Abstract
Posttranslational modifications (PTMs) can alter many fundamental properties of a protein. One or combinations of them have been known to regulate the dynamics of many cellular pathways and consequently regulate all vital processes. Understandably, pathogens have evolved sophisticated strategies to subvert these mechanisms to achieve instantaneous control over host functions. Here, we present the first report of modulation by intestinal pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) of host SUMOylation, a PTM pathway central to all fundamental cellular processes. Both in cell culture and in a mouse model, we observed that S. Typhimurium infection led to a dynamic SUMO-conjugated proteome alteration. The intracellular survival of S. Typhimurium was dependent on SUMO status as revealed by reduced infection and Salmonella-induced filaments (SIFs) in SUMO-upregulated cells. S. Typhimurium-dependent SUMO modulation was seen as a result of depletion of crucial SUMO pathway enzymes Ubc-9 and PIAS1, at both the protein and the transcript levels. Mechanistically, depletion of Ubc-9 relied on upregulation of small noncoding RNAs miR30c and miR30e during S. Typhimurium infection. This was necessary and sufficient for both down-modulation of Ubc-9 and a successful infection. Thus, we demonstrate a novel strategy of pathogen-mediated perturbation of host SUMOylation, an integral mechanism underlying S. Typhimurium infection and intracellular survival.
INTRODUCTION
Posttranslational modifications (PTMs) represent key regulatory mechanisms that can rapidly and reversibly affect the cumulative outcome of the cellular proteome at any given time. Modification(s) of target protein results in differential subcellular localization, activity, stability, and ability to interact with other molecules. These series of changes may impact the overall functional properties of the modified protein. Several pathogens, including Salmonella enterica serovar Typhimurium (S. Typhimurium), have been reported to use PTMs to achieve successful infection (1; also reviewed in reference 2), a strategy utilized to regulate several key aspects of host proteins (3, 4). Small ubiquitin-like modifier (SUMO), a PTM pathway, is now recognized as a universal signal transducer that is capable of affecting fundamental cellular processes such as DNA replication, transcription, RNA processing, and cell signaling (5). It is therefore likely that any perturbation of the SUMO system will result in profound changes within the host cells. Humans have three functional analogues of SUMO (SUMO1, SUMO2, and SUMO3) that could be linked to hundreds of different target proteins. Mature forms of SUMO2 and SUMO3 differ only in three amino acids and are often referred to interchangeably (SUMO2/3), while together they share only about 50% sequence similarity with SUMO1. The SUMOylation process is mediated by a common machinery that comprises an E1 activating enzyme (Sae1/Sae2), an E2 conjugating enzyme (Ubc-9), and several E3 ligating enzymes (reviewed in reference 5). SUMO is expressed in all eukaryotes but is absent in prokaryotes. However, recently there have been a growing number of examples of involvement of SUMOylation in the host-pathogen cross talk, revealing new layers of information in the biology of infection (2, 6, 7). Intracellular pathogen Listeria monocytogenes was demonstrated to alter host SUMOylation by targeting Ubc-9 (8) for successful infection, while in the case of Shigella flexneri, the host cell SUMOylation status was shown to be critical for the outcome of the infection (9). Effectors TRP120 of Ehrlichia chaffeensis and AmpA of Anaplasma phagocytophilum have been demonstrated to undergo SUMO modification specifically during infection (10, 11). Together, these findings highlight a novel strategy employed by bacterial pathogens involving SUMO conjugation of their effectors for successful infection. A report on axin1, a proinflammatory molecule, demonstrated the protein to be preferentially SUMOylated and ubiquitylated before undergoing degradation during Salmonella infection (12).
S. Typhimurium is the causative agent of several million cases of gastroenteritis in both developed and developing nations. The hallmark of this condition is an acute self-limited intestinal inflammatory response that helps in elimination of the infection but also leads to the characteristic manifestations of diarrhea, vomiting, and abdominal pain. S. Typhimurium is a facultative pathogen that gains entry into the host system via contaminated food and water. The first type of host cells that the bacteria invade is the intestinal epithelial cells (IECs) that not only play the role of physical barrier but are also the key signaling mediators of the host immune machineries (reviewed in reference 13). S. Typhimurium is able to invade these cells by using two sophisticated type III secretion systems (T3SSs) encoded by two Salmonella pathogenicity islands (SPIs), SPI1 (involved in the early part of the infection) (14, 15) and SPI2 (involved in the later part of the infection). The secretory components of the T3SS, also called effectors, are injected into the host cytoplasm by a supramolecular needle complex (15–17). The secreted effectors coordinately orchestrate the process of infection, which involves several changes in the host cell, including massive cytoskeletal rearrangement (18), reprogramming of the host cell transcriptome (19), and major signaling alterations such as mitogen-activated protein kinase (MAPK) and NF-κB activation (20, 21). After entering the host cell, the bacterium resides within a membranous pouch called a Salmonella-containing vacuole (SCV), where it multiplies and survives from a few hours to several days (22, 23). Effector proteins such as SifA ensure formation of Salmonella-induced filaments (SIFs), which are membranous extensions of the SCV and are crucial for its maintenance and positioning (24). Together, these changes lead to S. Typhimurium invasion and induced inflammation. How the pathogen is able to modulate these changes in the host has been a longstanding question. Understanding the role of SUMOylation machinery in S. Typhimurium infection may potentially help us to unravel these unanswered questions.
Here, using a multipronged approach, we provide evidence that S. Typhimurium utilizes a unique mechanism involving microRNAs (miRNAs) to alter the host SUMOylome for improving its capacity for intracellular survival.
MATERIALS AND METHODS
Materials.
All chemicals unless otherwise specified were obtained from Sigma-Aldrich. Antibodies directed against SUMO1 (catalog no. S8070), actin (catalog no. A3853), LAMP1 (catalog no. L1418), RANGAP1 (catalog no. R0155), Sae2 (catalog no. S7824), and Ubc-9 (catalog no. U2634) were obtained from Sigma-Aldrich. Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (catalog no. 39-8600) and anti-SUMO2/3 (catalog no. 51-9100) horseradish peroxidase (HRPO)-conjugated anti-rabbit (catalog no. 65-6120) and HRPO-conjugated anti-mouse (catalog no. 62-6520) antibodies were obtained from Invitrogen. Anti-p65 (catalog no. MA5-15563 and PA516545), anti-PIAS1 (catalog no. PA5-20951), anti-PIASy (catalog no. PA5-29146), and anti-peroxisome proliferator-activated receptor gamma (anti-PPARγ) (catalog no. MA5-15003) antibodies were procured from Thermo Scientific.
Methods. (i) Tissue culture and bacterial strains.
HCT-8 intestinal epithelial cells (ATCC, Manassas, VA) (passages 2 to 25) were cultured in RPMI medium supplemented with 14 mM NaHCO3, 15 mM HEPES buffer (pH 7.4), 2 mM glutamine, 1 mM sodium pyruvate, 40 mg/liter penicillin, 8 mg/liter ampicillin, 90 mg/liter streptomycin, and 10% fetal bovine serum. J774 and HeLa cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 14 mM NaHCO3, 15 mM HEPES buffer (pH 7.5), 40 mg/liter penicillin, 8 mg/liter ampicillin, 90 mg/liter streptomycin, and 10% fetal bovine serum. Cells were treated with different pharmacological inhibitors (20 μM cycloheximide, 20 μM MG132, or 5 μg/ml actinomycin D) 1 h before infection.
S. Typhimurium strain SL1344 is referred to as wild type throughout the study. All mutants are from the same background. Strains were grown in Luria broth (LB) at 37°C aerobically for 4 to 5 h followed by growth under stationary and hypoxic conditions overnight. These were then used to infect epithelial cells at a multiplicity of infection (MOI) of 1:40. Bacteria were heat killed by being heated at 95°C for 10 min. S. Typhimurium expressing mCherry was constructed by transforming S. Typhimurium with pFPV-mCherry procured from Addgene (Addgene plasmid 20956). SL1344 and Escherichia coli were obtained from Beth McCormick. pCDNA/Ubc-9 (Edward Yeh; Addgene plasmid 20082), pCDNA-SUMO2-HA (Addgene plasmid 48967), and pEYFP-SUMO1 (Addgene plasmid 13380) were procured from Addgene (25, 26).
(ii) In vivo S. Typhimurium infection model in mice.
Six- to eight-week-old female C57BL/6 mice were infected using the streptomycin pretreatment model (27, 28). Water and food were withdrawn 4 h prior to treatment with 3.75 mg of streptomycin intragastrically. The following day, water and food were again withdrawn for 4 h before the mice were infected with 5 × 107 CFU of SL1344 (500 μl of a suspension in phosphate-buffered saline [PBS] intragastrically by gavage). Mice were left for 24 h before euthanization and removal of the colon and cecum for examination. Experiments were carried out in the Small Animal Facility of the National Institute of Immunology (NII). The study was approved by the NII Institutional Animal Ethics Committee (approval no. IAEC#331/14).
(iii) Transfection of epithelial cells.
HCT-8 and HeLa cells were transfected as described earlier (29). One day before transfection, 2.5 × 105 cells were plated in 24-well plates to obtain 70 to 80% confluence and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. Briefly, 1 μg of plasmid or 25 pmol of small interfering RNA (siRNA) (Dharmacon) was diluted in Opti-MEM (Invitrogen, Carlsbad, CA). Separately, Lipofectamine 2000 or DharmaFECT (Dharmacon), respectively, was also diluted and incubated at room temperature for 5 min. Following incubation, the two mixtures were combined and incubated at room temperature for 20 min. This cocktail was added to cells with Opti-MEM and incubated without selection for 24 h.
(iv) Cellular global SUMOylation profiling assay.
S. Typhimurium (wild type [WT] or mutants) was grown and used to infect HCT-8 cells as described above. Following incubation of cells with bacteria for 1 h, the unbound bacteria were washed off and the infection was allowed to proceed for the appropriate time interval. At the end of the infection period, the cells were lysed in Laemmli's buffer separated through a 12% polyacrylamide gel (Bio-Rad, Hercules, CA) by electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Immunoblotting assays were carried out at optimized dilutions for various antibodies. Goat anti-rabbit and rabbit anti-mouse IgG labeled with horseradish peroxidase (Invitrogen) and diluted 1:10,000 was used to detect the bands, which were visualized by enhanced chemiluminescence using Clarity Western ECL substrate (Bio-Rad).
(v) Immunoprecipitation.
HCT-8 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and debris was removed by centrifugation at 13,000 rpm for 10 min at 4°C. The lysates obtained were incubated with protein A-agarose (Sigma) beads for 30 min at 4°C on an end-to-end rotor, followed by centrifugation to remove the beads and the non-specifically bound proteins. The precleared lysate was then used to immunoprecipitate SUMO1- or SUMO2/3-conjugated proteins by incubation with their respective antibodies overnight at 4°C on an end-to-end rotor. As controls, IgGs raised in mouse and rabbit were also used. The antibody-bound proteins were then captured using protein A-agarose beads and washed five times with RIPA buffer followed by boiling in Laemmli's buffer.
(vi) qPCR.
Total RNA from the HCT-8 cells was isolated using the NucleoSpin RNA II kit (Macherey-Nagel, Germany) according to the manufacturer's protocol. One microgram of each total RNA sample was used to synthesize cDNA using the i-Script cDNA synthesis kit (Bio-Rad, USA). The real-time PCR was performed using a 20-μl reaction volume on 96-well plates by using KiCq Start Sybr green quantitative PCR (qPCR) Ready mix (Sigma, USA) according to the manufacturer's instructions on the Bio-Rad CFX 96 real-time detection system. The PCRs were initiated by a denaturation step of 2 min at 95°C, followed by 40 cycles of amplification, which were performed according to the following thermocycling profiles: denaturation for 5 s at 95°C, annealing at 60°C for 10 s, and extension for 15 s at 72°C. Fluorescence data were acquired during the last step. A dissociation protocol with a gradient (0.5°C every 30 s) from 65°C to 95°C was used to investigate the specificity of the qPCRs and the presence of primer dimer. All reactions were done in triplicate, and all experiments were repeated at least three times. Levels of mRNA were quantified based on the ratio of the Ubc-9, PIAS1, Sae2, NF-κB, or SENP (sentrin-specific protease) gene mRNA to GAPDH and 18S rRNA using the cycle threshold (2−ΔΔCT) method as described in the detection system. Primer sequences are provided in the supplemental material.
(vii) Immunofluorescence.
Cells were grown in chambered slides (Eppendorf). For live-cell imaging of cells expressing yellow fluorescent protein (YFP)-SUMO1 and infected with mCherry-labeled bacteria, cells were directly visualized under the microscope. Quantitation was carried out manually for the number of bacteria present within each cell. For fixed-cell imaging postinfection, cells were washed two times in 1× PBS and fixed in 2.5% paraformaldehyde (15 min), pH 7.6, at room temperature (RT) followed by washing in PBS. Cells were permeabilized by incubation in PBS containing 0.1% saponin and 1% bovine serum albumin (PSB buffer) for 1 h. For immunostaining for LAMP1, cells were incubated with polyclonal rabbit anti-LAMP1 (1:800; Sigma; prepared in PSB buffer) for 1 h at RT and then washed three times and incubated with Cy5-conjugated goat anti-rabbit immunoglobulin G (1:800; Jackson Laboratory) for a further 1 h. Cells were washed three times and incubated in 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; Sigma) for 5 min. Coverslips were mounted using mounting medium (Sigma). The cells were observed in a Leica confocal SP5 microscope using a 63× oil objective. Quantitation was carried out manually for the presence of SIFs.
(viii) Gentamicin protection assay (GPA).
HCT-8 cells were infected with S. Typhimurium or its mutants either with or without perturbation of SUMOylation for 1 h. The unbound bacteria were then washed off with 1× Hanks balanced salt solution (HBSS) followed by incubation in HBSS containing 100 μg/ml of gentamicin to kill any extracellular bacteria for the requisite time period. Following incubation, the HCT-8 cells were lysed using 0.1% Triton X-100 in PBS. Samples were serially diluted in sterile PBS and plated onto LB agar plates. CFU were calculated by counting the colonies obtained the next day. A countable range of 30 to 300 was utilized. To achieve this, multiplicities of infection (MOIs) were slightly altered when cell treatments rendered the obtained bacterial colony counts beyond this range.
(ix) miRNA analysis.
The primers to detect mature miRNAs and miRNA mimics and inhibitors were obtained commercially (Qiagen). Total RNAs containing small noncoding RNA species were isolated using the miRNeasy kit (Qiagen) and converted to total cDNA using the miScript II reverse transcriptase kit. Quantitative real-time PCR (qRT-PCR) was carried out using the miScript SYBR green PCR kit (Qiagen) according to the manufacturer's instructions on the Bio-Rad CFX 96 real-time detection system (Bio-Rad, USA). Initial activation was carried out at 95°C for 15 min followed by 40 cycles of a 3-step cycle of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 70°C for 30 s. Transfection of mimics and inhibitors was carried out using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions. Briefly, 2 × 105 cells were seeded per well in 500 μl of culture medium containing serum. Mimics (30 nM) and inhibitors (50 nM) were diluted in 100 μl of Opti-MEM along with 3 μl of HiPerFect reagent per well and incubated at RT for 5 to 10 min. Following incubation, the complexes were added to cells plated on the wells dropwise and incubated for 24 h before experimentation.
(x) Statistics.
All results are expressed as the mean ± standard error from an individual experiment done in triplicate. P values were calculated according to Student's t test, and values of <0.05 were considered statistically significant.
RESULTS
Global alteration of host SUMOylation during S. Typhimurium infection.
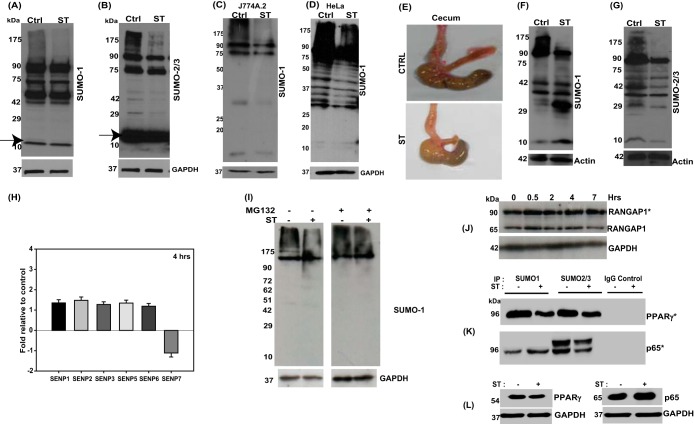
In our efforts to identify any role of PTM, such as SUMOylation, in Salmonella biology, we carried out cellular global SUMOylation profiling (CGSP) assays. In these assays, we infected human colonic cell line HCT-8 with a wild-type S. Typhimurium strain (SL1344) and allowed the infection to proceed for 4 h. This was followed by cell lysis in the presence of N-ethylmaleimide (NEM; deSUMOylase inhibitor) and immunoblotting using anti-SUMO1 (Fig. 1A) or anti-SUMO2/3 antibodies (Fig. 1B). The difference between control and infected cell lysates was apparent in the entire blot length but more pronounced at higher molecular weights. This reflected the fact that a large portion of SUMOylated proteome (here referred to as the SUMOylome) was affected. Similar patterns of alteration were also observed in other cell lines tested, including murine macrophage cell line J774A.2 and HeLa cells (Fig. 1C and D). In order to validate these findings in an in vivo context, we used a mouse model of Salmonella colitis (27). Streptomycin-pretreated C57BL/6 female mice were infected with S. Typhimurium for 24 h, after which they were euthanized and dissected. The ceca and the colons of infected animals were massively inflamed (Fig. 1E). Immunoblotting of the epithelial cell preparation (28) from colon (proximal) revealed, similarly to the cell culture experiment, a decrease in the global SUMOylomes. SUMO1 (Fig. 1F)- and SUMO2/3 (Fig. 1G)-conjugated proteomes (SUMO1ylome and SUMO2/3ylome, respectively) in the infected samples were lower than in the uninfected controls. Together, our in vitro and in vivo analyses reveal that S. Typhimurium alters both SUMOylomes but, however, to different extents. This observation is not unexpected, given that SUMO1 and SUMO2/3 are known to have their specific pools of target proteins with some overlaps (25).
FIG 1.
Salmonella infection leads to alteration in levels of SUMO-conjugated proteins in vitro and in vivo. (A and B) Comparison of cellular global SUMOylation profiling (CGSP) from uninfected human intestinal epithelial cells, HCT-8 (Ctrl), with those infected with S. Typhimurium SL1344 (ST) for 4 h. Lysates were analyzed by immunoblotting for SUMO1 (A) and SUMO2/3 (B) levels. GAPDH was used for normalization. The arrow indicates free SUMO. (C and D) CGSP assays were carried out for 4 h, and the lysates were immunoblotted using SUMO1 in J774A.2 macrophages (C) or HeLa cells (D). Numbers to the left are molecular masses in kilodaltons (also in panels F, G, I, and L). GAPDH was used for normalization. (E) Gross morphology of ceca of mice infected with S. Typhimurium (bottom) or untreated controls (Ctrl). (F and G) Immunoblot analysis of lysates prepared from mouse epithelial cells isolated from the proximal colon and probed for SUMO1 (F) and SUMO2/3 (G). Beta-actin was used for normalization. (H) Quantitative real-time PCR was carried out for all six known SENPs upon infection with S. Typhimurium for 4 h. GAPDH and 18S rRNA were used for normalization. (I) A CGSP assay was carried out in the presence of MG132, and the blot was probed for SUMO1. GAPDH was used for normalization. (J) SUMO1-conjugated and non-SUMOylated fractions of RANGAP1 over different time points of infection. GAPDH was used for normalization. (K) Samples immunoprecipitated for SUMO1- and SUMO2-conjugated proteins were probed with anti-PPARγ (top panel) and anti-RelA (bottom panel) antibodies. The asterisk indicates the SUMO-conjugated forms (J and K). Immunoprecipitation (IP) was also carried out using isotype control (IgG control). (L) Input lysates were probed with respective antibodies as well as GAPDH for normalization.
To understand the reason for downregulation of the SUMOylomes, we looked at the levels of known cellular deSUMOylases (SENPs) (26) using quantitative real-time PCR (qRT-PCR). We found no significant increase in any of the six SENPs tested in response to S. Typhimurium infection (Fig. 1H). This was further corroborated by the absence of increase in free SUMO in the immunoblots (Fig. 1A, lane 1 versus lane 2, arrow). Further, we examined the SUMOylomes in the presence of MG132, a well-known inhibitor of the proteasomal degradation machinery. We found that in the absence of proteasomal activity, S. Typhimurium mediated degradation of SUMOylome was compromised (Fig. 1I, lane 2 versus lane 4), indicating a role for the proteasomal machinery in SUMOylome depletion during S. Typhimurium infection. The phenomenon of S. Typhimurium-mediated SUMOylome downregulation was probed in greater detail, and we analyzed the SUMOylation status of SUMO-regulated proteins in the context of S. Typhimurium infection. RANGAP1, a nuclear pore-associated GTPase-activating protein known to be constitutively conjugated to SUMO1 (30), exhibited no change in its SUMO1-modified pools over several time points (Fig. 1J). On the other hand, p65, a component of the master regulator NF-κB, undergoes a depletion in its SUMO2- as well as an increase in the SUMO1-conjugated pool during S. Typhimurium infection (Fig. 1K, bottom panel). This implies a difference in the levels of the various SUMOylated pools of p65 which could potentially have different consequences. PPARγ, a nuclear receptor involved in inflammation, on the other hand exhibited a decrease in both its SUMO1- and its SUMO2/3-conjugated pool upon S. Typhimurium infection (Fig. 1K, top panel). The input lysates were probed for basal levels of both PPARγ and p65 (Fig. 1L), and GAPDH was used as a loading control (Fig. 1L). Together, these data indicate that the S. Typhimurium-mediated downregulation of the SUMOylome was a discriminatory process. This was further evident also from SUMO1 immunoblotting assays of infected animals versus control animals (Fig. 1F), in which we could see that a large fraction of the SUMOylome was reduced upon infection but that certain bands also displayed an increase in appearance. These results suggest that only a distinct subset of the SUMOylome was affected by S. Typhimurium. Keeping in view that even small quantities of substrate SUMO modification can have profound biological consequences (31), we went ahead and probed this process in greater detail.
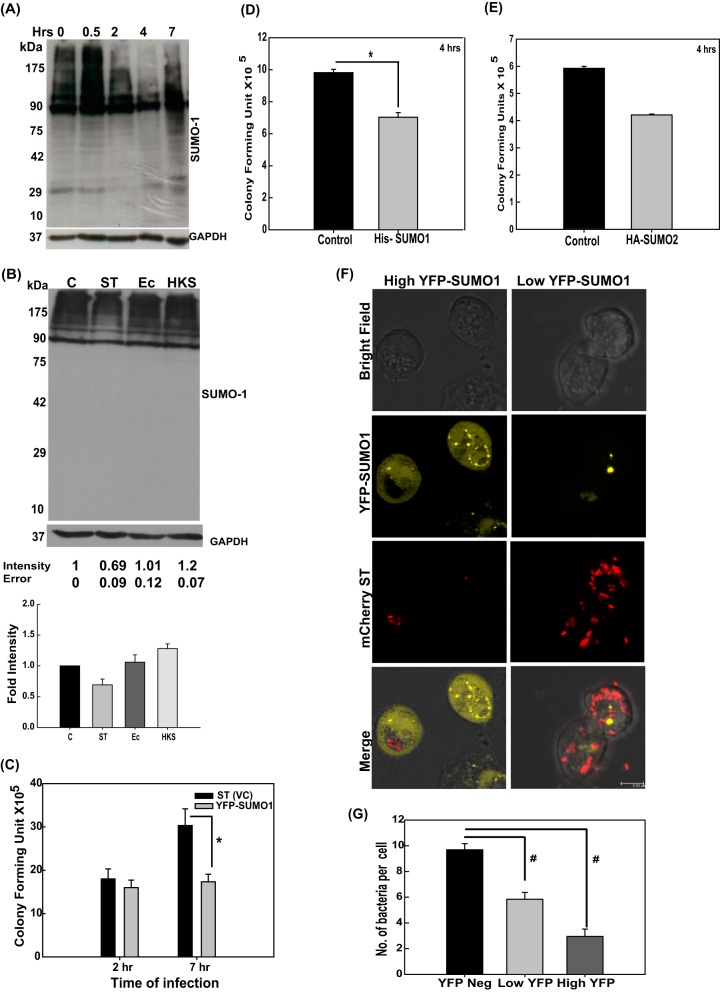
In order to explore the progression of the S. Typhimurium-mediated alteration of the SUMO1ylome in intestinal epithelial cells, a time course CGSP analysis was carried out. The results revealed that the modulation of the host SUMOylome was a dynamic process (Fig. 2A). At early stages of infection (30 min), the level of the SUMO1ylome was upregulated, being similar to the host response seen by others in the case of heat shock (32), followed by a gradual decrease, most significant at 4 h, indicating it to be a result of the complex host-pathogen cross talk. At 7 h postinfection (hpi), however, the global SUMO conjugation profile was very similar to that of the control untreated sample, revealing that normalcy returns at later stages of infection (Fig. 2A). Next, we investigated the specificity of the mechanism using heat-killed S. Typhimurium (HKS) and live E. coli in a CGSP assay (Fig. 2B). Interestingly, treatment with E. coli and HKS (Fig. 2B, lanes 3 and 4) failed to cause the same level of SUMO1ylome alteration of the host that we observed with live S. Typhimurium (Fig. 2B, lane 2 versus lane 1), implying that entry of the bacterium is essential for observed alteration of the SUMOylome.
FIG 2.
S. Typhimurium-mediated alteration of SUMOylation is a dynamic and active process that affects intracellular growth of the bacterium. (A) A CGSP assay was performed for the indicated time of infection, and the lysate was blotted for SUMO1. (B) CGSP assay upon 4-h infection with S. Typhimurium (ST), heat-killed S. Typhimurium (HKS), and E. coli (Ec). GAPDH was used for normalization. Means ± standard errors of the means of densitometric analysis from three independent experiments have been indicated as well as plotted. Representative blots from two to three independent experiments are depicted. (C) Gentamicin protection assays (GPAs) were performed after S. Typhimurium infection in HCT-8 cells transfected with either pEYFP vector control plasmid (VC) or pEYFP-SUMO1, and CFU scores were plotted. Efficient transfection was confirmed by YFP fluorescence imaging and immunoblot analysis (data not shown). Means ± standard errors of the means from three independent experiments are shown. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05. (D and E) HCT-8 cells overexpressing SUMO1 via the pCDNAHis-SUMO1 construct (D) and SUMO2 via the pCDNA-HA-SUMO2 construct (E) were infected for 4 h with S. Typhimurium, GPAs were performed, and CFU were plotted. (F) HCT-8 cells were transfected with pEYFP-SUMO1 plasmids followed by infection with mCherry-labeled S. Typhimurium for 4 h, and immunofluorescence confocal imaging was carried out. Cells with high (left panels) and low (right panels) YFP fluorescence were analyzed for bacterial load. Representative images of three independent experiments are shown. (G) Quantitative representation (means ± standard errors of the means from three independent experiments) of number of S. Typhimurium bacteria per cell plotted for low- and high-YFP-SUMO1-expressing as well as nonexpressing cells. Statistical analysis was carried out using Student's t test. The number sign indicates a P value of ≤0.01.
These data led us to conclude that S. Typhimurium infection leads to alteration of host cell SUMOylation, a process that was dynamic and specific to live S. Typhimurium. We thus hypothesized that S. Typhimurium-mediated alteration of host SUMOylation machinery could be a crucial strategy employed by this pathogen to manipulate host cells.
SUMOylation plays a crucial role in Salmonella-host cross talk.
We next set forth to examine if SUMO alterations seen during infection actually influence the biology of S. Typhimurium pathogenesis in any way. Using SUMO1 expression plasmids, we transiently upregulated the host SUMO machinery so as to counter the effect of S. Typhimurium on the host SUMO status (25). SUMO1 overexpression is a commonly used method to upregulate the SUMO status of the host (25). In the SUMO1-overexpressing cells, we carried out gentamicin protection assays (GPAs; see Materials and Methods) to score for the bacterial load. We observed a substantial decrease in CFU in the case of SUMO1 upregulation at 7 h postinfection (Fig. 2C), while only a marginal difference was observed at 2 h postinfection. This revealed that optimal SUMO1 status of the host is an important parameter for S. Typhimurium to cause successful infection, particularly at later time points. The same results were obtained with an alternate SUMO1-expressing construct, pCDNAHis-SUMO1, 4 h after infection (Fig. 2D). We also observed a similar restriction of bacterial infection at 4 h postinfection, upon upregulation of SUMO2 (using plasmid pCDNA-HA-SUMO2; Addgene catalog no. 48967), albeit less pronounced (Fig. 2E).
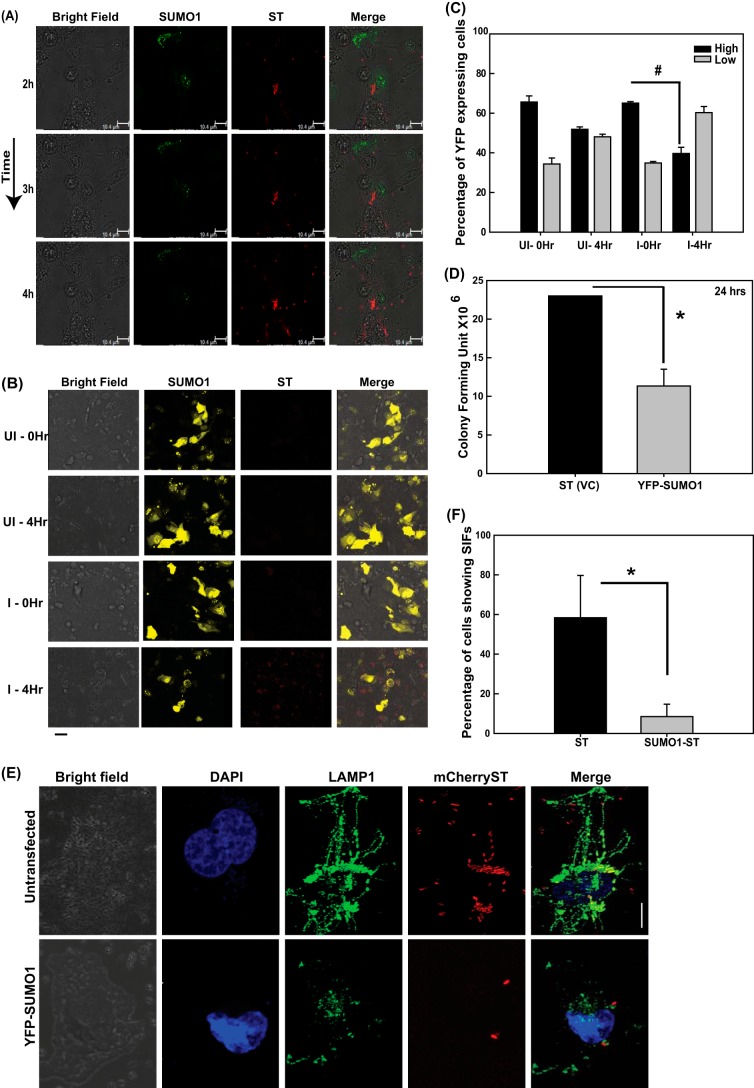
To further consolidate these observations, we carried out high-resolution confocal microscopy (HRCM) using IECs that have upregulated SUMO1 levels (using pEYFP-SUMO1) and that had been infected with S. Typhimurium expressing mCherry (using pFPV-mCherry plasmid). In our analysis, we deliberately focused on both cells with high levels of YFP-SUMO1 and those with very low levels of YFP-SUMO1 (Fig. 2F and G). As expected, a marked decrease in the number of Salmonella cells was observed in cells with upregulated SUMO1 levels (Fig. 2F, left panels). On the other hand, the cells with lower YFP-SUMO1 levels (Fig. 2F, right panels) had significantly larger numbers of S. Typhimurium cells. In order to understand the cause-versus-consequence relationship between alteration in SUMOylation levels and bacterial infection, we carried out time-lapse imaging of YFP-SUMO1-expressing cells infected with mCherry-labeled S. Typhimurium. Three well-spaced time points were chosen to observe the fluorescence of YFP and mCherry while negating photobleaching. We observed that YFP-SUMO1 fluorescence diminished with time in S. Typhimurium-infected samples, while that of mCherry expression by S. Typhimurium remained unchanged. This was accompanied by bacterial multiplication (Fig. 3A). To further address any issues regarding bleaching of YFP, we expressed YFP-SUMO1 in HCT-8 cells and analyzed fluorescence and cell number during the course of infection. Quantitation of cells with high YFP-SUMO1 levels in infected versus control samples at the start of the experiment and at 4 h revealed a significant decrease in YFP expression in infected cells but not in the control cells (Fig. 3B and C). This clearly implied that the bacterium lowers the host SUMOylation levels and promotes its own intracellular replication (Fig. 2C to G and 3A to C).
FIG 3.
S. Typhimurium modulates the SUMOylation status of the host for SCV maturation, thus affecting long-term intracellular growth. (A) Time-lapse confocal imaging was carried out at 2 h, 3 h, and 4 h postinfection in HCT-8 cells overexpressing pEYFP-SUMO1 upon infection with mCherry-labeled S. Typhimurium (ST). (B) Confocal microscopy was carried out before and 4 h after infection with mCherry-labeled S. Typhimurium of HCT-8 cells overexpressing YFP-SUMO1. Bar, 50 μm. Representative images of three or more independent experiments are shown. (C) Quantitative representation (means ± standard errors of the means from three independent experiments, depicted in panel B) of number of high-YFP-expressing (intense and all over the cell) and low-YFP-expressing (mild and localized only to the nucleus) HCT-8 cells transfected with pEYFP-SUMO1, before and after infection with S. Typhimurium for 4 h. I, infected; UI, uninfected. Statistical analysis was carried out using Student's t test; #, P ≤ 0.01. (D) HCT-8 cells transfected with pEYFP vector control (VC) plasmid or pEYFP-SUMO1 were infected with S. Typhimurium for 24 h, and CFU were plotted. Mean values ± standard errors of the means from three independent experiments are shown. Statistical analysis was carried out using Student's t test; the asterisk indicates a P value of ≤0.05. (E) Confocal microscopic analysis of HeLa cells with (bottom panel, YFP-SUMO1) or without (top panel, untransfected) SUMO1 upregulation infected with mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs). Bar, 5 μm. (F) Quantitative representation (means ± standard errors of the means from three independent experiments) of percentages of cells displaying intact SIFs. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05.
Further, we looked at the long-term effect of SUMOylome perturbation by carrying out GPA at 24 h postinfection upon upregulation of the host SUMO1ylome via the plasmid pEYFP-SUMO1. Similar to our observation at 7 hpi, even at later time points, lower CFU were obtained in SUMO1-overexpressing cells than in control cells (Fig. 3D), highlighting the significance of the alteration.
The intracellular multiplication of the bacterium occurs in the SCV, a membrane-bound compartment where S. Typhimurium thrives and multiplies within the host (22). We thus probed the maturation of the SCV to understand the mechanism of the observed compromised intracellular multiplication of S. Typhimurium in SUMO1-upregulated cells.
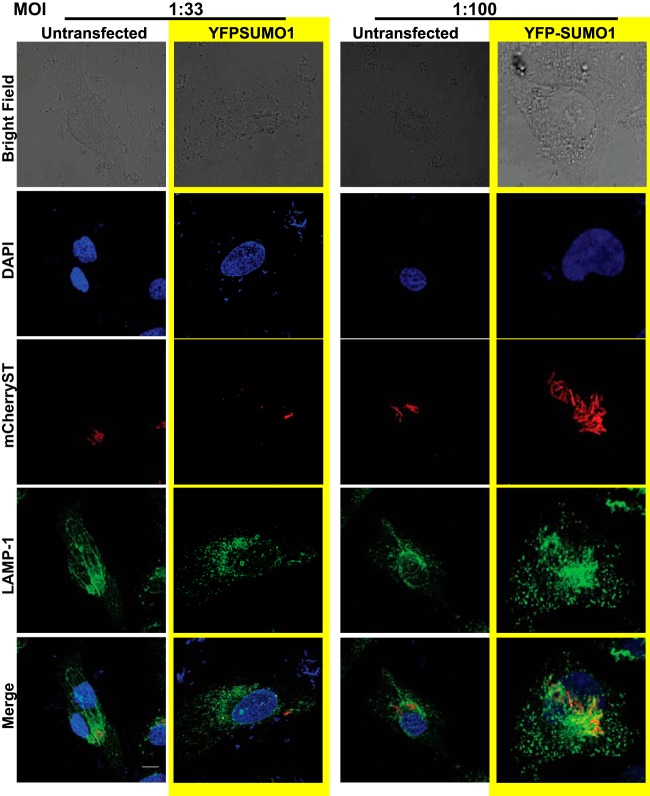
The SCVs are usually decorated by several lysosomal proteins at different phases of infection. LAMP1 is one such protein that is recruited during late stages of infection and is a crucial determinant of the fate of SCVs (33). Salmonella-induced filaments (SIFs), which are required for SCV stability and maturation, are extensions that arise from the SCVs and thereby are also decorated with LAMP1 (34). SUMO-perturbed and control cells were infected with mCherry-labeled S. Typhimurium for 7 h, and intracellular localization of LAMP1, as a marker of SIFs, was monitored by confocal microscopy. In control cells, LAMP1-stained SCVs and SIFs were distinctly visible (Fig. 3E, top panels). In contrast to this, in SUMO1-upregulated cells (as was done earlier in Fig. 2F), there were fewer S. Typhimurium cells in the LAMP1-stained compartment in concordance with our previous results, with a significantly compromised SIF formation (Fig. 3E and F, bottom panels). We inferred from the above imaging and GPA data that compromised intracellular survival of S. Typhimurium in SUMO-upregulated cells results from unstable SCVs that are deprived of SIFs (Fig. 3F). Since the YFP-SUMO1-expressing cells are seen with fewer S. Typhimurium cells, we wanted to address the obvious question of whether the low levels of SIFs were a result of the infection with fewer bacteria due to SUMO1 overexpression and not a direct consequence of the SUMO alteration. We therefore carried out infection at a higher multiplicity of infection (MOI) and looked at cells with higher numbers of S. Typhimurium cells to examine SIF formation. The data revealed that despite increasing the number of intracellular bacteria, the SIF formation was compromised in SUMO1-overexpressing cells, as there were no LAMP1-positive long tubular elongations (Fig. 4). Together, these data reveal that host SUMO status is critical for SIF formation and affects the overall course of infection by restricting intracellular bacterial multiplication.
FIG 4.
SUMOylation status of host affects SIF formation. Confocal microscopic analysis of HeLa cells with (YFP-SUMO1) or without (Untransfected) SUMO1 upregulation infected with different multiplicities of infection (MOIs) of mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs). Bar, 5 μm.
S. Typhimurium targets multiple components of the SUMO machinery to modulate host SUMOylation.
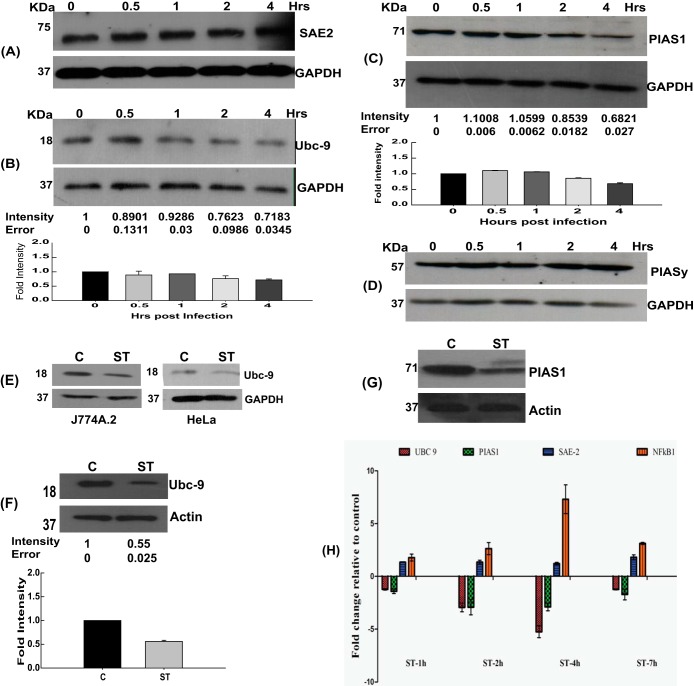
Having established the SUMO alteration to be an essential component of the host-pathogen interaction, it was important to understand how S. Typhimurium achieves it. Owing to the global scale of the remodeling of the SUMOylome, we examined the status of the SUMOylation machinery components, including Sae1/2, Ubc-9, and some crucial E3 ligases. HCT-8 cells infected for different time periods were lysed and immunoblotted for the components of the SUMOylation machinery. Remarkably, Ubc-9 was significantly decreased in infected samples (Fig. 5B), while other components of the SUMOylation machinery, such as Sae2 (Fig. 5A) and PIASy (Fig. 5D), remained unaltered. S. Typhimurium-dependent depletion of Ubc-9 levels was also seen 4 h postinfection in other cell lines, J774 and HeLa, again pointing toward a conservation in mechanism in a wide array of cell types (Fig. 5E). Levels of PIAS1, an E3 ligase and an inhibitor of STAT1 (35), were also decreased upon infection (Fig. 5C). When we investigated the expression of SUMOylation pathway enzymes in the animal model, we found that, similarly to the cell culture system, the in vivo model also revealed downregulation of Ubc-9 and PIAS1 pathway enzymes (Fig. 5F and G). We next set forth to probe the consequences of S. Typhimurium infection at the transcriptional level, if any, of the SUMO pathway genes. We observed a significant downregulation of the mRNA level of Ubc-9 along with PIAS1 but not that of the E1 enzyme-encoding Sae2 gene, which remained constant. NF-κB, known to be upregulated upon S. Typhimurium infection, was used as a positive control (Fig. 5H). Interestingly, the levels of Ubc-9 mRNA returned to near the control level at 7 h (Fig. 5H), further consolidating the dynamic modulation of host SUMOylation via the host SUMOylation machinery.
FIG 5.
Salmonella targets multiple genes of SUMOylation machinery. (A to D) Immunoblot analysis of HCT-8 cells with or without infection with S. Typhimurium at different time points, probing for Sae2 (A), Ubc-9 (B), PIAS1 (C), or PIASy (D). GAPDH was used for normalization. Representative blots of three or more independent experiments are shown. Means ± standard errors of the means of densitometric analysis from three or more independent experiments are shown numerically (middle) as well as graphically (bottom). (E) A CGSP assay was carried out for 4 h, and the lysates were immunoblotted for Ubc-9, in the J774A.2 macrophage cell line (left) or HeLa cells (right). GAPDH was used for normalization. Numbers at left in panels E to G are molecular masses in kilodaltons. (F and G) Immunoblot analysis of lysates prepared from mouse epithelial cells isolated from the proximal colon and probed for Ubc-9 (F) and PIAS1 (G). Beta-actin was used for normalization. Means ± standard errors of the means of densitometric analysis from three or more independent experiments are shown numerically (middle) as well as graphically (bottom). (H) mRNA expression levels as represented by relative fold differences against uninfected control cells were plotted for the indicated genes: the Ubc-9, PIAS1, Sae2, and NF-κB genes. GAPDH and 18S rRNA were used for normalization.
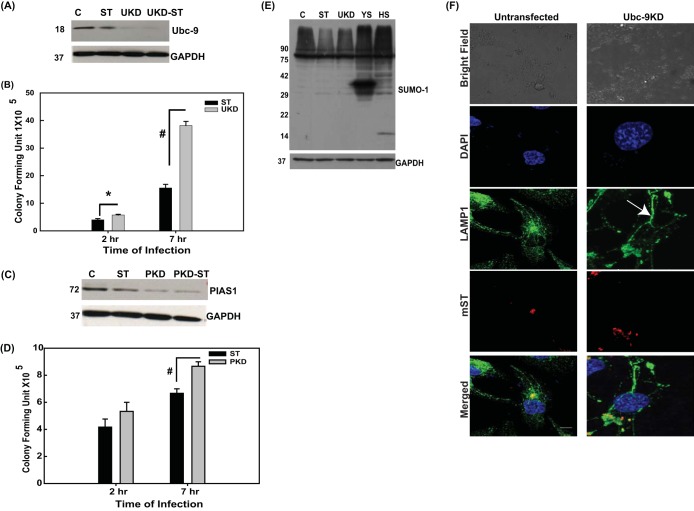
It was also important to see if downregulation of Ubc-9 actually accounts for SUMOylome downregulation and if that can lead to the corresponding effects on Salmonella infection observed above upon SUMO1 overexpression. Since Ubc-9 is an essential gene, in order to have minimal effect on overall fitness of the host cells, we transiently transfected cells with Ubc-9 siRNA, which led to significant downregulation of expression of the encoded protein as revealed by immunoblotting assays (henceforth called Ubc-9KD cells [Fig. 6A]). We observed that downregulation of Ubc-9 was sufficient to bring about a decrease in global SUMO1 proteome profile similar to that observed upon S. Typhimurium infection (Fig. 6E, lane 3). As expected in cells transfected with plasmid pEYFP-SUMO1 (YS) or pCDNAHis-SUMO1 (HS), we saw an increase in the SUMO1ylome profile in these samples (Fig. 6E, lanes 4 and 5). Infection of Ubc-9KD cells with S. Typhimurium resulted in better infection than that in control as revealed by GPA (Fig. 6A and B). CFU in Ubc-9KD samples at 7 h postinfection (Fig. 6A and B) were about 3-fold higher than that in the control samples. Upon PIAS1 knockdown (Fig. 6C and D), however, the increase in infectivity was not so pronounced. At 2 h after infection of the Ubc-9KD cells (Fig. 6A and B), the CFU were only slightly higher than the control samples, as seen with SUMO1 upregulation (Fig. 2C), again reiterating that the effect of SUMO perturbation mediated via Ubc-9 downregulation was more pronounced on intracellular bacterial multiplication than for the entry process per se. In addition, post-Ubc-9KD, the SIF formation was comparable to control, if not higher, as can be observed by the LAMP1-positive tubular structures (Fig. 6F). These data together point toward a role of Ubc-9 downregulation in concurrence with quick cellular turnover of SUMOylated proteins, in bringing down the host SUMOylation. These profound effects of Ubc-9 depletion made us probe deeper into the mechanism of S. Typhimurium-mediated Ubc-9 depletion.
FIG 6.
Downregulation of Ubc-9 is sufficient to alter S. Typhimurium infection. (A and C) Immunoblot analysis of HCT-8 cells with or without downregulation of Ubc-9 (A) and PIAS1 (C) using siRNA and either infected with S. Typhimurium or left untreated. Numbers at left in panels A, C, and E are molecular masses in kilodaltons). GAPDH was used for normalization. Representative blots from three or more independent experiments are depicted. (B and D) GPA was performed in infected cells that were downregulated for either Ubc-9 (B) or PIAS1 (D). CFU were plotted after 2 h or 7 h of infection. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05, and the number sign indicates a P value of ≤0.01. (E) Immunoblot analysis of HCT-8 cells infected with S. Typhimurium, upon downregulation of Ubc-9 with siRNA (UKD) and upregulation of SUMO1 via pEYFP-SUMO1 (YS) or pCDNAHis-SUMO1 (HS). (F) Confocal microscopic analysis of HCT-8 cells with or without downregulation of Ubc-9 using siRNA, infected with mCherry-labeled S. Typhimurium (mST) for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs; arrow) and Salmonella-induced filaments (SIFs). Representative images of three independent experiments are depicted. Bar, 5 μm.
To probe the stage of synthesis of Ubc-9 at which down-modulation was taking place, we carried out S. Typhimurium infection on cells treated with inhibitors of proteasomal degradation (MG132), eukaryotic protein synthesis (cycloheximide [CHX]), and eukaryotic transcription (actinomycin D [AcT]). MG132 and CHX had no effect on S. Typhimurium-mediated Ubc-9 downregulation (Fig. 7A and B); however, treatment with AcT rescued the Ubc-9 degradation during infection (Fig. 7C), indicating a role for the eukaryotic transcription machinery. Together, these data suggested that S. Typhimurium-mediated downregulation of host SUMO machinery is mediated by a process that requires active transcription but acts by downregulating the mRNA transcripts of Ubc-9. Literature available on Ubc-9 downregulation suggests a role for microRNAs, particularly those belonging to members of the miR30 family, in certain cancers (36, 37). MicroRNAs (miRNAs) are small noncoding RNAs, known to play a crucial role in eukaryotic gene regulation, which are transcribed in the nucleus precursors that get processed by Drosha and Dicer into mature miRNA, which acts as a regulator of transcription and translation. For the current study, we chose two miRNAs, miR30c and miR30e, both experimentally validated to be miRNAs targeting Ubc-9 (37).
FIG 7.
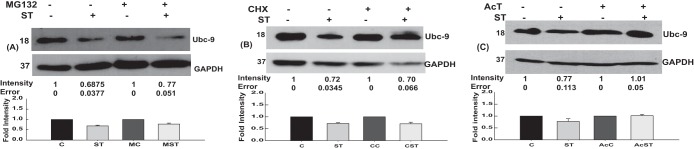
Mechanism of Ubc-9 downregulation. Mechanism of Ubc-9 downregulation was assessed via inhibitors of proteasome (MG132) (A), protein synthesis (cycloheximide) (B), and transcription (actinomycin D [AcT]) (C). Representative blots from three or more independent experiments are shown. Means ± standard errors of densitometric analysis results from three independent experiments have been indicated as well as plotted. Numbers at left of blots are molecular masses in kilodaltons.
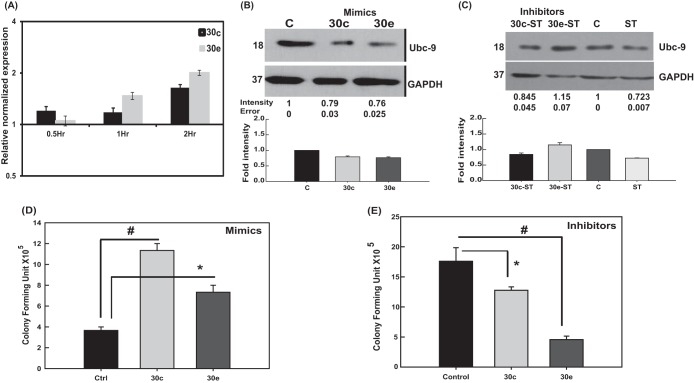
Control cells and cells infected with S. Typhimurium for different time points were quantified for the presence of miR30c and miR30e by qRT-PCR to check if they had any role in S. Typhimurium-induced Ubc-9 downregulation. S. Typhimurium infection resulted in upregulation of mature forms of miR30c and miR30e in a time-dependent manner (Fig. 8A). The miR30c and miR30e levels were upregulated from as early as 30 min following infection in comparison to control cells. To validate the findings, we transfected the cells with miRNA mimics specific for miR30c and miR30e and examined the expression levels of Ubc-9. We observed downregulation of the expression of Ubc-9, but not PIAS1 (data not shown), in the presence of miRNA mimics. The Ubc-9 downregulation was almost to the same extent as that seen during S. Typhimurium infection (Fig. 8B). In contrast, in the presence of the inhibitors, the S. Typhimurium-mediated Ubc-9 downregulation was significantly abrogated, especially for miR30e (Fig. 8C). These data suggested that miR30c and particularly miR30e were sufficient to downregulate Ubc-9. To test if the presence of miR30c and miR30e could actually alter the intracellular survival of S. Typhimurium, we examined the CFU by GPA in cells that were transfected with the miRNA mimics and inhibitors followed by S. Typhimurium infection. In the presence of miR30c and miR30e mimics, we observed more than a 2-fold increase in the CFU of S. Typhimurium at 7 h postinfection while the presence of inhibitors abrogated this effect (Fig. 8D and E). These experiments corroborated the idea that S. Typhimurium mediates Ubc-9 downregulation by upregulating members of the miR30 family of miRNAs.
FIG 8.
Salmonella engages members of the miR30 family to downregulate Ubc-9. (A) Quantitative real-time PCR analysis of mature miR30c and miR30e upon S. Typhimurium infection at different time points. SnoRNAs SNORD61 and RNU6 were used for normalization. A representative experiment of three replicates is depicted. (B) Immunoblot analysis of Ubc-9 was carried out in cells transfected with 30 nM concentrations of the mimics of miR30c and miR30e. GAPDH was used for normalization. Numbers at left of panels B and C are molecular masses in kilodaltons. (C) Ubc-9 levels were assessed in HCT-8 cells upon infection with S. Typhimurium with or without inhibitors of the miRNAs miR30c and miR30e. GAPDH was used for normalization. (D and E) GPA to assess bacterial load was performed after S. Typhimurium infection in HCT-8 cells transfected with mimics (D) or inhibitors (E) of miR30c and miR30e. CFU were plotted, and statistical analyses were carried out using Student's t test. The asterisk indicates a P value of ≤0.05, and the number sign indicates a P value of ≤0.01.
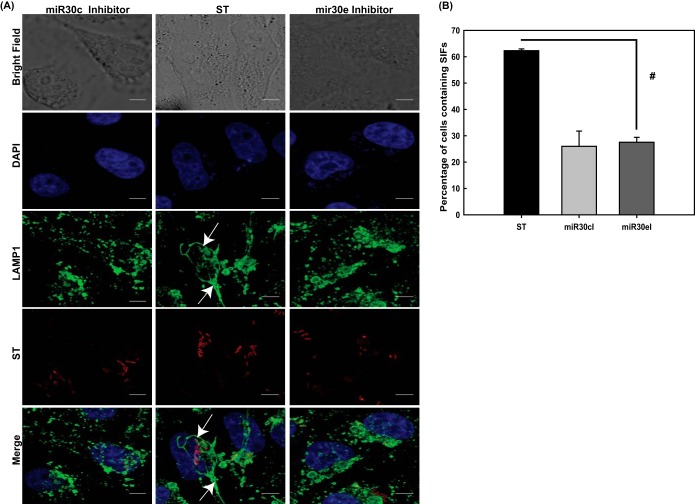
In order to validate the significance of the miRNA-mediated downregulation of Ubc-9 and its consequent effect on bacterial load within the cell, we treated HeLa cells with the inhibitors of these miRNAs and assessed the SIF formation 7 h after infection with S. Typhimurium. We observed that upon abrogation of Ubc-9 downregulation via inhibition of the miRNAs, the percentage of cells containing SIFs significantly decreased (Fig. 9). Thus, miRNA-mediated downregulation of Ubc-9 was essential for SIF formation and consequent multiplication of bacteria within them (Fig. 8B to E and 9).
FIG 9.
Inhibitors of miRNAs affect SIF formation. (A) Confocal microscopic analysis of control HeLa cells (middle panel) and HeLa cells transfected with inhibitors of miR30c (left panel) and miR30e (right panel) that were infected with mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs; arrows). Bars, 5 μm. (B) Quantitative representation (means ± standard errors of the means from three independent experiments) of percentages of cells displaying intact SIFs. Statistical analysis was carried out using Student's t test. The number sign indicates a P value of ≤0.01.
All these data point toward a concerted effort by the bacterium in altering the host SUMOylation during infection by targeting the key components of the machinery in order to promote its own intracellular proliferation.
DISCUSSION
Posttranslational modifiers such as ubiquitin and ubiquitin-like modifiers are related pathways that are involved in regulation of diverse cellular functions. The existence of intricate connections of host ubiquitination machinery with the infection process is well established in the case of S. Typhimurium infection (38). However, until our present work, detailed investigation of the role of SUMOylation in S. Typhimurium infection and associated cellular mechanisms somehow was left undone, while the cross talk of other pathogens with the host SUMOylation machinery has been shown to be a complex and recurring theme. For example, some viral proteins such as BZLF1 of Epstein-Barr virus (39) or ICP0 of herpes simplex virus (40) target specific host SUMOylated proteins. Other pathogens attack components of the SUMOylation machinery of the host, such as Gam1 of adenovirus, which targets and degrades E1 and E2 SUMO enzymes (41). Pore-forming toxins such as listeriolysin O (LLO) (Listeria monocytogenes), perfringolysin (PFO) (Clostridium perfringens), and pneumolysin (PLY) (Streptococcus pneumoniae) trigger the degradation of Ubc-9, altering the entire host SUMO status (2). In addition, some pathogen proteins like XopD of Xanthomonas euvesicatoria (6) or YopJ of Yersinia pestis (42) mimic host deSUMOylases. Thus, alteration of SUMOylation of the host protein/proteome is a phenomenon observed across the entire spectrum of pathogens that may help overcome cellular defenses or simply induce a more advantageous intracellular milieu. In order for the pathogens to cause an effective infection, complete control of several host machineries is required. Our studies point toward a large-scale alteration of the host cell SUMO proteome, a pool of which may be specifically required for its own needs, to promote a niche that is supportive of its intracellular replication and induced inflammation. This feature is a parallel of a recent elegant report which revealed a global host gene expression alteration during S. Typhimurium infection that led to better intracellular survival of the pathogen (19).
We observed that during infection with S. Typhimurium, the overall SUMOylome (both SUMO1 conjugated [SUMO1ylome] and SUMO2/3 conjugated [SUMO2/3ylome]) was reduced at early stages of infection, albeit specifically, since constitutively SUMO1-conjugated protein RANGAP1 was largely unaffected. Our observation that the SUMO alteration returns to normalcy fits with the characteristics of Salmonella infection. As seen in the case of other pathways, usually the damage caused during early stages of infection is repaired by the pathogen once it has invaded the host cell (14, 43). A pathway such as SUMOylation is central to all the fundamental pathways of the cell, and therefore, it is important for the pathogen to restore homeostasis once the invasion process is completed. The role of modulation of SUMOylation may be to set the stage for subsequent steps of infection. One of the most compelling aspects connected to S. Typhimurium-mediated SUMOylation alteration was the observation that these changes actually led to significant improvement in the bacterial survival inside the host cells. Our data revealed that the bacterial survival was inversely correlated with the host SUMO status. Notably, not much is known about the connection of SUMOylation and the biology of intracellular pathogens. In the case of Ehrlichia chaffeensis, however, a clear connection between the intracellular survival and host SUMOylation was described (10). However, in contrast to S. Typhimurium, Ehrlichia chaffeensis does not dramatically alter the host SUMOylome and the host SUMO1 is beneficial for intracellular survival. Importantly, the bacterial effector TRP120 was demonstrated to be SUMOylated in vitro and in human cells (10). Similarly, a pathogen protein undergoing SUMO conjugation was also seen in the case of Anaplasma phagocytophilum effector AmpA (11). S. Typhimurium infection of SUMOylation-upregulated cells and the failure of SIF formation point toward the fact that crucial molecules required for SCV maintenance may be regulated by SUMOylation. SUMOylation-dependent regulation of mechanisms that control the intracellular survival of pathogens is an unexpected finding that now opens up a whole new arena of research for S. Typhimurium and other intracellular pathogens.
The mechanism of S. Typhimurium alteration of the host SUMOylome involves primarily the downregulation of Ubc-9 and PIAS1. Ubc-9 being the sole conjugating enzyme within mammals and having the distinguishing characteristic of directly recognizing substrate proteins make it a crucial point of intervention for pathogens, as has been noted in the examples above. From an evolutionary perspective, proteins such as Ubc-9 are suitable targets for pathogens because of their multifunctional role. These proteins, in addition to their E2 enzyme activity, could potentially function as biological chaperones, transcriptional coregulators, and regulators of signaling pathways (44–47). The evolutionary conservation of Ubc-9 being at the center of many strategies to subvert host SUMO machinery is also mirrored in our observation regarding the mechanism of Ubc-9 degradation. S. Typhimurium-mediated downregulation of PIAS1, an E3 enzyme that has been known to be important for inflammatory pathways, is probably involved in the case of S. Typhimurium as an additional level of regulation required for a pathogen that causes inflammatory diarrhea. Unlike Ubc-9, PIAS1 has not been shown to be a target of any other SUMO-modulating pathogen as yet and is a novel finding of the present study. This modulation could be offering the pathogen a higher degree of control over the subset of the host SUMOylome that undergoes alteration. However, understanding its significance and implications in S. Typhimurium-host cross talk would require further studies.
We probed the mechanism of S. Typhimurium-mediated Ubc-9 downregulation in detail since Ubc-9 is the sole E2 enzyme in the SUMOylation pathway and can account for the global scale of changes observed in the SUMOylomes. Wu et al. (37), by utilizing DNA-demethylating agents such as 5-azadeoxycytidine and inhibitors of histone deacetylases such as trichostatin, did not observe a role of these epigenetic mechanisms in the regulation of Ubc-9 in cancers. They, however, did observe Ubc-9 to be a direct target of miR30c and miR30e, which were seen to be downregulated in tumor samples (37). Our results also revealed the involvement of miRNAs in downregulation of Ubc-9, a novel means of host SUMO enzyme regulation by any pathogen. Recently, the let-7 family of miRNAs were shown to be involved in S. Typhimurium-mediated cytokine production (48). An miRNA that was carried by S. Typhimurium was shown to suppress nitric oxide synthesis by the host and thereby facilitate invasion and intracellular survival (49). A high-content screening of miRNAs that could suppress S. Typhimurium growth in host cells identified the miR15 family (50). These and our study reveal that S. Typhimurium utilized the miRNA-mediated regulation as a potent weapon to combat host defense mechanisms and to alter host signaling for efficient infection.
Cells that were treated with inhibitors of miR30e displayed abrogation of S. Typhimurium-mediated Ubc-9 depletion, thus indicating that S. Typhimurium-mediated Ubc-9 depletion works via the miRNA machinery. The present study is the first to address the all-round role of host SUMOylation in Salmonella infection and induced inflammation. Moreover, it reveals considerable new insights about the cellular mechanisms, both bacterial and host driven, that are operational during complex host-pathogen cross talk.
Supplementary Material
ACKNOWLEDGMENTS
We thank Beth McCormick, University of Massachusetts Medical School, Worcester, MA, for all the S. Typhimurium strains and Bobby Cherayil, Harvard Medical School, Boston, MA, for all the discussions and feedback while the manuscript was being prepared. We thank Satyajit Rath, National Institute of Immunology, for his help.
Financial support was provided by a DBT Wellcome Trust fellowship and core funding from the UNESCO Regional Centre for Biotechnology, Faridabad, India.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00397-15.
REFERENCES
- 1.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. 2006. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol 62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 2.Ribet D, Cossart P. 2010. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desterro JM, Rodriguez MS, Hay RT. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2:233–239. doi: 10.1016/S1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 4.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 5.Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 6.Kim JG, Stork W, Mudgett MB. 2013. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13:143–154. doi: 10.1016/j.chom.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, Ozato K. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog 5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribet D, Hamon M, Gouin E, Nahori MA, Impens F, Neyret-Kahn H, Gevaert K, Vandekerckhove J, Dejean A, Cossart P. 2010. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature 464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritah S, Lhocine N, Golebiowski F, Mounier J, Andrieux A, Jouvion G, Hay RT, Sansonetti P, Dejean A. 2014. Sumoylation controls host anti-bacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep 15:965–972. doi: 10.15252/embr.201338386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyer AR, Truchan HK, May LJ, Walker NJ, Borjesson DL, Carlyon JA. 2015. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol 17:504–519. doi: 10.1111/cmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YG, Wu S, Xia Y, Chen D, Petrof EO, Claud EC, Hsu W, Sun J. 2012. Axin1 prevents Salmonella invasiveness and inflammatory response in intestinal epithelial cells. PLoS One 7:e34942. doi: 10.1371/journal.pone.0034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srikanth CV, Cherayil BJ. 2007. Intestinal innate immunity and the pathogenesis of Salmonella enteritis. Immunol Res 37:61–78. doi: 10.1007/BF02686090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 15.Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect 3:549–559. doi: 10.1016/S1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 16.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 17.Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis CL, Starnbach MN, Falkow S. 1992. Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol Microbiol 6:3077–3087. doi: 10.1111/j.1365-2958.1992.tb01765.x. [DOI] [PubMed] [Google Scholar]
- 19.Hannemann S, Gao B, Galan JE. 2013. Salmonella modulation of host cell gene expression promotes its intracellular growth. PLoS Pathog 9:e1003668. doi: 10.1371/journal.ppat.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen LM, Hobbie S, Galan JE. 1996. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 21.Patel JC, Galan JE. 2006. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol 175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrol ME, Jackett PS, Aber VR, Lowrie DB. 1979. Phagolysosome formation, cyclic adenosine 3′:5′-monophosphate and the fate of Salmonella typhimurium within mouse peritoneal macrophages. J Gen Microbiol 110:421–429. doi: 10.1099/00221287-110-2-421. [DOI] [PubMed] [Google Scholar]
- 23.Drecktrah D, Knodler LA, Howe D, Steele-Mortimer O. 2007. Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8:212–225. doi: 10.1111/j.1600-0854.2006.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Albert J, Yu XJ, Beuzon CR, Blakey AN, Galyov EE, Holden DW. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol 44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 25.Ayaydin F, Dasso M. 2004. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bekes M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. 2011. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem 286:10238–10247. doi: 10.1074/jbc.M110.205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pott J, Stockinger S, Torow N, Smoczek A, Lindner C, McInerney G, Backhed F, Baumann U, Pabst O, Bleich A, Hornef MW. 2012. Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS Pathog 8:e1002670. doi: 10.1371/journal.ppat.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srikanth CV, Wall DM, Maldonado-Contreras A, Shi HN, Zhou D, Demma Z, Mumy KL, McCormick BA. 2010. Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science 330:390–393. doi: 10.1126/science.1194598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matunis MJ, Coutavas E, Blobel G. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hay RT. 2005. SUMO: a history of modification. Mol Cell 18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. 2009. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-del Portillo F, Finlay BB. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J Cell Biol 129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson LK, Nawabi P, Hentea C, Roark EA, Haldar K. 2008. The Salmonella virulence protein SifA is a G protein antagonist. Proc Natl Acad Sci U S A 105:14141–14146. doi: 10.1073/pnas.0801872105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Shuai K. 2008. Targeting the PIAS1 SUMO ligase pathway to control inflammation. Trends Pharmacol Sci 29:505–509. doi: 10.1016/j.tips.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan J, Qiang B, Peng X. 2012. microRNA-214-mediated UBC9 expression in glioma. BMB Rep 45:641–646. doi: 10.5483/BMBRep.2012.45.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu F, Zhu S, Ding Y, Beck WT, Mo YY. 2009. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res 15:1550–1557. doi: 10.1158/1078-0432.CCR-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel JC, Hueffer K, Lam TT, Galan JE. 2009. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol 75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol 74:10006–10017. doi: 10.1128/JVI.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. 2004. A mechanism for inhibiting the SUMO pathway. Mol Cell 16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 43.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA, Yang C, Emili A, Philpott DJ, Girardin SE. 2012. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Liu LB, Omata W, Kojima I, Shibata H. 2007. The SUMO conjugating enzyme Ubc9 is a regulator of GLUT4 turnover and targeting to the insulin-responsive storage compartment in 3T3-L1 adipocytes. Diabetes 56:1977–1985. doi: 10.2337/db06-1100. [DOI] [PubMed] [Google Scholar]
- 45.Kurihara I, Shibata H, Kobayashi S, Suda N, Ikeda Y, Yokota K, Murai A, Saito I, Rainey WE, Saruta T. 2005. Ubc9 and protein inhibitor of activated STAT 1 activate chicken ovalbumin upstream promoter-transcription factor I-mediated human CYP11B2 gene transcription. J Biol Chem 280:6721–6730. doi: 10.1074/jbc.M411820200. [DOI] [PubMed] [Google Scholar]
- 46.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. 2010. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene 29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uemura A, Taniguchi M, Matsuo Y, Oku M, Wakabayashi S, Yoshida H. 2013. UBC9 regulates the stability of XBP1, a key transcription factor controlling the ER stress response. Cell Struct Funct 38:67–79. [DOI] [PubMed] [Google Scholar]
- 48.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. 2011. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J 30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu H, Zhang T, Li D, Liu F, Zhang CY, Zen K. 2014. A Salmonella-encoded microRNA-like RNA facilitates bacterial invasion and intracellular replication via suppressing host cell inducible nitric oxide synthase (MPF3P.809). J Immunol 192:132. [Google Scholar]
- 50.Maudet C, Mano M, Sunkavalli U, Sharan M, Giacca M, Forstner KU, Eulalio A. 2014. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun 5:4718. doi: 10.1038/ncomms5718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.