Abstract
The classic nonhomologous end-joining (c-NHEJ) pathway is largely responsible for repairing double-strand breaks (DSBs) in mammalian cells. XLF stimulates the XRCC4/DNA ligase IV complex by an unknown mechanism. XLF interacts with XRCC4 to form filaments of alternating XRCC4 and XLF dimers that bridge DNA ends in vitro, providing a mechanism by which XLF might stimulate ligation. Here, we characterize two XLF mutants that do not interact with XRCC4 and cannot form filaments or bridge DNA in vitro. One mutant is fully sufficient in stimulating ligation by XRCC4/Lig4 in vitro; the other is not. This separation-of-function mutant (which must function as an XLF homodimer) fully complements the c-NHEJ deficits of some XLF-deficient cell strains but not others, suggesting a variable requirement for XRCC4/XLF interaction in living cells. To determine whether the lack of XRCC4/XLF interaction (and potential bridging) can be compensated for by other factors, candidate repair factors were disrupted in XLF- or XRCC4-deficient cells. The loss of either ATM or the newly described XRCC4/XLF-like factor, PAXX, accentuates the requirement for XLF. However, in the case of ATM/XLF loss (but not PAXX/XLF loss), this reflects a greater requirement for XRCC4/XLF interaction.
INTRODUCTION
The last bona fide classic nonhomologous end-joining (c-NHEJ) factor discovered was XLF (XRCC4-like factor) or Cernunnos, discovered by two laboratories in ∼2006 (1–3). Although XLF lacks primary sequence homology with XRCC4 (X-ray cross complementing 4), it shares remarkable structural similarity with it, including a globular head, a C-terminal helix, and an unstructured C-terminal tail (4, 5). XLF dimers can interact with XRCC4 dimers, and structural studies reveal an extended filament composed of alternating XRCC4/XLF dimers (6–9). These filaments bridge and stabilize DNA ends in vitro, implicating XRCC4/XLF in bridging broken ends prior to repair (4, 6). Invoking a bridging function for XLF would be attractive, because XLF promotes the ligation of mismatched ends, an activity that might require a factor to hold DNA ends together while end processing progressed (10–13). Alternatively, XLF's ability to stimulate ligation might be associated with its ability to stimulate adenylation of Lig4 (14).
Patients with XLF mutations have microcephaly, growth retardation, and progressive lymphopenia and are radiosensitive and immunodeficient (1, 15). Additionally, cells from these patients display defective responses to DNA replication stress (16). In contrast, XLF-deficient mice are immunocompetent and have no neurologic phenotype or growth retardation; however, different cell types from these mice are impacted differently by XLF deficiency (17, 18). For example, the progressive lymphopenia observed in XLF−/− patients can be recapitulated in ageing XLF−/− mice and is explained by a failure of hematopoietic stem cells (19). Alt and colleagues have observed that mice deficient in both XLF and either ATM (ataxia telangiectasia mutated), H2AX (histone 2A variant X), 53BP1 (p53 binding protein 1), or DNA-dependent protein kinase catalytic subunit (DNA-PKcs) have much more profound deficits than animals deficient in just XLF or any one of these other factors alone (20–24). Importantly, some of these factors have been suggested to have functional roles in bridging or synapsing DNA ends. Thus, these findings provide a potential explanation for why different cell types have different requirements for XLF.
Here, we characterize two XLF mutants that cannot form filaments with XRCC4 in vitro. One of these mutants still is proficient in stimulating XRCC4/Lig4 activity in vitro while the other is not. We find that all cell types tested are dependent on XLF's ability to stimulate XRCC4/Lig4; however, only certain cell types are dependent on XLF's stable interaction with XRCC4 (and potential DNA end bridging). Cells that lack XLF or express an XLF that cannot interact with XRCC4 display defective DNA-PK autophosphorylation, suggesting that at least one function of XRCC4/XLF complexes in living cells is to stabilize DNA-PK at synapsed DNA ends. Loss of either ATM or the newly described XRCC4/XLF-like factor, PAXX (paralog of XRCC4 and XLF) (25, 26), accentuates the cellular requirement for XLF. However, in the case of ATM/XLF loss (but not PAXX/XLF loss), this reflects a greater requirement for XRCC4/XLF stable interaction. Intriguingly, disruption of XLF in XRCC4-deficient cells enhances cellular sensitivity to DNA-damaging agents, suggesting a function for XLF that is independent of XRCC4. Finally, as with other c-NHEJ factors (27) (28), some XLF-deficient cells display increased sensitivity to replication stress; surprisingly, neither XLF's ability to stimulate XRCC4/Lig4 nor its ability to stably interact with XRCC4 is absolutely required for XLF in abating replication stress. These data suggest that XLF has multiple functions in DNA repair, and they offer potential explanations for the pleiotropic phenotypes associated with XLF deficiency.
MATERIALS AND METHODS
Plasmids.
The expression constructs for wild-type (wt) and mutant XRCC4 have been described (29). Wild-type and mutant XLF cDNAs were cloned into the pMSCV-neo vector plasmid (Clontech Laboratories, Palo Alto, CA). Rag1 and Rag2 expression plasmids were a gift from David Roth. Construction of the I-SceI expression plasmid was described already (30). The PAXX (C9orf142) expression construct was generated by PCR using IMAGE clone 3161564 as a template, engineering an NcoI restriction enzyme site at the ATG and an XhoI site after the STOP codon, and subcloned into the NcoI-XhoI sites of pHIS parallel I. The sequence is identical to that reported recently (26). DNA sequence analysis indicated that the sequence was identical to that of UniProt entry Q9BUH6. The I-SceI-trex2 fusion expression plasmid was obtained from Jeremy Stark. Fluorescent VDJ substrate plasmids have been described by others (J. Neal and K. Meek, unpublished data). Briefly, the red fluorescent protein (RFP) coding sequence was inserted between 12RSS and 23RSS (where RSS indicates recombination signal sequence), replacing the oop transcription terminator in both pJH290 and pJH289. The recombination cassette was cloned upstream of the cyan fluorescent protein (CFP) coding sequence in peCFP-N1 (which includes the simian virus 40 [SV40] origin of replication). In these plasmids, 12RSS and 23RSS are flanked by SalI and BamHI restriction enzyme sites, respectively. RSS with the unique coding/RSS junctures depicted were generated by inserting the following oligonucleotides (and their complements) into those sites: 12RSS, CTAGCCCGCCCCACAGTGCTACAGACTGGAACAAAAACCCTGCAG; 23RSS mismatch, GATCCCACGGGTTTTTGTACAGCCAGACAGTGGAGTACTACCACTGTGTTTATTTA; 23 RSS match, GATCCCACGGGTTTTTGTACAGCCAGACAGTGGAGTACTACCACTGTGGGGCGGGA.
Proteins.
Recombinant XRCC4, XLF, XRCC4/Lig4, and Ku were obtained using procedures described previously (4, 31–33). PAXX was produced in Escherichia coli Rosetta/pLysS cells and purified by nickel-nitrilotriacetic acid (Ni-NTA), followed by Sepharose Q chromatography using the same method as that for XRCC4 purification (31). Bridging assays were performed as described previously (6).
Electrophoretic mobility shift assay (EMSA).
The DNA substrate was obtained by annealing the oligonucleotide 5′-GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC to its reverse complement strand. DNA (25 nM final concentration) and the indicated final concentrations of proteins were mixed in a 10-μl reaction adjusted to 75 mM KCl, 10 mM Tris, pH 7.5, 0.5 mM EDTA, 0.5 mM dithiothreitol (DTT), and 5% glycerol. After incubation for >1 h at room temperature, reaction mixtures were fractionated by 6% PAGE in Tris-borate-EDTA buffer, followed by staining with ethidium bromide. Gel images were collected with a ChemiDoc XRS+ system and processed with the Image Lab software (Bio-Rad).
Ligation assays.
Reaction mixtures (10 μl) contained 100 ng of linearized pUC19 plasmid (digested with XbaI for cohesive-end ligation or with SmaI for blunt-end ligation), 2 mM MgCl2, 1 mM ATP, 75 mM KCl, 10 mM HEPES, pH 8.0, 0.5 mM EDTA, 5% glycerol, and the indicated final concentrations of proteins. T4 DNA ligase (New England BioLabs) was used at a final concentration of 8 U/μl. After a 30-min incubation at room temperature, the samples were deproteinized by the addition of pronase (1.25 μg/μl final concentration) and Sarkosyl (1.25% final concentration) and incubated at 55°C for 30 min. The reaction mixtures were fractionated by agarose gel electrophoresis using Tris-borate-EDTA buffer and stained with ethidium bromide. Gel images were acquired as indicated for the EMSAs.
Cell culture and cell strains.
293, HCT116, and 2BN cells were cultured in Dulbecco's modified Eagle medium (Life Technologies) supplemented with 10% fetal bovine serum (Atlanta Biologicals, GA), 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin (Life Technologies), and 10 μg/ml ciprofloxacin. Cells were maintained at 37°C with 5% CO2. Embryonic stem (ES) cells were cultured in the medium described above further supplemented with β-mercaptoethanol and 103 U/ml ESGRO (Millipore). Abl pre-B cells were grown in RPMI medium (Life Technologies) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin.
For the generation of cell strains stably expressing wild-type and mutant constructs of XLF in 293 cells, 5 μg of plasmid DNA was transfected with polyethyleneimine (PEI; 1 μg/ml; Polysciences) at 2 μl/1 μg DNA. Forty-eight hours posttransfection, cells were selected with medium containing 800 μg/ml G418 (Life Technologies), and single clones were analyzed for expression by immunoblotting. Similarly, ES and 2BN cells were transfected using Lipofectamine (Invitrogen) and FuGENE (Roche), respectively, according to the manufacturers' instructions. Stable XLF expression in HCT116 cells and Abl pre-B cells was achieved by retroviral transduction. Briefly, viruses were prepared by transient transfection of 1 μg of a pMSCV-XLF construct and 1 μg of helper plasmids (pCGP pVSVG ratio, 4:1) in 293T cells. Forty-eight hours after transfection, the medium was collected and passed through a 0.45-μm filter to remove cellular debris. The filtrate containing viral particles was treated with 20 mM HEPES and 8 μg/ml Polybrene (Sigma) for 10 min. The medium containing viruses then was laid over a monolayer of HCT116 cells for 4 h and then replaced with complete medium. Forty-eight hours after infection, cells were selected with medium containing 800 μg/ml G418. Similarly, Abl pre-B cells were plated at a concentration of 105 cells/100 ml medium in 24-well plates. One milliliter of viral supernatant was added to each well and spun at 1,800 rpm for 90 min at room temperature. After centrifugation, the medium was replaced with fresh complete medium. Forty-eight hours after infection, cells were selected with 2 mg/ml G418 at a concentration of 1,000 cells/100 μl medium in a 96-well plate. Stable transfectants were screened by immunoblotting.
Immunoblot analyses.
Immunoblotting was performed as described previously (29). Antibodies used in this study are rabbit polyclonal anti-XLF (Abcam), rabbit polyclonal anti-XRCC4 (Abcam), goat anti-C9ORF142 (Santa Cruz Biotechnology) for PAXX, rabbit anti-ATM (Serotec), and rabbit polyclonal anti-DNA-PKcs-phospho-S2056 (Abcam). The DNA-PKcs antibody (42–27) was the generous gift of Tim Carter. To detect DNA-PKcs autophosphorylation, cells (plated 18 h earlier) were treated in complete media with zeocin (2 mg/ml) for 3 h. Cells were harvested and whole-cell extracts prepared for immunoblotting.
Ni-agarose pulldown.
Ten micrograms of pEF vectors expressing either the wild-type or mutant forms of C-terminal His-tagged XLF was transfected into 293 cells as described above. Forty-eight hours after transfection, cells were harvested and washed with phosphate-buffered saline (PBS). Pellets were lysed with 1 ml lysis buffer (50 mM Tris, pH 7.5, 120 mM sodium chloride, 0.5% NP-40, 1 mM sodium fluoride, 1 mM sodium orthovanadate, protease inhibitor cocktail) and rocked for 30 min on ice. Lysates were centrifuged and the supernatant used for pulldown assays. Fifty microliters of Ni-NTA–agarose beads (Qiagen) was added to 1 ml cell lysate containing 10 mM imidazole and rocked for 3 h at 4°C. Beads were collected by centrifugation and washed three times in the same buffer containing 50 mM imidazole. After washing, beads were resuspended in 30 μl 4× SDS-PAGE buffer and analyzed by immunoblotting.
Cas9-mediated gene disruption.
Cas9-targeted gene disruption was performed using methods similar to those reported by Mali et al. (34). Briefly, guide RNAs (gRNAs) specific for XLF, ATM, XRCC4, or PAXX were synthesized as 455-bp fragments (Integrated DNA Technologies). The synthesized fragments were cloned into pCR2.1 using a TOPO TA cloning kit according to the manufacturers' instructions (Life Technologies). Cells were transfected with 1 μg gRNA plasmid and 1 μg Cas9 expression plasmid (Addgene). In some cases, cells were cotransfected with 0.2 μg of pcDNA6 (Life Technologies) or pSuper-Puro to confer blasticidin or puromycin resistance. Western blotting was used to identify clones with deletions in each of these factors; in all cases, deletion also was confirmed by PCR amplification that revealed deletions at the target site. The 19-mers specific for each factor synthesized into the 455-bp fragments are the following: ATM, TCTTTCTGTGAGAAAATAC; XRCC4, CCTGCAGAAAGAAAATGAA; XLF, GGCCTGTTGATGCAGCCAT; PAXX-1, CTGGCCTTTGACCTCTCCA; PaXX-2, TGCTTCACGCCGGACAGCCT.
Survival assays.
Clonogenic survival assays were performed for ES cells, 2BN cells, and HCT116 cells. Briefly, a hundred cells were plated for each transfectant into complete medium containing the indicated dose of zeocin or hydroxyurea (HU) in 60-mm-diameter tissue culture dishes. For HU assays, cell culture medium was changed after 24 h. After 7 to 10 days, cell colonies were stained with 1% (wt/vol) crystal violet in ethanol to measure relative survival. 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) staining was performed to assess cell viability for 293 cells and Abl pre-B cells. Thirty thousand to 50,000 cells were plated in each well of a 24-well plate containing medium with various concentrations of zeocin. After 5 to 7 days of zeocin treatment, cells were treated with 1 mg/ml MTT (Sigma) solution for 1 h. Medium containing MTT then was removed, and the formazan crystals produced were solubilized in acidic isopropanol. Absorbance was read at 570 nm to determine relative survival.
VDJ recombination assays.
Extrachromosomal VDJ recombination assays utilizing the signal joint substrate (pJH201) and coding joint substrate (pJH290) were performed as described previously (35). Briefly, cells plated at 20 to 40% confluence in 60-mm-diameter dishes were transiently transfected with 1 μg substrate, 4 μg each of RAG1 and RAG2, and 4 μg of the indicated expression construct or empty vector using the Lipofectamine 2000 transfection reagent according to the manufacturers' instructions. Forty-eight hours after transfection, substrate plasmids were isolated by alkaline lysis and subjected to DpnI restriction enzyme digestion for 1 h. DpnI-digested DNA was transformed into competent DH5α cells (Invitrogen) according to the manufacturers' instructions. Transformed cells were spread onto LB agar plates containing 100 μg/ml ampicillin only or with 100 μg/ml ampicillin and 22 μg/ml chloramphenicol. The percentage of recombination was calculated as the number of colonies resistant to ampicillin and chloramphenicol divided by the number of colonies resistant to ampicillin.
The fluorescent VDJ substrates (diagramed in Fig. 3) and VDJ assays have been described recently (Neal and Meek, unpublished). These substrates (derived from pECFP-N1; Clontech) contain the SV40 origin of replication; thus, they are efficiently replicated episomally in all primate cell strains; it has been reported that this origin replicates to very low copy numbers in rodent cells (36). We have directly compared VDJ proficiency in CHO cell strains with pJH290/pJH201 and the fluorescence-based substrates with completely analogous results (Neal and Meek, unpublished). Briefly, extrachromosomal fluorescent VDJ assays were performed on cells plated at 20 to 40% confluence into 24-well plates in complete medium. Cells were transfected with 0.125 μg substrate, 0.25 μg RAG1 per well, and 0.25 μg RAG2 per well using PEI (1 μg/ml; Polysciences) at 2 μl/1 μg DNA. In experiments with additional expression plasmids, 0.25 μg of the expression plasmid or vector control was included. Cells were harvested 72 h after transfection and analyzed for CFP and RFP expression by flow cytometry. The percentage of recombination was calculated as the percentage of live cells expressing CFP divided by the percentage expressing RFP. Data presented represent at least three independent experiments, each of which includes triplicate transfections.
FIG 3.
XRCC4/XLF interaction is variably required in different cell types to support VDJ recombination of episomal substrates. The fluorescent substrates (top left) were utilized to detect coding and signal joints in VDJ assays in 293 cells, whereas substrates pJH290 and pJH201 (top right) were used to detect coding and signal joints in VDJ assays in ES cells. The lower panel shows percent recombination of episomal fluorescent coding and signal joint substrates in XLF-deficient 293 cells transiently expressing full-length Rag1, Rag2, and wild-type and mutant forms of XLF. Note that the decrease in percent recombination observed by L115A and L115D are statistically significant for signal joint substrate (wt versus L115A, P = 0.021; wt versus L115D, P = 0.0002) and coding joint substrate (wt versus L115A, P = 0.0001; wt versus L115D, P < 0.0001) according to a two-tailed Mann-Whitney test. Error bars indicate SEM from five independent experiments. The lower panel shows percent recombination of pJH290 (left) and pJH289 (right) in XLF-deficient mouse ES cells transiently expressing full-length Rag1, Rag2, and wt and mutant forms of XLF. Note that the decrease in recombination percentage observed by L115D compared to that of the wt is statistically significant (P = 0.0019 for coding joint assay and P = 0.0047 for signal joint) according to two-tailed Mann-Whitney test. Error bars indicate SEM from four independent experiments.
RESULTS
XLF L115A does not interact with XRCC4 or bridge DNA in vitro but is fully sufficient to stimulate XRCC4/Lig4-mediated ligation in vitro.
We and others have implicated L115 as being an XLF residue that is critical for the molecule's interaction with XRCC4 (4, 37). Whereas our biochemical data show that an L115A substitution completely disrupts the XRCC4/XLF interaction (4), Fattah et al. have demonstrated end joining of plasmid substrates and VDJ intermediates of this mutant in living cells (38); the relative ability of L115A to restore radioresistance was not studied by these authors. In contrast, de Villartay and colleagues found that an L115D substitution (which also does not interact with XRCC4 in vitro) does not reverse the c-NHEJ deficits or reverse radiosensitivity associated with XLF deficiency (37). To understand the basis for these differences, the biochemical function of these two mutants in vitro were directly compared. Previously, we developed a DNA bridging assay that measures end bridging between a free 500-bp DNA fragment and a 1,000-bp biotin-labeled fragment that can be immobilized onto streptavidin beads (6). If bridging occurs (by protein-DNA interaction), the untagged DNA will be pulled down onto the streptavidin beads. In the presence of wild-type XLF and XRCC4 together but not with either alone and not with either XLF L115A or XLF L115D (and wild-type XRCC4), the free 500-bp fragment could be efficiently pulled down (Fig. 1A). As another test of DNA bridging in vitro, the ability of XRCC4 and XLF to promote ligation and formation of concatemers by T4 DNA ligase was assessed. Neither XRCC4 nor XLF alone significantly enhance ligation of either blunt or cohesive ends by T4 ligase (Fig. 1B). However, ligations of both blunt and cohesive DNA ends are markedly enhanced by wild-type XLF and XRCC4 together. Similar to the results with the DNA bridging assay, neither the L115A nor the L115D mutant stimulates ligation of either blunt or cohesive DNA ends by T4 DNA ligase (Fig. 1B). We conclude that neither mutant can efficiently bridge DNA in vitro.
FIG 1.
XLF L115A does not interact with XRCC4; thus, it does not bridge DNA in vitro but is fully sufficient to stimulate XRCC4/Lig4. (A, left) Schematic of the DNA bridging assay. (Right) Agarose gel showing recovery of DNA fragments bound to streptavidin beads by ethidium bromide staining. Molecular size markers are indicated (kilobases). (B to D) Ethidium bromide staining of agarose gels showing ligation products obtained from in vitro ligation reactions as described in Materials and Methods. Molecular size markers are indicated (kilobases). (B) T4 DNA ligase is utilized. (C) XRCC4/Lig4 complexes (0.4 μM) are utilized. Four different concentrations of XLF were utilized: 0.25 μM, 0.5 μM, 1 μM, and 2 μM. (D) XRCC4/Lig4 complexes (0.2 μM) are utilized, with wild-type or mutant XRCC4 as indicated and with wild-type or mutant XLF (0.5 μM). (E, top) Immunoblot analyses of lysates from 293 cells transiently transfected with His-tagged wt and mutant forms of XLF probed with antibodies to XRCC4, XLF, or Lig4. (Bottom) Immunoblot analyses of pulldown fractions recovered from Ni-NTA–agarose beads after 3 h of incubation of cell lysates with beads and subsequent washing. The immunoblot was probed with antibodies to XRCC4, XLF, or Lig4.
XRCC4 interacts with and supports the stability of Lig4 in living cells; from this perspective, it is unclear whether the stimulation of T4 ligase activity in vitro genuinely recapitulates XRCC4/XLF's effect on Lig4. Thus, recombinant human XRCC4/Lig4 complexes were prepared, and the ability of XLF (wild type or mutant) to stimulate ligation was tested. As expected, wild-type XLF markedly stimulates the activity of XRCC4/Lig4 (Fig. 1C). To our surprise, the L115A mutant was just as proficient as wild-type XLF in stimulating intermolecular ligation by XRCC4/Lig4, whereas L115D did not stimulate ligation by XRCC4/Lig4 (Fig. 1C and D). Moreover, no differences were observed at any ratio of XLF to XRCC4 tested (0.25, 0.5, 1, or 2:0.4). These in vitro results accurately mirrored and corroborated the conflicting in vivo reports (4, 37, 38) and allowed us to conclude that XLF's capacity to stimulate XRCC4/Lig4 in vitro is independent of its DNA bridging capacity in vitro.
Previously we studied DNA bridging using XRCC4 mutants that could not interact with XLF (29); however, XLF's ability to stimulate XRCC4/Lig4 with these mutations was not assessed. Thus, XRCC4/Lig4 complexes were prepared with two XRCC4 mutants. Both wild-type and L115A XLFs stimulate wild-type XRCC4/Lig4 (Fig. 1D), but neither wild-type XLF nor L115A significantly stimulates the ligase activity of either mutant XRCC4/Lig4 complex, although minimal stimulation is observed with both. We also considered that the presence of Lig4 stabilizes a potential weak interaction with XRCC4. Thus, we also tested (by pulldown assays) whether these mutants transiently expressed in 293 cells interact with XRCC4 and Lig4; neither mutant interacts with either XRCC4 or Lig4 (Fig. 1E). Although other explanations are possible, we suggest that XLF (either wild type or L115A) can stimulate XRCC4/Lig4 without stably interacting with the complex. The failure of XLF L115D to stimulate ligations is less clear. The more dramatic alteration of L115D might impede the ability of XLF to access XRCC4/Lig4. The fact that the two mutant XRCC4/Lig4 complexes (i.e., with XRCC4 K65/99E and K72/90/99E) show markedly reduced stimulation by either wild-type XLF or the L115A mutant suggests that these mutants also partially exclude XLF from stimulating XRCC4/Lig4.
Stimulation of XRCC4/Lig4 is intrinsic to XLF's function in living cells; XRCC4/XLF interaction is variably required.
As noted above, XLF deficiency has a variable impact on different cell types and in different organisms. We considered that a bridging function of XRCC4/XLF in vivo (but not XRCC4/Lig4 stimulation) might be variably required in different cell types depending on the relative expression or activity of factors that share functional redundancy with XLF (ATM, 53BP1, H2AX, DNA-PKcs, and potentially others) (20–22). XLF L115A appears to represent a separation-of-function mutant that could be useful in discerning whether XRCC4/XLF interaction and potential filament formation is important in living cells. Thus, we next derived stable clonal transfectants of wild-type and mutant XLF in a panel of different XLF-deficient cell strains (Fig. 2A). These include a human fibroblast cell strain (2BN) derived from an XLF-deficient patient (39), XLF-deficient HCT116 cells (a human colon cancer cell strain from which XLF was deleted by gene targeting) (38), human 293 cells that are XLF deficient via a Cas9/CRISPR-mediated mutation, mouse XLF-deficient embryonic stem cells (17), and mouse v-Abl-transformed XLF-deficient pre-B cells (18). Wild-type XLF substantially reverses the zeocin (a bleomycin analogue that induces double-strand breaks [DSBs])-sensitive phenotype in all five cell strains (Fig. 2B to F). The complementation of the zeocin-sensitive phenotype in XLF-deficient 293 cells demonstrates that the zeocin-sensitive phenotype is the effect of XLF deletion and not off-target effects of Cas9/CRISPR (Fig. 2F). Expression of the XLF mutant L115D that cannot stimulate XRCC4/Lig4 does not reverse zeocin sensitivity in any of the five cell strains. L115A that is proficient in stimulating XRCC4/Lig4 (but deficient in DNA end bridging in vitro) completely reverses the zeocin sensitivity in the human patient fibroblast cells (2BN) and in XLF-deficient ES cells and partially reverses zeocin sensitivity in human 293 cells and murine Abelson-transformed B cells. In contrast, L115A does not reverse zeocin sensitivity in human XLF-deficient HCT116 cells. From these data, we conclude that XLF's capacity to stimulate XRCC4/Lig4 is required in all cell types for maximal survival following DNA DSBs, whereas XLF's ability to stably interact with XRCC4 is variably required in different cell types. This variation in the cellular requirement for XRCC4/XLF interaction is not readily attributable to differences in expression of other DNA repair factors (see Fig. S1 in the supplemental material), although HCT116 cells express less ATM than 2BN or 293 cells. Of note, HCT116 cells have been shown to have defects in both mismatch repair and in one allele of Mre11 (40, 41). Moreover, HCT116 cells display the most sensitivity to zeocin of all the cell strains studied here.
FIG 2.
Stimulation of XRCC4/Lig4 is intrinsic to XLF's function in living cells; XRCC4/XLF interaction is variably required. (A) Immunoblot showing stable expression of actin or tubulin and wild-type or mutant XLF in various XLF-deficient cell strains, as indicated. vect, vector. (B to F) Zeocin sensitivity of the indicated cell strains stably expressing equivalent levels of wild-type or mutant XLF. Error bars indicate standard errors of the means (SEM) from at least three independent experiments.
Stable XRCC4/XLF interaction is variably required in different cell types to support VDJ recombination of episomal substrates.
XLF initially was discovered by studies of a patient with immunodeficiency (1, 39). Although XLF deficiency does not result in substantial VDJ deficits in murine lymphocytes or murine B cell lines (18), XLF is required in episomal VDJ recombination assays in mouse ES cells (17), mouse embryonic fibroblasts (17, 18), human patient fibroblasts (37, 39, 42), and XLF-deficient HCT116 cells (38). Although VDJ rearrangements have not been examined in lymphocytes from human XLF-deficient patients, the SCID phenotype of these patients (1) intuits that XLF also is required for VDJ recombination in developing human lymphocytes. We performed episomal VDJ assays in both XLF-deficient murine ES cells and XLF-deficient 293 cells. In ES cells, the standard Gellert assay (35, 43) was utilized, whereas in 293 cells a fluorescent substrate (Fig. 3, top) and a flow-cytometric assay were utilized (described in Materials and Methods; also see the supplemental material). Importantly, L115A, but not L115D, supports substantial levels of both coding and signal end joining in XLF-deficient ES cells (Fig. 3). Similarly, albeit not as robustly, L115A but not L115D restores both coding and signal end joining in XLF-deficient 293 cells.
The proficient coding end joining by XLF L115A (that does not interact with XRCC4 and cannot form XRCC4/XLF filaments in vitro) is at odds with our previous study showing that XRCC4 mutants that could not interact with XLF (and could not bridge DNA or form XRCC4/XLF filaments in vitro) had modest VDJ recombination deficits that asymmetrically impacted coding joint formation (44). These previous studies were performed in the XRCC4-deficient CHO (Chinese hamster ovary) cell strain XR-1. Given the substantial variability in cellular phenotypes that may reflect differences in requirements for stable XRCC4/XLF interaction, the ability of these XRCC4 mutants to complement VDJ deficits was assessed in XRCC4-deficient ES cells (generated by conventional gene targeting [45]) and in XRCC4-deficient 293 cells that were prepared for these studies by a Cas9/CRISPR-mediated mutation. Neither XRCC4 mutant support wild-type levels of coding end joining in 293 cells, consistent with the partial complementation of XLF−/− 293 cells by the L115A mutant (see Fig. S2 in the supplemental material). In contrast, coding joint formation is substantially complemented by both mutants in XRCC4-deficient ES cells, again consistent with L115A's capacity to complement VDJ joining in XLF-deficient ES cells. ES clones stably expressing wild-type XRCC4 or K65/99E were established. As with L115A in XLF-deficient ES cells (Fig. 2D), K65/99E complements the zeocin-sensitive phenotype of XRCC4-deficient ES cells (see Fig. S2). These data underscore the variable dependence on XRCC4/XLF interaction; 293 cells show a strong dependence on stable XRCC4/XLF complexes compared to that of mouse ES cells, where stable XRCC4/XLF interactions apparently are dispensable.
XLF L115A is more defective in ligation of incompatible DNA ends than of compatible DNA ends.
Two DNA ends must be closely juxtaposed for ligation to occur, although emerging data suggest that repair of the two strands occurs independently (12, 46, 47). It is also well appreciated that DSBs can be maintained in a spatiotemporal manner in large DNA repair centers (48). There is evidence that XRCC4/XLF filaments affect both the precise alignment of DNA ends, in that XLF promotes ligation of noncohesive ends in vitro (10, 11) (Fig. 1), as well as the maintenance of ends in “repair centers,” in that XLF deficiency is exacerbated when factors that function in assembling γH2AX foci are disrupted (20–24). We first assessed, in living cells, whether L115A's deficit in resolving coding ends was exacerbated if the ends are not compatible. The RAG endonuclease generates blunt-ended signal ends and hairpinned coding ends (49). It has been shown that coding hairpins are nicked to generate 4-bp 3′ overhangs (50). Two additional fluorescence-based coding joint substrates were prepared. One substrate generates 4-bp cohesive ends and also provides additional pairing opportunities even if nucleotide loss (up to 7 bp at each end) occurs at either coding end (Fig. 4, matched coding end). A second substrate generates overhangs with no microhomology, even if nucleotide loss should occur. Analyses of these substrates is complicated by the fact that the RAG nuclease is exquisitely sensitive to the sequences of the coding end-recombination signal sequence (RSS) junctures (51) (which are fairly optimal in the parental substrate but cannot be optimized in the matched/mismatched substrates). Thus, these coding/RSS substrates are not optimal and recombine 3- to 4-fold less efficiently in wild-type cells than the original 290/RFP/CFP substrate (not shown). Hence, comparison of recombination levels for each substrate must be normalized to the efficiency of recombination in cells that are c-NHEJ proficient. Although L115A and wild-type XLF are similarly proficient in joining the substrate that generates matched ends, L115A is less proficient in joining coding ends with completely mismatched termini (3.2-fold) compared to the optimal substrate (2.0-fold) (Fig. 4). We conclude that XRCC4/XLF interaction facilitates end joining when DNA termini lack terminal microhomology.
FIG 4.
XLF L115A is more defective in ligation of incompatible DNA ends than in that of compatible DNA ends. (Top) Schematic showing altered coding end sequences to generate perfectly matched or mismatched overhangs in the fluorescent coding joint substrate depicted in Fig. 3. (Bottom) Comparison of recombination rate of episomal substrates with matched and mismatched coding ends in XLF-deficient 293 cells transiently expressing Rag1, Rag2, and wild-type and mutant forms of XLF. Error bars indicate SEM from four independent experiments. Note that the decrease in recombination rate observed by XLF mutant L115A compared to that of the wild type is statistically significant for 290 (P = 0.0021) and mismatched (P = 0.0404) termini according to two-tailed Mann-Whitney test. There is no significant difference observed between recombination rate of wt XLF and XLF mutant L115A in the presence of matched termini.
XRCC4/XLF interaction is required for robust DNA-PK autophosphorylation.
Calsou and colleagues demonstrated that DNA damage-induced autophosphorylation of DNA-PKcs is defective in cells deficient in Lig4 (32). Moreover, a catalytically inactive mutant of Lig4 reversed this defect, suggesting that the XRCC4/Lig4 complex facilitates autophosphorylation. The 293, HCT116 (Fig. 5), and 2BN (not shown) isogenic cell strain panels were exposed to zeocin, and DNA-PKcs autophosphorylation at serine 2056 was assessed by immunoblotting. In all three cell types, although phosphorylation of γH2AX is similar, S2056 phosphorylation is markedly reduced in cells lacking XLF; neither L115D nor L115A reverses the defect in autophosphorylation, suggesting that XRCC4/XLF complexes promote DNA-PKcs autophosphorylation (Fig. 5). Although other explanations are possible, since S2056 phosphorylation can occur in trans (52), these data suggest that XRCC4/XLF complexes help stabilize synapsed DNA-PK bound ends, enhancing trans-autophosphorylation. However, this defect in autophosphorylation does not explain differences in the cellular requirement for XRCC4/XLF interaction, since L115A completely complements the zeocin sensitivity of 2BN cells but not of HCT116 cells (Fig. 2). It should be noted that DNA-PKcs can be phosphorylated on many residues (likely more than 60) (53), it has been shown that some autophosphorylations can occur in cis (54, 55), and phosphorylation at 2056 has been shown to occur in trans (52).
FIG 5.
XRCC4/XLF interaction is required for robust DNA-PK autophosphorylation. Immunoblot analysis showing levels of DNA-PKcs autophosphorylation at S2056, total DNA-PKcs, XLF expression, and γH2AX phosphorylation with and without zeocin treatment of 293 cells (left) and XLF-deficient HCT116 cells (right) stably expressing wild-type and mutant forms of XLF.
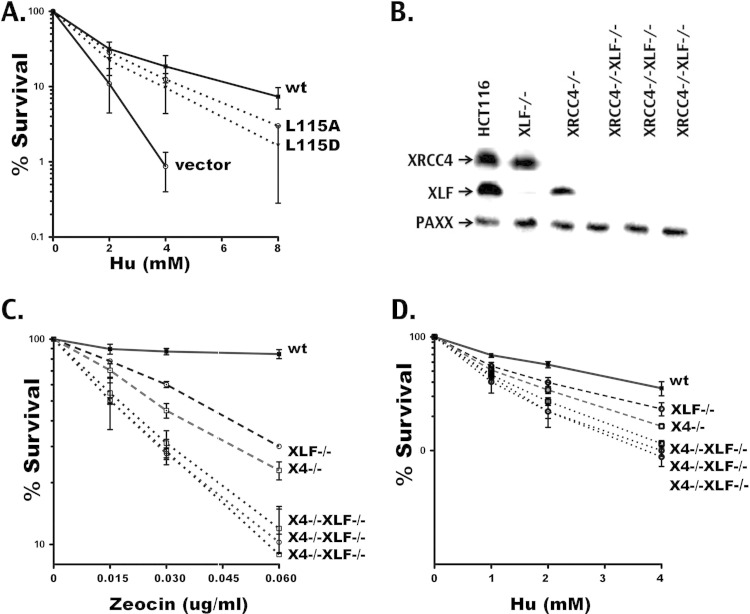
An ATM or PAXX deficiency accentuates the c-NHEJ deficits of L115A.
As noted above, the different cellular phenotypes could be due to the functional redundancy of XLF/XRCC4 complexes with other proteins in certain cell types. Thus, we next generated 293 cells that lack both XLF and ATM (see Fig. S3A in the supplemental material). In addition, two laboratories recently have described another XRCC4/XLF-like factor, PAXX (25, 26). PAXX interacts with DNA-bound Ku and can be targeted to sites of DNA damage. Thus, given the DNA bridging capacity of XRCC4 and XLF, 293 and HCT116 cells lacking PAXX or both PAXX and XLF also were generated (see Fig. S3A). Both HCT116 and 293 cells lacking only PAXX were resistant to zeocin to an extent similar to that for wild-type cells at the zeocin doses used to study zeocin sensitivity in XLF-deficient cells, although modest sensitivity was observed using higher zeocin doses (data not shown). These data are consistent with the mild radiosensitivity observed in PAXX-deficient cells by others (25, 26). However, the loss of PAXX expression in XLF-deficient cells (either 293 cells or HCT116) exacerbated zeocin sensitivity in both HCT116- and 293 XLF-deficient cells (Fig. 6A). We conclude that for zeocin resistance in both HCT116 cells and 293 cells, PAXX and XLF are not epistatic. Although Jackson and colleagues concluded that PAXX is epistatic with XLF for repair of infrared-induced damage, XLF was depleted with short interfering RNA and residual XLF was present (26). Moreover, Xing and colleagues observed a lack of epistasis between PAXX and XLF in chicken DT40 cells for the repair of some types of DNA damage but not others (25). Thus, different cell types may have distinct requirements for XLF in response to different types of DNA damage.
FIG 6.
ATM and PAXX deficiency accentuates the c-NHEJ deficits of L115A. (A) Zeocin sensitivity of parental wild-type cells, XLF-deficient and XLF−/− PAXX−/− 293 cells (left), and HCT116 cells (right). Error bars indicate SEM from three independent experiments. (B) Recombination percentage of episomal fluorescent coding and signal joining substrates comparing XLF-deficient, XLF/ATM doubly deficient, and XLF−/− PAXX−/− 293 cells transiently expressing Rag1, Rag2, and wild-type and mutant forms of XLF as indicated. Error bars indicate SEM from at least 3 independent experiments. Note that XLF L115A shows a significantly lower coding end joining rate in XLF-deficient (P = 0.0001) and XLF−/− ATM−/− (P = 0.0022) 293 cells, unlike XLF−/− PAXX−/− cells. Similarly, XLF L115A performs significantly reduced signal joining in XLF-deficient (P = 0.0210) and XLF−/− ATM−/− (P = 0.0022) cells, unlike XLF−/− PAXX−/− 293 cells. (C) Zeocin sensitivity of the XLF−/− ATM −/− 293 cell strain stably expressing equivalent levels of wt or mutant XLF. Data from three clones expressing L115A are presented. Error bars indicate SEM from three independent experiments. (D) Immunoblot analysis depicting XLF expression levels in XLF−/− ATM −/− cells stably transfected with wt or mutant XLF or vector control. (E) Immunoblot showing levels of DNA-PKcs autophosphorylation at S2056 with and without zeocin (Zeo) treatment of XLF/ATM doubly deficient cells stably expressing wt and mutant forms of XLF.
Episomal VDJ assays also were performed on doubly deficient 293 cells. Loss of ATM or PAXX accentuates the requirement for XLF in rejoining both coding and signal ends (Fig. 6B). Cotransfection of either PAXX or XLF partially reverses the VDJ deficits in XLF−/− PAXX−/− cells, whereas cotransfection of both results in more substantial complementation (see Fig. S3B in the supplemental material). In addition, whereas the wild type and L115A mutant similarly reverse the deficits in XLF−/− PAXX−/− cells, the VDJ deficits associated with the L115A mutant are exacerbated in XLF−/− ATM−/− cells (Fig. 6B). Consistent with these data, the VDJ deficits associated with the XRCC4 mutants K65/99E and K72/90/99E also are exacerbated by the loss of ATM (see Fig. S4). Wild-type and mutant XLF were stably integrated into the XLF−/− ATM−/− 293 cells. Whereas wild-type XLF substantially reverses the zeocin hypersensitivity, neither the L115A nor L115D mutant reverses zeocin sensitivity whatsoever (Fig. 6C). This is in marked contrast to the significant level of complementation by L115A in XLF-deficient (but ATM-proficient) 293 cells (Fig. 2F). Of note, the lower ATM expression levels in HCT116 cells provide a potential explanation for the complete lack of complementation of zeocin hypersensitivity by L115A (see Fig. S1). In addition, loss of both ATM and XLF results in a further deficiency in DNA-PKcs autophosphorylation (Fig. 6E). In sum, these data suggest that it is the ability of XLF to complex with XRCC4 (and potentially form filaments that bridge DNA) that is functionally redundant with ATM, as proposed by others (20).
PAXX interacts with Ku-DNA complexes but does not bridge DNA or cooperate with XRCC4/XLF to bridge DNA in vitro.
In contrast to XLF−/− ATM−/− cells, in XLF−/− PAXX−/− cells, the L115A (but not the L115D) mutant was as proficient as wild-type XLF in partially reversing both coding and signal joining defects. Thus, the loss of PAXX does not accentuate the cellular requirement for XRCC4/XLF interaction. To understand the biochemical basis of these differences, recombinant PAXX was generated (Fig. 7A). Consistent with recent reports (25, 26), although it does not itself bind DNA, PAXX interacts with Ku-bound DNA (Fig. 7B). As reported by Xing et al. (25), we find that PAXX interacts only with Ku/DNA complexes containing one Ku heterodimer, whereas Ochi et al. (26) found that PAXX interacts exclusively with complexes containing two Ku heterodimers. One potential explanation for these differences is expression strategies (baculovirus for Ku expression for these studies and those of Xing et al. versus bacterial expression for the studies of Ochi et al.). Moreover, PAXX by itself does not promote DNA end bridging, nor does it alter the DNA end-bridging capacity of XRCC4/XLF (Fig. 7C). From data presented here, we conclude that although PAXX can interact with Ku-bound DNA, it does not contribute to DNA end bridging in vitro; moreover, loss of PAXX does not accentuate the cellular requirement for XRCC4/XLF interaction (and potential DNA end bridging) in living cells.
FIG 7.

PAXX interacts with Ku-DNA complexes but does not itself bridge DNA or cooperate with XRCC4/XLF filaments to bridge DNA. (A) Polyacrylamide gel electrophoresis under reducing and denaturing conditions stained with Coomassie blue showing recombinant Ku (1 μg) and PAXX (5 μg). (B) A native polyacrylamide gel stained with ethidium bromide showing the electrophoretic mobility shift of a 61-bp double-stranded DNA (dsDNA) after incubation with PAXX, Ku, or Ku and PAXX together. (C) Agarose gel stained with ethidium bromide showing recovery of DNA fragments bound to streptavidin beads after DNA bridging assays (as described for Fig. 1).
XLF promotes cell survival after replication stress; its role in this process is independent of its interaction with XRCC4.
Cells deficient in various c-NHEJ factors are more sensitive to agents that induce replication stress than c-NHEJ-proficient cells (27, 28, 53). Schwartz et al. have shown previously that fibroblasts derived from XLF-deficient patients are sensitive to low-dose aphidicolin, display increased fragile site instability, and display cell cycle disruptions consistent with an inability to resolve DNA damage associated with replication stress (16). Thus, we next assessed cell survival of these isogenic cell strains after exposure to hydroxyurea, which depletes nucleotide pools, resulting in replication stress. Whereas XLF-complemented and vector control 2BN cells are similarly sensitive to HU (data not shown), XLF-deficient HCT cells are remarkably sensitive to HU; this sensitivity is reversed by wild-type XLF (Fig. 8A). To our surprise, both L115A and L115D substantially reverse the HU hypersensitivity, albeit slightly less so than wild-type XLF. Of note, complementation by L115A and L115D is equivalent, suggesting that XLF's unexpected role in reversing HU hypersensitivity is at least partially independent of its ability to interact with XRCC4 or stimulate ligase IV activity.
FIG 8.
XLF promotes cell survival after replication stress; its role in this process is independent of its interaction with XRCC4. (A) HCT116 transfectants expressing wild-type XLF, XLF L115A, XLF L115D, or no XLF (vector) were plated at cloning densities into complete medium with increasing doses of hydroxyurea. Colonies were stained after 7 days, and percent survival was calculated. Error bars represent the SEM. (B) XLF and XRCC4 protein expression levels were examined by Western blotting of whole-cell extracts obtained from the indicated cell strains. (C and D) HCT116 cells and HCT116 cells deficient in XLF, XRCC4, or both XLF and XRCC4 were plated at cloning densities into complete medium with increasing doses of zeocin (C) or hydroxyurea (D). Data from three clones deficient in both XLF and XRCC4 are presented.
To test this conclusion more stringently, cells deficient in both XRCC4 and XLF were derived and tested for their capacity to survive HU-induced replication stress and zeocin-induced DSBs. Three independent clones targeting XLF in XRCC4-deficient HCT116 cells were isolated (Fig. 8B). The loss of XLF in each cell line exacerbated the sensitivity of XRCC4-deficient cells to both HU and zeocin (Fig. 8C and D). Implicit in these data is an XRCC4-independent function for XLF.
DISCUSSION
We conclude from these studies that XLF has multiple cellular functions that are biochemically distinct. Recent biochemical studies have proposed that one of XLF's functions in living cells is to tether or bridge DNA ends with XRCC4 (potentially as filaments or filament bundles) (reviewed in reference 56). However, evidence was lacking for formation of these DNA end-bridging filaments in living cells. During the final revision of the manuscript, elegant studies from Rothenberg and colleagues unequivocally demonstrated that XRCC4/XLF form filaments in living cells (with or without Lig4). Superresolution microscopy demonstrated the presence of these filaments adjacent to Ku-bound DSBs, in some cases spanning Ku-bound DSBs (57). What is still unclear is precisely how DNA bridging impacts end joining. Given that the homodimerization of DNA-PK also can tether/bridge ends together (58, 59, 60), it may seem redundant to postulate the need for a second tethering system. However, the use of two discrete tethering systems may have distinct mechanistic advantages. The presence of a second, XRCC4/XLF-mediated tethering process could solve a significant topological problem. The DNA-PK complex is able to tether broken DNA ends together; this requires both the catalytic subunit and the DNA binding subunit, Ku, a circular-shaped protein through which the DNA is threaded (61, 62). Numerous investigators have noted that Ku's unique DNA binding mode would leave Ku topologically locked onto the DNA after ligation. Such an occurrence would prohibit strand separation and jeopardize subsequent transcription and replication in that region. It has been suggested that such events can occur and that the cell's solution to this problem is to proteolytically degrade the trapped Ku (63). The presence of an ancillary and independent XRCC4/XLF-mediated tethering system might obviate such pathways. If the Ku/DNA-PKcs tethering temporally occurs first, the presence of a subsequent XRCC4/XLF-mediated tether would permit DNA-PKcs to undergo the significant conformational changes that it is known to undergo upon becoming activated (64–66) and would permit the release of Ku from the DNA by sliding without the broken DNA ends becoming separated from one another. Thus, a requirement for XRCC4/XLF-mediated bridging might particularly impact the efficacy of joining noncompatible ends, since these ends presumably need to be synapsed long enough to appropriately process the mismatched ends. Consistent with this prediction, the absence of XLF is clearly deleterious for the joining of mismatched ends (Fig. 4) (10–12, 67, 68). Our data demonstrate that XRCC4/XLF complexes have the ability to bridge DNA in vitro, facilitating DNA-PKcs autophosphorylation, suggesting that DNA ends are bridged by both synapsed DNA-PK and XRCC4/XLF concurrently, physically promoting DNA-PK autophosphorylation at 2056, a site which can be phosphorylated in trans (52). It also is possible that XRCC4/XLF bridging occurs prior to DNA-PK assembly; in fact, it is known that XLF requires Ku (but not DNA-PKcs) to be appropriately targeted to DNA damage in living cells (65, 66). Our previous studies suggest that DNA-PK phosphorylation of XRCC4/XLF disrupts filaments (29). Work is ongoing to define how XRCC4/XLF filaments promote repair in living cells.
Our cellular studies show that only some cell types require XLF's function of forming DNA end-bridging, XRCC4/XLF complexes. Although the mechanistic basis for this cell type-specific requirement still eludes us, it is clear that it is the ability to form these XRCC4/XLF complexes that is overlapping with ATM. Thus, in ATM-proficient cells, ATM compensates for L115A's defect in XRCC4 stable interaction and potential DNA end bridging (Fig. 6). In contrast, although loss of PAXX accentuates the cellular requirement for XLF, this effect does not accentuate the cellular requirement for XRCC4/XLF interaction and potential DNA end bridging. This is entirely consistent with in vitro studies showing that PAXX does not itself bridge DNA ends or cooperate with XRCC4/XLF in bridging DNA ends (Fig. 7). Thus, although these studies do not define how PAXX affects c-NHEJ, our studies seem to exclude DNA end bridging as a function for PAXX in c-NHEJ.
A second function for XLF is in stimulating Lig4 catalysis. While we noted above that XLF indirectly augments catalysis by facilitating synapsis, we believe that this function of XLF is independent of a stable interaction with XRCC4. Thus, XLF that only forms homodimers and does not form XRCC4 multimers (i.e., the XLF L115A variant) nonetheless substantially reverses NHEJ deficits in many cell types. We attribute this to XLF's capacity to promote Lig4 catalysis without forming a stable XRCC4/XLF complex. Our cellular studies demonstrate that unlike XLF's stable interaction with XRCC4 and potential DNA end-bridging activity, all cell types have a requirement for XLF's function in stimulating Lig4.
In 2009, Schwartz and colleagues demonstrated that fibroblasts derived from XLF-deficient patients are sensitive to agents that induce DNA replication stress (16). These authors suggested that this role for XLF in DNA replication explains the phenotypes observed in XLF-deficient patients that cannot be attributed to defective VDJ recombination. Recent studies demonstrate that hematopoietic stem cells from XLF-deficient mice fail at an early age (19). Although this defect was not directly attributed to premature replicative failure, other DNA repair defects phenocopy this. We show here that a subset of XLF-deficient cells are remarkably sensitive to HU-induced replication stress (Fig. 8). Surprisingly, XLF's role in abating replication stress was in large part independent of XLF's interaction with XRCC4. In sum, these data imply that XLF has at least a third function that is nonoverlapping with its other two functions. This demonstration underscores the need for further investigation into the distinct functions of XLF and to delineate why different cell types and different organisms have such variable requirements for this multifunctional factor.
In summary, we propose that XLF has (at least) two distinct functions: (i) DNA end bridging (likely in filaments with XRCC4) and (ii) stimulation of XRCC4/Lig4 as a homodimer. Recently, the current dogma that c-NHEJ functions in a well-defined stepwise manner (69–71) has been challenged. Emerging data using both biochemical and genetic (12, 46, 72, 73) approaches suggest that c-NHEJ is much more flexible. Instead, these reports propose models whereby XRCC4/Lig4 organizes how c-NHEJ proceeds based upon the degree of end processing that is required for repair. Consistent with these new models, it seems intuitive that an XRCC4/XLF bridging function occurs at an early step in DNA DSB repair, whereas XLF's role in stimulating ligation would be a later step.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Public Health Service grant AI048758 (K.M.), by the USDA National Institute of Food and Agriculture and Michigan AgBioResearch (K.M.), by project number SFI20121205867 from the Fondation ARC pour la Recherche sur le Cancer (M.M.), and by grant PLBIO13-103 from the Institut National du Cancer (M.M.). E.H. was supported by a grant from the National Institutes of Health (GM088351) and the National Cancer Institute (CA154461).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01503-14.
REFERENCES
- 1.Buck D, Moshous D, de Chasseval R, Ma Y, le Deist F, Cavazzana-Calvo M, Fischer A, Casanova JL, Lieber MR, de Villartay JP. 2006. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur J Immunol 36:224–235. doi: 10.1002/eji.200535401. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, Malivert L, de Villartay JP. 2006. Cernunnos-XLF, a recently identified non-homologous end-joining factor required for the development of the immune system. Curr Opin Allergy Clin Immunol 6:416–420. doi: 10.1097/01.all.0000246623.72365.43. [DOI] [PubMed] [Google Scholar]
- 3.Ahnesorg P, Smith P, Jackson SP. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. 2007. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol Cell 28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. 2008. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J 27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. 2012. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res 40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. 2011. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem 286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, Le Cam E, Revy P, de Villartay JP, Charbonnier JB. 2011. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci U S A 108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL. 2011. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem Soc Trans 39:1387–1392. doi: 10.1042/BST0391387. [DOI] [PubMed] [Google Scholar]
- 10.Tsai CJ, Kim SA, Chu G. 2007. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A 104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akopiants K, Zhou RZ, Mohapatra S, Valerie K, Lees-Miller SP, Lee KJ, Chen DJ, Revy P, de Villartay JP, Povirk LF. 2009. Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res 37:4055–4062. doi: 10.1093/nar/gkp283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. 2007. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res 35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. 2007. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J 26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. 2009. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res 37:482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodbine L, Gennery AR, Jeggo PA. 2014. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 16:84–96. doi: 10.1016/j.dnarep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz M, Oren YS, Bester AC, Rahat A, Sfez R, Yitzchaik S, de Villartay JP, Kerem B. 2009. Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS One 4:e4516. doi: 10.1371/journal.pone.0004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zha S, Alt FW, Cheng HL, Brush JW, Li G. 2007. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci U S A 104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. 2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell 31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avagyan S, Churchill M, Yamamoto K, Crowe JL, Li C, Lee BJ, Zheng T, Mukherjee S, Zha S. 2014. Hematopoietic stem cell dysfunction underlies the progressive lymphocytopenia in XLF/Cernunnos deficiency. Blood 124:1622–1625. doi: 10.1182/blood-2014-05-574863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. 2011. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature 469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oksenych V, Kumar V, Liu X, Guo C, Schwer B, Zha S, Alt FW. 2013. Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining. Proc Natl Acad Sci U S A 110:2234–2239. doi: 10.1073/pnas.1222573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C. 2012. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc Natl Acad Sci U S A 109:2455–2460. doi: 10.1073/pnas.1121458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. 2008. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc Natl Acad Sci U S A 105:9302–9306. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S. 2012. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc Natl Acad Sci U S A 109:3903–3908. doi: 10.1073/pnas.1120160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, Yan Z, Li W, Wang Q, Hou M, Dong C, Guo R, Gao G, Ji J, Zha S, Lan L, Liang H, Xu D. 2015. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat Commun 6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, Blundell TL, Jackson SP. 2015. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol 22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy S, Andres SN, Vergnes A, Neal JA, Xu Y, Yu Y, Lees-Miller SP, Junop M, Modesti M, Meek K. 2012. XRCC4's interaction with XLF is required for coding (but not signal) end joining. Nucleic Acids Res 40:1684–1694. doi: 10.1093/nar/gkr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. 2011. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol 31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. 2000. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J 19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. 2013. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 200:173–186. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, Tucker PW, Capra JD. 1994. Production and characterization of recombinant human Ku antigen. Nucleic Acids Res 22:3918–3924. doi: 10.1093/nar/22.19.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hesse JE, Lieber MR, Gellert M, Mizuuchi K. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell 49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 36.Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ. 1999. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res 27:426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, Callebaut I, de Villartay JP. 2010. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J Biol Chem 285:26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fattah FJ, Kweon J, Wang Y, Lee EH, Kan Y, Lichter N, Weisensel N, Hendrickson EA. 2014. A role for XLF in DNA repair and recombination in human somatic cells. DNA Repair 15:39–53. doi: 10.1016/j.dnarep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. 2003. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci U S A 100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. 1994. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res 54:4308–4312. [PubMed] [Google Scholar]
- 41.Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M. 2008. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol Biol Cell 19:1693–1705. doi: 10.1091/mbc.E07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malivert L, Callebaut I, Rivera-Munoz P, Fischer A, Mornon JP, Revy P, de Villartay JP. 2009. The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo. Mol Cell Biol 29:1116–1122. doi: 10.1128/MCB.01521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieber MR, Hesse JE, Lewis S, Bosma GC, Rosenberg N, Mizuuchi K, Bosma MJ, Gellert M. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell 55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 44.Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP. 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem 279:86–94. doi: 10.1074/jbc.M306841200. [DOI] [PubMed] [Google Scholar]
- 45.Bennardo N, Stark JM. 2010. ATM limits incorrect end utilization during non-homologous end joining of multiple chromosome breaks. PLoS Genet 6:e1001194. doi: 10.1371/journal.pgen.1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters CA, Strande NT, Pryor JM, Strom CN, Mieczkowski P, Burkhalter MD, Oh S, Qaqish BF, Moore DT, Hendrickson EA, Ramsden DA. 2014. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat Commun 5:4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Y, Lieber MR. 2006. In vitro nonhomologous DNA end joining system. Methods Enzymol 408:502–510. [DOI] [PubMed] [Google Scholar]
- 48.Scully R, Xie A. 2013. Double strand break repair functions of histone H2AX. Mutat Res 750:5–14. doi: 10.1016/j.mrfmmm.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983–991. doi: 10.1016/0092-8674(92)90248-B. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Pannicke U, Schwarz K, Lieber MR. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 51.Gerstein RM, Lieber MR. 1993. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev 7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 52.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. 2007. Trans-autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol 27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neal JA, Sugiman-Marangos S, VanderVere-Carozza P, Wagner M, Turchi J, Lees-Miller SP, Junop MS, Meek K. 2014. Unraveling the complexities of DNA-dependent protein kinase autophosphorylation. Mol Cell Biol 34:2162–2175. doi: 10.1128/MCB.01554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma Y, Lu H, Schwarz K, Lieber MR. 2005. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle 4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 55.Yoo S, Dynan WS. 1999. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res 27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. 2013. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol 91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, Oksuz BA, Fenyo D, Lieber MR, Ramsden DA, Rothenberg E. 2015. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci U S A 112:E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeFazio LG, Stansel RM, Griffith JD, Chu G. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. 2002. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 60.Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC. 2003. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res 31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grundy GJ, Moulding HA, Caldecott KW, Rulten SL. 2014. One ring to bring them all–the role of Ku in mammalian non-homologous end joining. DNA Repair 17:30–38. doi: 10.1016/j.dnarep.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 62.Walker JR, Corpina RA, Goldberg J. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 63.Postow L. 2011. Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett 585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. 2005. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13:243–255. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 65.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. 2008. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yano K, Morotomi-Yano K, Lee KJ, Chen DJ. 2011. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett 585:841–846. doi: 10.1016/j.febslet.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lieber MR. 2008. The mechanism of human nonhomologous DNA end joining. J Biol Chem 283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 68.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. 2004. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 69.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weterings E, Verkaik NS, Keijzers G, Florea BI, Wang SY, Ortega LG, Uematsu N, Chen DJ, van Gent DC. 2009. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol 29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. 2004. The DNA-dependent protein kinase: the director at the end. Immunol Rev 200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 72.Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. 2014. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair 21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lieber MR, Lu H, Gu J, Schwarz K. 2008. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res 18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.