic model, to Ser 1012 in an arbitrary but stereochemically reasonable conformation, and residues 1015 to 1024 were modeled in an extended conformation. All free termini were capped with N-acetyl or amide groups. To roughly optimize the bond geometry, this model was subjected to simulated annealing in CNS (Crystallography & NMR System) (34) with the standard CNS protein force field, and with the positions of all atoms from the crystallographic model and all backbone atom positions for residues 1012 to 1014 fixed. This model was then prepared for simulation in Amber as follows. Hydrogens were added with the tleap module, and the model was placed in an orthorhombic box filled with TIP3P water molecules, at a closeness of 0.75 Å, such that the minimum distance from the protein to the box edge was 15.0 Å, and 3 Na+ ions were added to neutralize the charge of the system. After an initial energy minimization in Amber, equilibration was carried out with three rounds of molecular dynamics, with Cartesian harmonic restraints on the protein atoms. For equilibration and production molecular dynamics, long-range electrostatic interactions were calculated with particle-mesh Ewald summation, and a nonbonded interaction cutoff was set at 10.0 Å. Hydrogen-containing bond lengths were constrained with the SHAKE algorithm (35). The time step size was 2 fs. The system was first heated to 300 K during 20 ps of constant number and volume equilibration. This was followed by 10 ps of constant number, temperature, and volume equilibration, and then 20 ps of constant number, temperature, and pressure equilibration with a Berendsen barostat (36). Unrestrained production molecular dynamics was carried out at 300 K with the Berendsen weak temperature coupling algorithm and a Monte Carlo barostat (31), with initial atom velocities carried over from the equilibration. Coordinates from production runs were saved every picosecond. The starting model for the simulation that produced the model described in Results and Discussion was an instantaneous structure at ∼30 ns from a simulation that was started with the manually built model. For each replicate simulation, the same starting structure was assigned new random velocities and reequilibrated for 20 ps under constant number, temperature, and pressure conditions with harmonic restraints before production simulation as described above. Hydrogen bond distances (amide nitrogen to carbonyl oxygen) and helix formation were calculated with the cpptraj module of Amber 14.
Protein purification and reconstitution in vesicles.
A construct of human EGFR spanning the transmembrane segment and intracellular module (TM-ICM; residues 618 to 1186) was cloned into the pFastBac HT vector (Invitrogen) using the NcoI and HindIII restriction sites. The construct also includes an N-terminal 6×His tag, a TEV protease cleavage site, and three linker lysine residues to assist membrane insertion. These components are followed by the transmembrane, juxtamembrane, and kinase domains and the full-length tail. A C-terminal GB1 domain (B1 immunoglobulin-binding domain of streptococcal protein G residues 1 to 56) (37) is added to prevent protein degradation.
Protein was expressed using a recombinant baculovirus expression system in Sf9 cells. The cells were lysed by a French press after the cells were resuspended in lysis buffer (50 mM Tris [pH 8], 5% glycerol, 5 mM β-mercaptoethanol supplemented with protease inhibitor cocktail [Roche]). Cell debris was cleared by centrifugation at 12,000 rpm for 30 min. The membrane fraction was isolated by centrifuging the supernatant at 44,000 rpm for 2 h. The isolated membrane pellet was redissolved in membrane solubilization buffer (50 mM Tris [pH 8], 20% glycerol, 5 mM β-mercaptoethanol, 4% Triton X-100, 0.5% n-dodecyl-β-d-maltopyranoside [DDM], 0.5 M NaCl, 20 mM imidazole supplemented with protease inhibitor cocktail [Roche]) by gentle agitation for 4 h at 4°C. The solubilized membrane fraction was incubated with nickel-nitrilotriacetic acid (Ni-NTA) beads (GE Healthcare) overnight at 4°C. The Ni-NTA beads were extensively washed with Ni-A buffer (50 mM Tris [pH 8], 10% glycerol, 5 mM β-mercaptoethanol, 0.03% DDM, 0.5 M NaCl, 40 mM imidazole), and the protein was eluted with 250 mM imidazole. Protein was further purified by passing though a gel filtration column preequilibrated with 50 mM Tris (pH 8), 10% glycerol, 2 mM DΤΤ, 0.03% DDM, and 50 mM NaCl. The quality of the protein fractions was assessed by SDS-PAGE, and pure fractions were pooled, concentrated to 1 to 2 mg/ml, and stored at −80°C.
Small unilamellar 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipid, Inc.) vesicles were prepared by rehydrating DOPC film in dialysis buffer (20 mM Tris [pH 7.5], 50 mM NaCl, 10% glycerol, and 1 mM DTT). The lipid mixture was resuspended to form multilamellar vesicles by vortexing followed by repeated freeze-thaw cycles. A procedure that normally yields homogenous unilamellar vesicles of approximately 100-nm diameter was employed, in which the lipid mixture was extruded through a miniextruder (Avanti Polar Lipid, Inc.). To reconstitute the EGFR TM-ICM construct in the vesicles, the protein was mixed with the extruded lipid preparation at a molar ratio of 1:1,000. The mixture was then dialyzed against the dialysis buffer for 40 h at 4°C. The lipid preparations of various constructs were examined by negative-stain electron microscopy as described in the supplemental material.
Kinetic analysis of EGFR autophosphorylation by Western blotting.
The kinetics of EGFR autophosphorylation were measured in buffer containing 20 mM Tris (pH 7.5) and 10 mM MgCl2. The protein concentration (EGFR TM-ICM incorporated into DOPC vesicles) was set at 1 μM. The reaction was started by adding 0.5 mM ATP, and the phosphorylation reaction was allowed to proceed at 20°C for different lengths of time (0, 10, 25, 40, 60, 90, and 120 s and 3, 5, 8, 10, and 12 min). The reaction was stopped by adding quenching solution (SDS-PAGE loading dye supplemented with 20 mM EDTA and 1 mM Na3VO4) and immediately boiling the resulting solution.
Samples were run on 12% SDS-polyacrylamide gels, blotted using semidry transfer (Bio-Rad), and blocked in 5% dry milk for 1 h. The blots were probed with rabbit primary antibodies directed against specific tyrosine phosphorylation sites (listed in “Flow cytometry” above) at 1:1,000 dilution overnight at 4°C. The blots were probed with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (catalog no. 7074; Cell Signaling Technology) at 1:1,000 dilution for 1 h at room temperature. For chemiluminescence detection, the enhanced chemiluminescence (ECL) reagent of GE Healthcare was used. Images were acquired using the Bio-Rad Chemidoc XRS system, and quantitative densitometry analysis was performed using ImageJ (38).
Mass spectrometry.
EGFR TM-ICM constructs phosphorylated for different lengths of time were resolved by SDS-PAGE followed by in-gel trypsin digestion and liquid chromatography coupled to mass spectrometry (LC-MS). Gel slices containing the TM-ICM construct were excised from the gel and processed according to the in-gel digestion protocol established by Shevchenko et al. (39). Desalted TM-EGFR peptide samples in 0.1% formic acid were directly injected, onto a reversed-phase capillary column (75 μm by 130 mm) packed in-house with Magic C18AQ stationary phase (Michrom Bioresources, Auburn, CA) at 1 μl/min with 98% mobile phase A (0.1% formic acid in water) for 12 min before eluting at 300 nl/min over a 1-h gradient from 2% to 60% mobile phase B (0.1% formic acid in acetonitrile). Peptides were eluted and detected by electrospray ionization at 2.5 kV into an LTQ-OrbitrapXL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The mass spectrometer duty cycle consisted of seven scan events: a precursor ion scan from 300 to 2,000 m/z with 60,000 resolving power (at m/z = 400) followed by six data-dependent scans in the ion trap isolating and fragmenting the six most abundant precursor ions from scan 1. The minimum signal for precursor selection was 500, and undefined and 1+ charge states were excluded. Dynamic exclusion was enabled for precursors with a repeat count of 2 within a 12-s duration. Up to 500 precursors were excluded at any given time with an exclusion duration of 30 s. Precursor ions were isolated within a 2 m/z window and fragmented at a normal collision energy of 35 or 30 ms at an isolation q of 0.25. Mass spectrometry data in RAW format were searched using the SEQUEST algorithm within Proteome Discoverer 1.1 (Thermo) against a FASTA formatted database containing the intracellular domain of EGFR, with background sequences consisting of the Escherichia coli proteome. Search parameters consisted of a precursor mass tolerance of ±10 ppm and a fragment ion tolerance of 0.8 Da. Variable modifications were allowed for oxidation of methionine (+15.995 Da), asparagine/glutamine deamidation (+0.984 Da), and serine/threonine/tyrosine phosphorylation (+79.996 Da) with a fixed modification for cysteine carbamidomethylation (+57.021 Da). The relative extents of tyrosine phosphorylation for each identified site were determined using the integrated peak areas of the precursor ions' extracted ion chromatograms as follows: ExtentpY =AreapY/(AreaUnmodified + AreapY), where ExtentpY is the extent of tyrosine phosphorylation (pY), AreapY is the area of pY, and AreaUnmodified is the area of unphosphorylated tyrosines.
Dynamic light scattering.
Dynamic light scattering of TM-EGFR was performed on a DynoPro instrument from Wyatt Technology Corporation, Santa Barbara, CA. Measurements were acquired at 25°C in the presence of 2 mM ATP or ADP and 5 mM MgSO4. Each data point is an average of 10 measurements.
Stochastic simulations.
EGFR autophosphorylation was simulated using a continuous-time Monte Carlo method (Gillespie algorithm). In brief, the system is described by a series of unimolecular elementary reactions and bimolecular elementary reactions. At time t = t0, all the reaction rates (Gillespie rate) r are calculated based on the law of mass action, using the absolute molecule number N as the mass descriptor. For a unimolecular elementary reaction involving reactant A, the law of mass action states the following:
| (1) |
where [A] indicates the concentration of A and [A] = NA/V, in which NA is the absolute number of molecule A and V is the effective reaction volume. Equation 1 can be rewritten as follows:
| (2) |
where rg and kg denote the Gillespie rate and Gillespie rate constant, respectively. Similarly, for a bimolecular elementary reaction involving reactants A and B, rg can be obtained by the following:
| (3) |
Next, the algorithm selects the time point for the next reaction event. Because the time between the reaction events follows a Poisson process (assuming reactions occur continuously and independently), a random time Δt is then generated by an exponential probability distribution function (PDF) with a mean value the inverse of the total rate R:
| (4) |
At time t = t0 + Δt, a second random number γ is generated by a uniform distribution function in the range [0, R], and the reaction i is chosen to happen for which .
The process iterates until t + Δt > T, where T is the total reaction time.
Protein structure accession number.
The atomic coordinates and structure factors of V924R 658–998 EGFR crystals and I682Q 672–1018 EGFR crystals have been deposited in Protein Data Bank under PDB accession no. 5CNO and 5CNN, respectively.
RESULTS AND DISCUSSION
The EGFR tail plays dual roles in regulating autophosphorylation.
We began by studying two constructs of EGFR in which large segments of the tail were deleted. One construct, named Δ(vIVb) for EGFR variant vIVb, has a deletion of a proximal segment spanning residues 958 to 1030 (see Fig. 1B for schematic representations of the constructs used in this work). The other, denoted Δ(999–1186), has a deletion of the distal residues 999 to 1186, retaining only the portion that is visualized in the published crystal structures (residues 958 to 998).
As expected based on the earlier analysis by Pines and colleagues (22), the Δ(vIVb) construct shows higher levels of phosphorylation for two tyrosine residues in the tail that we monitored, Tyr 1068 and Tyr 1173 (Fig. 2A; see Fig. S2 in the supplemental material for an analysis of the specificity of the antibodies used in this work). We also monitored phosphorylation of the tyrosine residue located in the activation loop, Tyr 845 (Fig. 2C). Here, too, the Δ(vIVb) construct shows a level of activity in the absence of EGF that is comparable to that of the wild-type receptor in the presence of EGF.
FIG 2.
Effects of deletions in the EGFR tail on EGFR phosphorylation. (A) The vIVb deletion in the proximal region of the EGFR tail enhances autophosphorylation on Tyr 1068 and Tyr 1173, even in the absence of EGF stimulation. The top panels represent the whole data set (A.U., arbitrary unit). Data for the wild type (WT) and deletion mutants are shown. Phosphorylation levels with EGF stimulation (+) and without EGF (−) stimulation are shown. The bar graphs in the bottom panels present the data from cells expressing intermediate levels of EGFR. Phosphorylation levels are normalized to the unstimulated, wild-type EGFR-expressing cells. (B) Same as in panel A, but showing data for cells expressing the Δ(999–1186) EGFR mutant with a deletion in the distal region of the tail. The levels of phosphorylation detected for Tyr 974 and Tyr 992 are substantially lower than those for the wild type. (C) Effects of the same deletion constructs as in panels A and B on the activation loop tyrosine (Tyr 845). The vIVb deletion in the proximal region leads to enhanced phosphorylation, whereas deletion of the distal region of the tail is necessary for activation loop tyrosine phosphorylation. Data are presented as in panels A and B.
A quite different, and unexpected, result is obtained when looking at phosphorylation of the Δ(999–1186) construct. In contrast to the enhanced phosphorylation seen with the proximal deletion, when the 999–1186 segment of the tail is deleted, the levels of phosphorylation of the remaining tyrosine residues are reduced substantially. We monitored the phosphorylation of two of the remaining tyrosine residues in the tail, Tyr 974 and Tyr 992 (Fig. 2B). For both residues, the phosphorylation levels in the deletion mutant are substantially lower than for the wild-type receptor, both with and without EGF. The effect is particularly marked for the activation loop tyrosine (Tyr 845), for which phosphorylation is almost completely suppressed in the Δ(999–1186) mutant (Fig. 2C).
The autoinhibitory effect of the proximal segment of the tail is distributed over the entire proximal region and involves both the activator kinase and the receiver kinase.
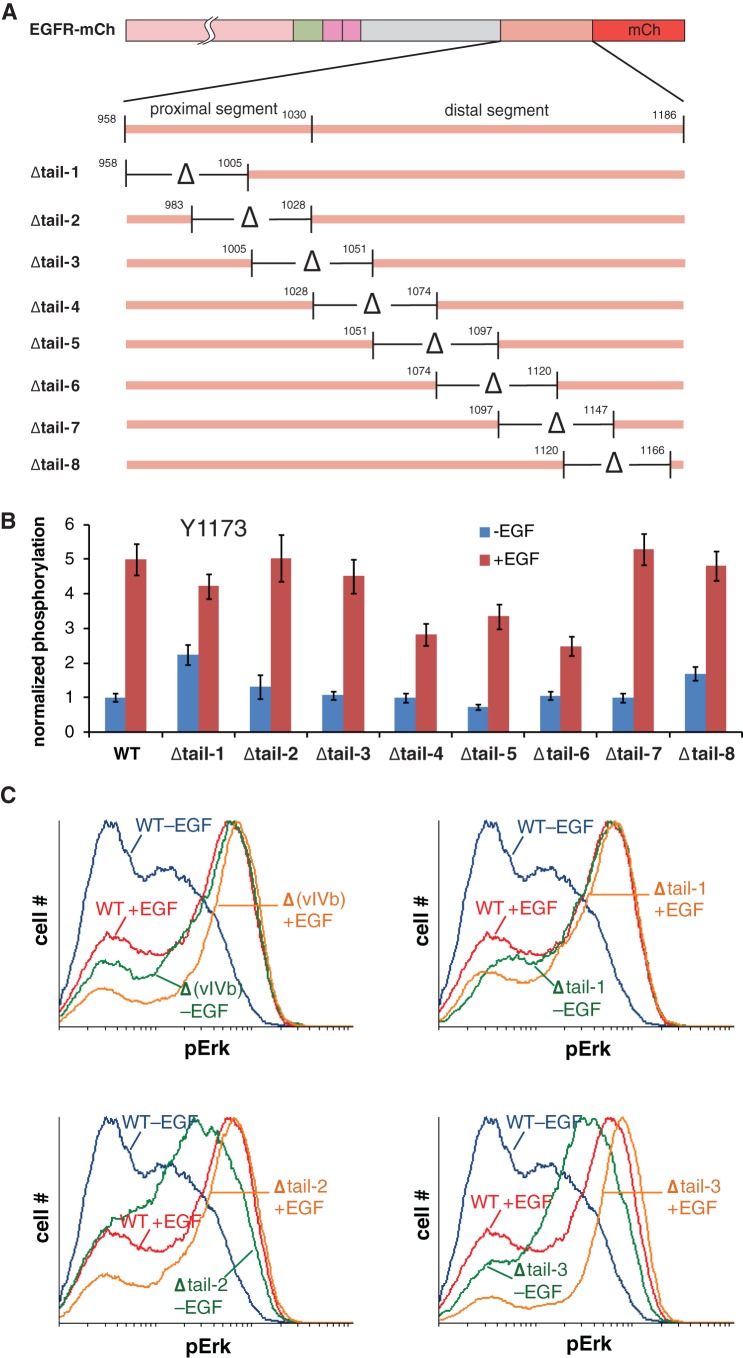
To better delineate the effects of different regions of the tail, we made eight EGFR deletion constructs, with an ∼50-residue region of the tail deleted in each of the constructs. These constructs are named Δtail-1 to Δtail-8, and the deletions are nested so that ∼25 residues overlap between adjacent deletions (Fig. 3A). To compare the effects of these deletions, we monitored phosphorylation on Tyr 1173, which is present in all of the constructs. For ease of presentation, we show bar graphs in which the phosphorylation levels in the middle range of EGFR expression in the flow cytometry data are presented (flow cytometry data as a function of EGFR expression level are shown in Fig. S3 in the supplemental material). As can be seen in Fig. 3B, none of the tail deletions leads to a dramatic change in the level of phosphorylation for Tyr 1173. In particular, the three individual deletions that span the proximal or vIVb segment (Δtail-1 to Δtail-3) have a smaller effect or no detectable effect on Tyr 1173 phosphorylation compared to the vIVb deletion.
FIG 3.
An autoinhibitory function of the EGFR C-terminal tail maps to the entire proximal region deleted in the vIVb mutant. (A) Illustration of overlapping deletion mutants scanning the EGFR C-terminal tail. (B) Flow cytometry analysis of Tyr 1173 phosphorylation with EGFR tail mutants, with and without EGF stimulation. The analysis was performed as described in the legend to Fig. 2, and phosphorylation levels are normalized to unstimulated, wild-type EGFR-expressing cells. (C) Flow cytometry analysis of phospho-Erk1/2 (pErk) in cells expressing the vIVb deletion mutant [Δ(vIVb)] or selected deletion mutants depicted in panel A. Histograms of pErk signal are shown for cells expressing moderate amounts of EGFR constructs, with or without EGF stimulation. The data for each mutant are separately plotted, overlaid on the data for wild-type EGFR.
These data suggest that each of the three regions of the tail corresponding to the deletions in the constructs Δtail-1 to Δtail-3 make some contribution individually to autoinhibition, so that deleting one region and not the others still leaves a degree of autoinhibition intact. In order to detect the effect on activation of the smaller deletions, we turned to a more sensitive assay, which is to measure the output of the mitogen-activated protein (MAP) kinase pathway in terms of the accumulation of phosphorylated Erk (pErk) by flow cytometry (Fig. 3C; see Fig. S4 in the supplemental material). The MAP kinase pathway, which is activated by EGFR through the Grb2/SOS-dependent activation of Ras, behaves like an ultrasensitive switch (40), and is therefore expected to be more responsive to small changes in the levels of EGFR phosphorylation.
We stimulated the cells with EGF as we did before and then stained for phosphorylated Erk using an antibody that detects phosphorylation of Erk1/2 (see Materials and Methods for antibody details), followed by flow cytometry. For these experiments, we pooled data from cells expressing medium to high levels of EGFR and generated histograms that show the distribution of pErk levels in the population of cells (more-detailed plots of these data are provided in Fig. S4 in the supplemental material). A comparison of data for the wild-type receptor and the vIVb variant is shown in Fig. 3C. For the wild-type receptor, there is a marked shift of the pErk distribution from low levels to high levels upon stimulation with EGF. In contrast, for the Δ(vIVb) construct, the pErk distribution without EGF stimulation looks similar to the distribution for the wild-type receptor after EGF stimulation. These results are consistent with the data for EGFR phosphorylation shown in Fig. 2.
The important result from the analysis of pErk levels is that each of the smaller deletions within the proximal segment of the tail are markedly activating (Fig. 3C). Results for the Δtail-1 construct, in which residues 958 to 1005 are deleted, are quite similar to those for the full vIVb deletion in this assay. Smaller, but clearly activating, effects are also seen for the Δtail-2 and Δtail-3 constructs, in which residues 983 to 1028 and residues 1005 to 1051 are deleted, respectively. We conclude, therefore, that the inhibitory effect of the proximal part of the tail is distributed over the whole region, but with the most important effects coming from the most proximal region, spanning residues 958 to 1005.
As EGFR requires the formation of an asymmetric dimer for activation, we wondered whether the inhibitory effect of the proximal region would be manifested on both the receiver tail and activator tail. To test this, we made receiver-impaired and activator-impaired EGFR constructs, the former with an I682Q mutation that blocks docking of the activator and the latter with a V924R mutation that disrupts the activator interface. Although both constructs are inactive on their own, when they are cotransfected, they form asymmetric dimers and respond to EGF (12). The receiver-impaired construct (I682Q) is tagged with Cerulean, and the activator-impaired one (V924R) is tagged with mCherry, allowing the expression levels of both proteins to be monitored separately. We then made a further modification to each construct and introduced the vIVb deletion in each tail. We cotransfected these two constructs and their variants into HEK-293T cells as before and monitored phosphorylation on Tyr 845 (Fig. 4A) and pErk levels (Fig. 4B). By combining receiver-impaired and activator-impaired constructs with the tail on one and not the other, we were able to assess whether the vIVb deletion is required on both the activator and receiver or whether its presence on either one suffices for activation.
FIG 4.
The vIVb deletion increases EGFR phosphorylation only when present in both the activator and receiver of the active asymmetric dimer. (A) Flow cytometry analysis of activation loop phosphorylation using cotransfected EGFR mutants. Pairs of EGFR constructs consisting of activator-impaired, mCherry-tagged EGFR, and receiver-impaired, Cerulean-tagged EGFR were cotransfected into HEK-293T cells. After EGF stimulation, cells were analyzed for mCherry, Cerulean, and FITC fluorescence, reflecting the expression levels of each construct and antiphosphotyrosine staining for Tyr 845. Data were binned according to mCherry and Cerulean intensity and represented as a two-dimensional histogram, with the color of each bin corresponding to the intensity of phosphotyrosine staining for cells within that bin. Phosphorylation at Tyr 845 is compared between EGFR pairs with none (wild type), one, or both dimer partners containing the vIVb deletion [Δ(vIVb), X symbol], as indicated. (B) Flow cytometry analysis of phospho-Erk1/2 (pErk) in cells expressing mutant EGFR asymmetric dimer pairs. Cells containing the pairs of constructs shown in panel A were treated with EGF or without EGF and stained for pErk. Each Δ(vIVb) combination (deletion in the activator-impaired construct, receiver-impaired construct, or both) is plotted separately and overlaid on the data for the intact-tail pair (wild type).
Examination of two-dimensional histograms of the phosphorylation level of the activation loop tyrosine (Tyr 845) versus expression levels of the two constructs shows that no activation occurs if the vIVb deletion is made in only one of the two proteins in the pair (when the vIVb deletion is made in either the activator-impaired receptor or the receiver-impaired receptor but not both). In contrast, when the vIVb deletion is made in both the activator and receiver, a substantial increase in phosphorylation is observed (Fig. 4A). To confirm that the vIVb deletion is necessary on both partners in the asymmetric dimer, we examined pErk levels for these cotransfection experiments (Fig. 4B). These data confirm that the presence of an intact tail on either the activator or the receiver suffices to block activation.
An Asn-Pro-Val-Tyr (NPXY) motif spanning Tyr 1086 in the distal segment of the tail is critical for phosphorylation of Tyr 845 in the activation loop.
As noted earlier, deleting residues 999 to 1186 in the tail results in a suppression of activation loop phosphorylation (Fig. 2). We monitored the effects of overlapping deletions in the tail on the phosphorylation of Tyr 845 (Fig. 5A). The most striking result is that the suppression of Tyr 845 phosphorylation is recapitulated by just two deletion constructs, Δtail-5 (with residues 1051 to 1097 deleted) and Δtail-6 (with residues 1074 to 1120 deleted) (Fig. 5A). Importantly, neither Δtail-4 (with residues 1028 to 1074 deleted) nor Δtail-7 (with residues 1097 to 1147 deleted) show substantial suppression of Tyr 845 phosphorylation.
FIG 5.
The NPXY motif encompassing Tyr 1086 of EGFR is required for Tyr 845 phosphorylation in HEK-293T cells. (A) Flow cytometry analysis of Tyr 845 phosphorylation with EGFR tail deletion mutants, with and without EGF stimulation. Phosphorylation at Tyr 845 is greatly diminished relative to wild-type EGFR in constructs lacking residues 1051 to 1097 (Δtail-5) or residues 1074 to 1120 (Δtail-6). (B) Flow cytometry analysis of Tyr 845 phosphorylation with EGFR mutated either at Tyr 1086 or with the deletion of residues 1083 to 1086 (ΔNPXY). (C) Western blot analysis of Tyr 845 phosphorylation with and without EGF stimulation for Y1086A and ΔNPXY mutants. Mutants lacking an intact phosphotyrosine recognition motif at Tyr 1086 have greatly diminished phosphorylation at Tyr 845. αEGFR, anti-EGFR antibody; αpY845, antibody against phosphorylated tyrosine at position 845. (D) Flow cytometry analysis of Tyr 974 and Tyr 992 phosphorylation for Y1086A and ΔNPXY mutants. Phosphorylation of proximal tail tyrosines is reduced by mutation of the Tyr 1086 site, but to a lesser degree than that of Tyr 845. (E) Flow cytometry analysis of Tyr 845 phosphorylation with fine-scale scanning deletion mutants. Three residues were deleted in each DS construct listed below the x axis, spanning residues 1075 to 1098. Mutants missing residues 1084 to 1086 (DS4) and 1087 to 1098 (DS5) have reduced phosphorylation at Tyr 845 relative to wild-type EGFR upon EGF stimulation.
Taken together, these data suggest that the region of the tail spanning residues 1074 to 1097 is important for phosphorylation of Tyr 845 in the activation loop. This region contains a single tyrosine residue, Tyr 1086, which is part of an NPXY motif. Indeed, a single point mutation that removes Tyr 1086 (Y1086A) or the deletion of the NPXY motif (ΔNPXY) reduces phosphorylation of Tyr 845 significantly (Fig. 5B). This striking effect can be clearly detected by Western blotting experiments as well (Fig. 5C),and can be demonstrated, although more weakly, for other proximal tyrosines such as Tyr 974 and Tyr 992 (Fig. 5D).
To analyze more finely the region of the tail surrounding Tyr 1086, we made eight deletion constructs, denoted DS1 to DS8, in which three residues at a time were deleted in the region spanning residues 1075 to 1098 (Fig. 5E). Of these, only two (DS4 and DS5, in which residues 1084 to 1086 and 1087 to 1089 are deleted, respectively) led to a suppression of activation loop phosphorylation. This localizes the critical region to the PVYHNQ sequence, flanking Tyr 1086.
The two kinases in one EGFR asymmetric dimer cannot access the activation loops of each other. Thus, phosphorylation of the activation loop of EGFR might require the formation of a higher-order oligomer of asymmetric dimers (41), as seen for the MuSK receptor (42, 43). Alternatively, when phosphorylated, Tyr 1086 might recruit a tyrosine kinase that phosphorylates the activation loops, such as a Src family kinase (44). The identity of the effector is yet to be determined and is under current investigation.
Crystal structures of activator-impaired and kinase-impaired EGFR kinase domains with portions of the proximal tail segment.
We have determined two new structures of the kinase domain of EGFR. One is a structure of the activator-impaired form with the V924R mutation and is similar to the structure we reported earlier (15) (PDB code 3GT8), except that the resolution has been improved substantially, from 2.95 Å previously to 1.55 Å in the current analysis. The crystallization construct is the same as before, with the tail extending to residue 998, but the new structure is in a different space group (C2, a = 155.4 Å, b = 71.8 Å, c = 76.4 Å and, β = 113.3°) from that of the previously reported structure (P21, a = 61.8 Å, b = 72.4 Å, c = 143.4 Å and, β = 101.7°). Despite a different crystallographic lattice, the inactive kinase domain forms a dimer very similar to the dimer reported previously, mediated primarily by interactions between the AP2 helix in the C-terminal tail in one kinase and the N-lobe of the other. The asymmetric unit of the crystal contains two molecules that are related by pseudotranslational symmetry, and each of these form an AP2 helix-mediated dimer through crystallographic symmetry (see Fig. S5A in the supplemental material). In both of the molecules, strong electron density is present in the tail until residue Glu 991.
The second structure we have determined is for a receiver-impaired construct of the kinase domain, with the I682Q mutation in the N-lobe of the kinase domain, determined at 1.9-Å resolution. The crystallization construct includes 23 additional residues from the tail and extends to Ser 1021. The crystals are in the same space group, with similar unit cell dimensions, as the structure of the V924R variant described above. As a consequence, the same dimer of inactive kinase domains is seen in this structure of the I682Q mutant as well.
The observation, discussed above, that deletion of residues 1005 to 1051 in the tail activates EGFR but that deletion of residues 1028 to 1074 does not implies that residues 1005 to 1028 span a region with an inhibitory role (Fig. 3). An obvious model for how such a peptide segment might interact with the kinase domain is provided by the structure of a fragment (residues 337 to 360) of the EGFR inhibitor Mig6, which binds to the C-lobe of EGFR and blocks the activator interface (33). Since the EGFR residues up to residue 1021 are included in the crystallization construct for the I682Q variant, we examined electron density maps for any evidence for interactions made by this region but found no features corresponding to an extension of the tail beyond residue Asp 990. However, lattice contacts in the crystals of the I682Q EGFR variant block the path that would be taken by the tail were it to mimic the binding mode of residues Ser 337 to Asn 343 of Mig6 (see Fig. S5E in the supplemental material).
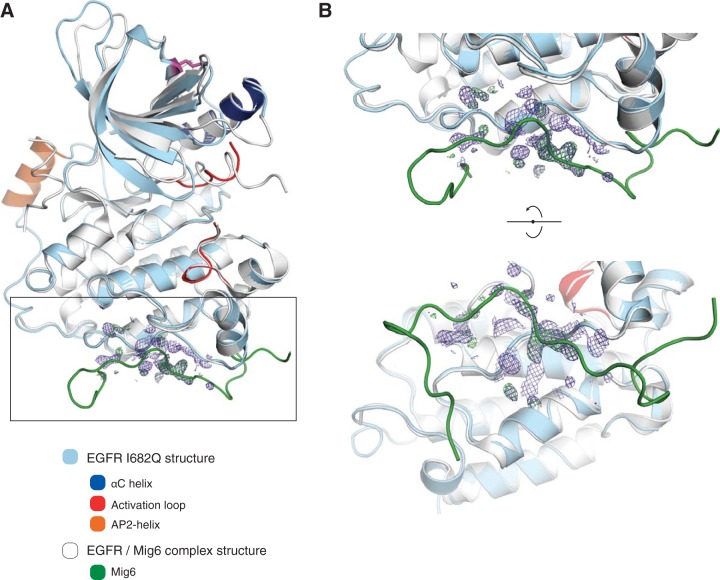
A difference electron density map calculated at a stage of the refinement when the tail had been built up to residue Asp 990 was then examined with the structure of Mig6 superimposed on the model. It was apparent that there is disconnected electron density in the map in the region where Mig6 residues 345 to 352 contact the C-lobe of the kinase domain (Fig. 6). This segment anchors Mig6 by forming a short antiparallel β sheet with a strand in the C-lobe, comprising residues Gly 906 to Arg 908, and it appears that this aspect of Mig6 is mimicked by the C-terminal tail, most likely by a conserved LLSSL segment (residues 1010 to 1014) that could establish an inhibitory interaction in cis with the C-lobe of the kinase domain. In this context of the EGFR I682Q mutant crystal lattice however, this interaction is more likely to be established in trans, due to the crystal contacts discussed above (see Fig. S5E in the supplemental material).
FIG 6.
Crystal structures of EGFR V924R and I682Q mutants. (A) Superposition of the EGFR I682Q structure here presented with the EGFR structure bound to Mig6 (PDB code 2RFE; chain A). The electron density at the base of the C-lobe of monomer A in the EGFR I682Q structure is shown, for a map calculated with the coefficients 2mFo-DFc (purple) and mFo-DFc (green positive peaks) and phases obtained from a model at the late stages of refinement. (B) Orthogonal zoomed views of the region delimited in panel A by a rectangle.
Model of the LLSSL motif in the tail docked on the C-lobe of the kinase domain.
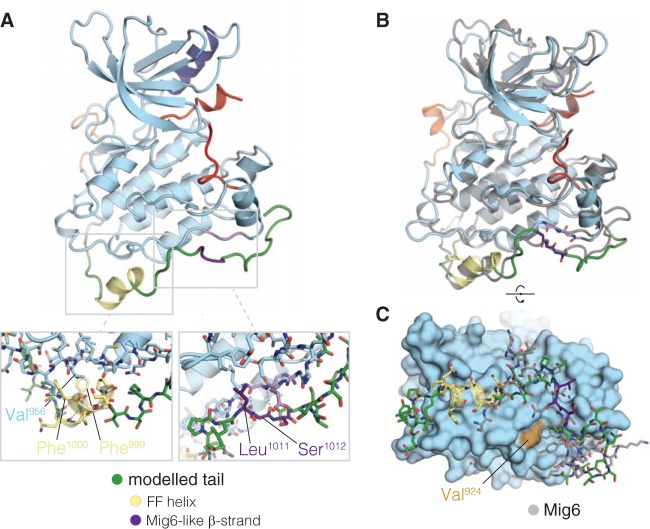
We used molecular dynamics simulations to evaluate plausible models of docking the tail segment comprising the LLSSL motif to the kinase domain (see Materials and Methods). One simulation produced a particularly compelling conformation of the tail (Fig. 7A), in which an α-helix is formed from Gln 996 to Ser 1002. In this helix, here referred to as the FF helix, residues Phe 999 and Phe 1000 pack against a small hydrophobic surface in the C-lobe centered on Val 956, and the polar side chains of Gln 996, Ser 1001, and Ser 1002 in this helix are exposed to solvent. Additional simulations were started from this model, and this helix was stable in five out of six replicate simulations and for more than 100 ns in two of these (Fig. 7B; see Fig. S6A in the supplemental material). Phe 999 and Phe 1000 remained largely buried while the helix is formed. Importantly, in all of these simulations, the Mig6-like β-sheet interaction made by the LLSSL motif was maintained for the entire length of the trajectories (Fig. 7C; Fig. S6B).
FIG 7.
Proposed model for activator interface occlusion by the C-terminal tail. (A) Molecular dynamics simulation snapshot ∼5 ps after spontaneous formation of an α-helix, including Phe 999 and Phe 1000 of the tail (FF helix). Structural features are colored as follows: α-helix C in dark blue, activation loop in red, AP2 helix in orange, α-helix comprising Phe 999 and Phe 1000 in yellow, β-sheet forming a Mig6-like interaction in purple, and other modeled tail residues (991 to 1024) in green. The left inset shows the packing of residues Phe 999 and Phe 1000 against the C-lobe, as allowed by the helical conformation of the tail. The right inset shows the details of the β-sheet interaction formed between Ser 1012 and Leu 1014 of the tail and Gly 906 and Arg 908 of the C-lobe. (B) Overlaid snapshots at 7 ps and 307 ps after formation of the FF helix. The structures from the trajectory were aligned to the backbone atoms of residues 853 to 959 of PDB 2RFE, chain A. (C) Comparison of the conformation of Mig6 and the EGFR tail model after simulation. The snapshot 307 ps after formation of the FF helix is shown with the kinase domain as a surface and the modeled tail in stick representation. Mig6 (chain E from PDB 2RFE) is shown in gray. The simulation snapshot is aligned to the crystal structure as in panel B.
This modeling exercise illustrates the possibility that residues in the tail can link the end of the crystallographic models presented above and the docking site used by Mig6. Given the potential for the proximal tail to form a stable interaction with the C-lobe during simulations, as well as the increased autophosphorylation upon deletion of this segment (Fig. 2), we propose that residues Tyr 992 to Leu 1014 of EGFR make specific docking interactions at the asymmetric dimer interface and thus contribute to autoinhibition.
To test this hypothesis, we employed the pErk assay described above (Fig. 3C; see Fig. S4 in the supplemental material) to detect the effects of mutations in the LLSSL region on receptor activation. We replaced all three leucines in the LLSSL segment with either negatively charged glutamate or polar glutamine residues, mutating it to EESSE and QQSSQ. For wild-type receptor, the pErk distribution shifts from low to high levels upon EGF stimulation (see Fig. S7A). In contrast, mutating the LLSSL segment to EESSE causes roughly half of the cells to have a higher level of pErk, in the absence of EGF (see Fig. S7A). However, there is no further shift of pErk distribution for this construct after EGF stimulation. Interestingly, substitution of leucine with glutamine in this LLSSL segment failed to activate the receptor, as shown by the similar distributions of pErk in QQSSQ and wild-type EGFR constructs (see Fig. S7B). These results are consistent with the anchorage of the LSSLL segment to the kinase domain principally by backbone interactions. We suggest that the introduction of negative charge into the segment breaks this anchorage but that the neutral substitution of leucine by glutamine does not.
Asymmetry in EGFR autophosphorylation in cells.
The formation of an asymmetric dimer of kinase domains upon activation means that the two tails in the dimer have different dispositions. To address whether this asymmetry translates into a difference in the phosphorylation of the two tails, we cotransfected cells with receiver-impaired (I682Q) and activator-impaired (V924R) EGFR variants, tagged with Cerulean and mCherry, respectively. These two variants had either the full-length tail or a short tail (up to residue 998), and we monitored phosphorylation of Tyr 1068 and Tyr 1173, which are present only in the construct with the full-length tail (Fig. 8).
FIG 8.
Tail phosphorylation in the activator and receiver kinases. (A and B) Flow cytometry analysis of Tyr 1173 phosphorylation by cotransfected EGFR mutants. mCherry- and Cerulean-tagged versions of EGFR (EGFR-mCh and EGFR-Cer, respectively) were cotransfected into Cos7 cells and were analyzed for mCherry, Cerulean, and FITC fluorescence, reflecting the expression levels of each construct and antiphosphotyrosine staining for pTyr 1173 after EGF stimulation. Data were binned according to mCherry and Cerulean intensities and represented as a two-dimensional histogram, with the color of each bin corresponding to the intensity of phosphotyrosine staining for cells within that bin. In panel A, phosphorylation at Tyr 1173 is compared between a pair of wild-type EGFR constructs (top row) and a pair of constructs consisting of activator-impaired (V924R) and receiver-impaired (I682Q) EGFR (lower left panel), as well as a pair with the Cerulean-labeled construct truncated at residue 998 (lower right panel). The activator- and receiver-impaired pair exhibit lower phosphorylation levels than the wild-type pair, and phosphorylation of the one-tailed pair depends only on the expression level of the tail-containing (and the only Tyr 1173-containing) construct. In panel B, the wild-type pair (top tow) is compared to V924R/I682Q pairs in which the tail is truncated at residue 998 in the receiver-impaired construct (tail on the receiver [lower left]) or truncated in the activator-impaired construct (tail on the activator [lower right]). The different number of bins in panels A and B reflects the difference in the number of cells that were analyzed in each set of experiments. (C) Mean phosphorylation level plotted against expression level for bins on the diagonal in panel B, reflecting cells expressing similar levels of mCherry- and mCerulean-tagged EGFR. Values are means ± standard errors of the means (SEM) (error bars). Phosphorylation at Tyr 1173 increases with expression level significantly more when the EGFR tail is on the activator than when it is on the receiver.
There is a substantial reduction in tail phosphorylation in the cotransfection experiments where activator-impaired and receiver-impaired constructs are combined, relative to the wild-type receptor (Fig. 8). There are two components to this reduction in phosphorylation. First, there is the expected decrease in phosphorylation levels when only one of the receptors has a tail (Fig. 8A). Second, even when both receptors have the full-length tail, we see a reduction in phosphorylation levels for the activator/receiver-impaired (VR/IQ) combination (Fig. 8A). This reduction might arise from scrambling of kinase dimers due to dimerization driven by the extracellular and transmembrane domains, resulting in unproductive VR/VR, IQ/IQ, and IQ/VR combinations.
Given the reduction in phosphorylation levels when the activator-impaired/receiver-impaired combinations are studied, quantification of the effects of differential tail positioning requires careful comparison of matched experiments done on the same day. We carried out such comparisons multiple times and found a small but significant preference for phosphorylation of the tail presented by the activator, compared to the tail presented by the receiver (Fig. 8B).
We wondered whether the asymmetry in tail phosphorylation arises from features in the sequence of the tail that determines the preference for activator or receiver positions. To address this, we turned to the HER3 tail, which bears no significant sequence similarity to the EGFR tail in either the proximal or distal segments (see Fig. S1C in the supplemental material). As before, we carried out experiments in which we cotransfected activator-impaired (V924R) and receiver-impaired (I682Q) EGFR constructs. We replaced the entire EGFR tail (after residue 957) with the entire HER3 tail in both the V924R and I682Q backgrounds and measured phosphorylation using an antibody that is specific for Tyr 1289 in the HER3 tail. This antibody does not detect any significant phosphorylation when cells expressing only EGFR are probed with it (see Fig. S8).
We first tested the effect of EGF on the EGFR/HER tail chimera with no mutations at the asymmetric dimer interface (that is, the kinase domain is the wild-type EGFR kinase domain). We observe an increase in phosphorylation when the EGFR/HER3 tail chimera is stimulated with EGF, showing that the EGFR kinase domain can phosphorylate the HER3 tail in these experiments, as shown previously (24) (Fig. 9). We then studied the effect of cotransfection with activator-impaired (V924R) and receiver-impaired (I682Q) constructs in which the HER3 tail is on one kinase or the other kinase. We observed a higher level of phosphorylation when the HER3 tail is in the activator position (Fig. 9).
FIG 9.
An EGFR-Her3 tail chimera produces higher phosphorylation levels when it takes the activator position in an asymmetric dimer. (A) Schematic illustrating the combinations of constructs shown in panels B and C. Pairs of constructs consisting of EGFR (residues 1 to 957) and the EGFR or Her3 C-terminal tail were cotransfected. The Her3 tail-containing constructs were constrained to take the activator or receiver position with the I682Q or V924R mutation, respectively. (B) Phosphorylation of Her3 Y1289 analyzed by flow cytometry. Phosphorylation level with (+) or without (−) EGF is plotted versus protein expression level for bins of cells expressing similar levels of each construct (mean bin fluorescence ± SEM). (C) Relative phosphotyrosine signal for each pair of constructs for intermediate expression levels (500 to 600 mCherry fluorescence units as shown in panel B), normalized to the unstimulated EGFR-Her3 tail signal (mean ± SEM [error bars]). The pair in which the Her3 tail is attached to an activator-only EGFR kinase shows reduced phosphorylation upon EGF stimulation compared to the pair in which the Her3 tail is attached to the receiver-only kinase domain.
The results with the EGFR/HER tail chimeras show that asymmetry in tail phosphorylation does not depend on the sequence of the tail. In particular, this suggests that the interactions made by the tail in structures of active EGFR kinase domains, in which the tail runs along the distal surface of the kinase domains, may not account for the asymmetry in tail phosphorylation. The asymmetry may arise from a higher-order oligomeric form of EGFR that is generated after activation (41) in which the relative positions of the activator and receiver kinases within a dimer control their access to activated kinases in other dimers. Such a mechanism has been suggested for the phosphorylation of the tails in HER2/HER3 heterodimers (45).
Asymmetry in phosphorylation within heterodimers of EGFR family members.
There is only one kind of kinase domain in the studies reported above (that of EGFR), so asymmetry in tail phosphorylation arises solely from the position of the tail. Recent analyses of heterodimers of the EGFR family have shown that there is a preference among these family members for which kinase takes the receiver position in various heterodimers (46, 47). For example, based on Western blot data, it has been concluded that in heterodimers formed by EGFR and HER2, the HER2 kinase exhibits little or no kinase activity when it is forced to take the receiver position in a heterodimer with EGFR in the activator position. We used our flow cytometry assay to evaluate tail phosphorylation in the HER2/EGFR heterodimer.
We constrained either EGFR or HER2 to be in the activator or receiver position by using two strategies. We generated activator-impaired or receiver-impaired combinations, as described above for EGFR. In addition, we made catalytically inactive variants of EGFR and HER2 by introducing the D813N mutation (EGFR numbering), which inactivates the kinase by replacing the catalytic base (Fig. 10A). First, we monitored phosphorylation of the HER2 tail, using an antibody that specifically recognizes phosphorylated tyrosine at position 1221 (pTyr 1221) (Fig. 10B). The phosphorylation of the HER2 tail is stronger when HER2 is in the activator position, either through mutation of the activator/receiver interfaces in both EGFR and HER2, or through making a catalytically inactive form of HER2. A quite different result is obtained, however, when the same experiments are repeated but with phosphorylation monitored on the EGFR tail (pTyr 1068) (Fig. 10B). In this case, stronger phosphorylation is seen when EGFR is in the receiver position. These results can be rationalized by the previously described preference for HER2 to be in the activator position with respect to EGFR. Our results show that this combination has stronger kinase activity than the opposite one, which has EGFR in the activator position and HER2 in the receiver position (Fig. 10C).
FIG 10.
C-terminal tail phosphorylation by EGFR/Her2 heterodimers is greater when EGFR takes the receiver position. (A) Schematic illustrating the combinations of constructs shown in panels B and C. EGFR and Her2 constructs, labeled with either mCherry or Cerulean and bearing either activator-impaired, receiver-impaired, or kinase-dead mutations, were cotransfected. (B) Phosphorylation of Her2 Tyr 1221 and EGFR Tyr 1173 upon EGF stimulation analyzed by flow cytometry. Mean bin phosphorylation level for the indicated tyrosine is plotted versus receptor expression level for bins of cells expressing similar levels of each construct. (C) Phosphotyrosine signal for each pair of constructs for intermediate expression levels (500 to 600 mCherry fluorescence units), as shown in panel B (mean ± SEM [error bars]). Mutation of the catalytic aspartate in EGFR or forcing EGFR to take the activator position reduces phosphorylation significantly.
Tail phosphorylation in membrane-reconstituted TM-ICM construct of EGFR shows biphasic kinetics.
The enzymatic properties of the full-length receptor have been studied by reconstitution in detergents (48, 49), but kinetic analysis of samples with the full-length receptor embedded in lipid bilayers is biochemically intractable at present. We therefore studied a construct that omits the extracellular domain but includes the transmembrane helices and the entire intracellular module (juxtamembrane segment, kinase domain, and the intact tail). This construct, denoted TM-ICM (for transmembrane-intracellular module), was purified from baculovirus-infected insect cells, and the detergent-solubilized protein was incorporated into DOPC vesicles and used for measurements of the kinetics of autophosphorylation of several tyrosine residues in the tail. To our surprise, we discovered that the TM-ICM construct leads to a peculiar membrane-stacking phenomenon when it is reconstituted in lipid bilayers using a procedure that normally results in small unilamellar vesicles with an average diameter of 100 nm (see Fig. S10 in the supplemental material). We describe this membrane-stacking phenomenon in more detail in the supplemental material.
The rates of phosphorylation of individual tyrosine residues in a soluble EGFR construct that contains the kinase domain and the full-length tail have been measured previously, using mass spectrometry and Western blot analyses (50). These studies revealed that there is a temporal order to the phosphorylation of tail residues in this construct, with distal residues being phosphorylated faster than the proximal ones. To monitor tail phosphorylation in the membrane-reconstituted TM-ICM samples, we initiated the reaction by adding ATP and then quenched the reaction at various time points by adding SDS-PAGE buffer and boiling the samples. The quenched samples were run on SDS-polyacrylamide gels, blotted, and probed with antibodies against several phosphorylation sites on the EGFR tail (tyrosines at positions 845, 974, 992, 1068, and 1173) (Fig. 11A). We found a surprising biphasic pattern for the time dependence of phosphorylation for all the tyrosine residues that we monitored. In each case, a fast phosphorylation step that reaches a plateau after 1 to 2 min is observed, but then a second phase of increasing phosphorylation takes place, resulting in a second plateau at a higher level.
FIG 11.
Vesicle-reconstituted TM-ICM EGFR exhibits biphasic autophosphorylation kinetics. (A) Time courses of EGFR autophosphorylation by TM-ICM EGFR reconstituted into vesicles determined by Western blotting with the indicated antibodies. Integrated band intensities for five replicate experiments (values are means ± SEM [error bars]) are plotted versus time and fit to two separate single exponential functions and offset by a fixed amount of time. Images for representative blots are shown below each graph. The table at bottom right lists the half-life for the first exponential function (mean ± SEM). (B) Time courses of phosphorylation reactions as quantified by tandem mass spectrometry. The graphs show the ratio of the area under the extracted ion chromatogram for the indicated phosphorylated peptide versus the total area for phosphorylated and unphosphorylated peptides for each sample.
The increase in phosphorylation in the first phase is roughly equal to that observed for the second step, as if ∼50% of the sites are phosphorylated in the first phase and the remaining 50% in the second phase. This biphasic phosphorylation pattern is more apparent for certain tyrosine residues than for others, but close examination of the first 2 min of the reaction makes it evident that there is indeed an initial plateau value in the phosphorylation for all the residues that we monitored (Fig. 11A, insets).
We estimated the half-times for phosphorylation of each of the monitored residues by fitting the first phase with an exponential function (Fig. 11A). We also measured phosphorylation kinetics for the same set of residues using mass spectrometry, and this validated the biphasic phosphorylation pattern (Fig. 11B). Phosphorylation at site Tyr 992 was not detected robustly, but for Tyr 845, Tyr 974, Tyr 1068, and Tyr 1173, phosphorylation clearly followed a biphasic pattern. The biphasic pattern to the phosphorylation kinetics is reversible. Treatment with phosphatase removes the phosphorylation, and the biphasic pattern is recovered upon reinitiating the reaction by adding phosphatase inhibitor and ATP (see Fig. S9 in the supplemental material).
The biphasicity in the phosphorylation kinetics requires the presence of the distal portion of the tail. This is demonstrated by experiments in which we monitored tail phosphorylation for a truncated TM-ICM construct, in which the tail is truncated at residue 998. This construct forms vesicle stacks similar to those of the wild-type protein (Fig. 12A), but the time dependence of the phosphorylation does not show a biphasic pattern and can be fit by single-exponential kinetics (Fig. 12B).
FIG 12.
A short-tailed EGFR TM-ICM construct incorporated into vesicles does not exhibit biphasic autophosphorylation kinetics. (A) Negative-stained electron micrograph of vesicles reconstituted with EGFR TM-ICM truncated after residue 998 (left) and illustration of the location of phosphorylated tyrosines in this construct (right). The electron microscopy (EM) sample was prepared as for the samples shown in Fig. 11. (B) Time courses of autophosphorylation on the indicated tyrosines for the short-tail TM-ICM incorporated into vesicles, as quantified by Western blotting. Integrated band intensities for five replicate experiments (mean ± SEM) are plotted versus time and fit to a single exponential function. The time scale and reaction conditions are the same as in panel A. The reaction progress curves fit well to a single exponential decay, unlike the full-length tail constructs shown in Fig. 11.
The biphasic time dependence of the tail phosphorylation cannot be fit satisfactorily by single-exponential functions or with double-exponential functions with a common origin in time. A more satisfactory fit to the data can be obtained by using two exponential functions, with one shifted later in time. While we have not carried out an exhaustive analysis, we find that a physically reasonable explanation for the biphasic character is provided by a kinetic model in which one of the two tails in an asymmetric dimer is sequestered and cannot be phosphorylated until a “gating” phosphorylation occurs. In this simple model, we consider only two states of EGFR, phosphorylated EGFR (pEGFR) and unphosphorylated EGFR, without specifying individual phosphorylation sites. One EGFR molecule can be activated by another EGFR molecule via the formation of an asymmetric dimer (EGFR)2. A key assumption in the model that is required to generate biphasic character is that only one of the two molecules in the asymmetric dimer is phosphorylated. The dimer then needs to dissociate for the other molecule to be phosphorylated. Kinetics simulated using this simple model captures the essential features of the experimentally observed biphasic character (see Fig. S12 in the supplemental material).
Concluding remarks.
As an ∼230-residue-long disordered region, the tail of EGFR has thus far eluded definitive structural and mechanistic characterization. Here, we demonstrate that the proximal region of the tail (residues 958 to 1030) participates in autoinhibitory interactions, as first described by Pines and colleagues (22). We show that this whole region is responsible for these interactions and that they are important on both the activator kinase and the receiver kinase. On the basis of these findings and a higher-resolution crystal structure of the EGFR kinase domain, we propose that the autoinhibitory interactions made by these parts of the tail are the following. (i) The AP2 helix stabilizes an inactive dimer. (ii) The E-hook and the following β-strand block formation of the juxtamembrane latch by which the activator kinase is held in place by the juxtamembrane segment of the receiver. (iii) The segment from positions 992 to 1014 covers the C-lobe of the kinase at the asymmetric dimer interface. All of these interactions could interfere with asymmetric dimer formation.
Our deletion scanning analysis of the tail also revealed that region in the vicinity of Tyr 1086 is critical for activation loop phosphorylation. This effect is most likely due to the recruitment of an adapter to this site that results in clustering or the recruitment of a tyrosine kinase that phosphorylates the activation loop.
As the active EGFR dimer is asymmetric, another question we asked is whether this asymmetry translates to any difference in the phosphorylation of the activator or receiver tail. Our data clearly demonstrate that there is preference for phosphorylation of the activator tail, and this preference is apparent in homo- and heterodimers of EGFR as well. The asymmetry in tail phosphorylation is amplified in membrane preparations of a construct in which the extracellular module is deleted, presumably because the membrane stacks that form trap the asymmetric dimer and slow down the dissociation of the activator and receiver kinases. The asymmetry in the rate of phosphorylation of the receiver and activator tails suggests that EGFR has evolved to most efficiently phosphorylate the activator tail. This has important functional consequences for the HER2/HER3 heterodimer, in which the catalytically impaired HER3 bears the phosphorylation sites with the strongest impact on cellular signaling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael L. Gross and Henry Rohrs for mass spectrometry instrument access and support and Hector Nolla at the Flow Cytometry Core Facility (UC Berkeley) for assistance.
Mass spectrometer instrument support was provided by the National Center for Research Resources of the NIH (grant 2P41RR000954 to M. L. Gross). Ron Bose is supported by NIH grant R01CA161001. T.S.C. is supported by NIH T32 training grant 2T32HL007088-36. This work was partially supported by a grant from the National Cancer Institute to J.K. (grant 2R01CA09650406).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00248-15.
REFERENCES
- 1.Arteaga CL, Engelman JA. 2014. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. 2015. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem 84:739–764. doi: 10.1146/annurev-biochem-060614-034402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. 2001. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Endres NF, Barros T, Cantor AJ, Kuriyan J. 2014. Emerging concepts in the regulation of the EGF receptor and other receptor tyrosine kinases. Trends Biochem Sci 39:437–446. doi: 10.1016/j.tibs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon MA, Schlessinger J, Ferguson KM. 2014. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol 6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner MJ, Stacey MM, Liu BA, Pawson T. 2013. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb Perspect Biol 5:a008987. doi: 10.1101/cshperspect.a008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. 2012. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 8.Leahy DJ. 2004. Structure and function of the epidermal growth factor (EGF/ErbB) family of receptors. Adv Protein Chem 68:1–27. doi: 10.1016/S0065-3233(04)68001-6. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. 2003. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell 11:507–517. doi: 10.1016/S1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 10.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. 2002. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell 110:763–773. doi: 10.1016/S0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 11.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. 2002. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110:775–787. doi: 10.1016/S0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. 2013. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. 2009. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell 34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. 2009. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. 2013. Architecture and membrane interactions of the EGF receptor. Cell 152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. 2012. Bipartite tetracysteine display reveals allosteric control of ligand-specific EGFR activation. ACS Chem Biol 7:1367–1376. doi: 10.1021/cb300216f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. 2008. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem 283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. 2009. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys J 96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mineev KS, Bocharov EV, Pustovalova YE, Bocharova OV, Chupin VV, Arseniev AS. 2010. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J Mol Biol 400:231–243. doi: 10.1016/j.jmb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Dankort DL, Wang Z, Blackmore V, Moran MF, Muller WJ. 1997. Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol Cell Biol 17:5410–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pines G, Huang PH, Zwang Y, White FM, Yarden Y. 2010. EGFRvIV: a previously uncharacterized oncogenic mutant reveals a kinase autoinhibitory mechanism. Oncogene 29:5850–5860. doi: 10.1038/onc.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorkin A, Mazzotti M, Sorkina T, Scotto L, Beguinot L. 1996. Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem 271:13377–13384. doi: 10.1074/jbc.271.23.13377. [DOI] [PubMed] [Google Scholar]
- 24.Bublil EM, Pines G, Patel G, Fruhwirth G, Ng T, Yarden Y. 2010. Kinase-mediated quasi-dimers of EGFR. FASEB J 24:4744–4755. doi: 10.1096/fj.10-166199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koushik SV, Chen H, Thaler C, Puhl HL III, Vogel SS. 2006. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys J 91:L99–L101. doi: 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Part A 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. 2005. The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. 2007. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- 35.Ryckaert JP, Ciccotti G, Berendsen HJC. 1977. Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Physics 23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 36.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 37.Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. 1997. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci 6:2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 40.Huang CY, Ferrell JE Jr. 1996. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A 93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. 2008. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors 26:316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- 42.Bergamin E, Hallock PT, Burden SJ, Hubbard SR. 2010. The cytoplasmic adaptor protein Dok7 activates the receptor tyrosine kinase MuSK via dimerization. Mol Cell 39:100–109. doi: 10.1016/j.molcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbst R, Burden SJ. 2000. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J 19:67–77. doi: 10.1093/emboj/19.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tice DA, Biscardi JS, Nickles AL, Parsons SJ. 1999. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci U S A 96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Park E, Kani K, Landgraf R. 2012. Functional isolation of activated and unilaterally phosphorylated heterodimers of ERBB2 and ERBB3 as scaffolds in ligand-dependent signaling. Proc Natl Acad Sci U S A 109:13237–13242. doi: 10.1073/pnas.1200105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. 2012. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci U S A 109:137–142. doi: 10.1073/pnas.1111316109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward MD, Leahy DJ. 2015. Kinase activator-receiver preference in ErbB heterodimers is determined by intracellular regions and is not coupled to extracellular asymmetry. J Biol Chem 290:1570–1579. doi: 10.1074/jbc.M114.612085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. 2011. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol 18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA. 2011. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol 18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y, Li Z, Apetri M, Luo B, Settleman JE, Anderson KS. 2012. Temporal resolution of autophosphorylation for normal and oncogenic forms of EGFR and differential effects of gefitinib. Biochemistry 51:5212–5222. doi: 10.1021/bi300476v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.