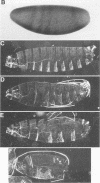
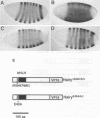
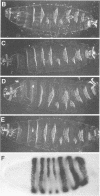
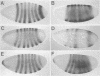
Abstract
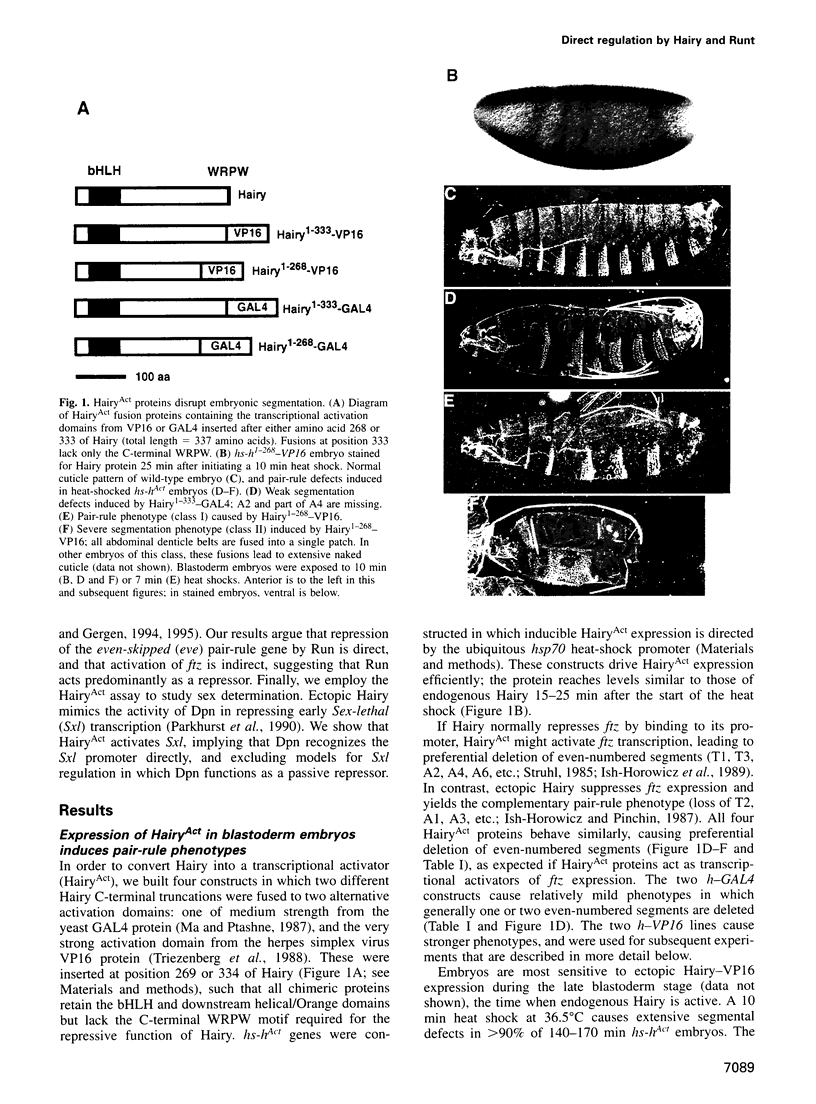
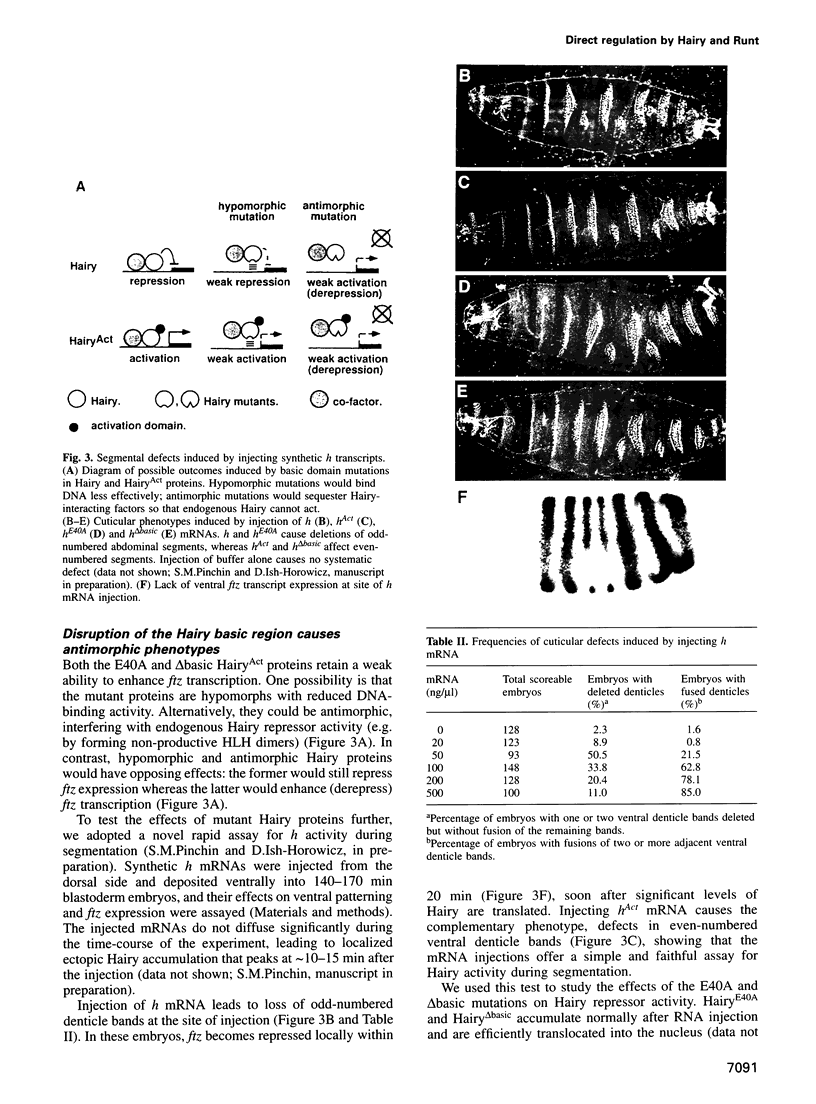
The Hairy and Runt pair-rule proteins regulate Drosophila segmentation by repressing transcription. To explore the ability of these proteins to function as promoter-bound regulators in vivo, we examined the effects of Hairy and Runt derivatives containing heterologous transcriptional activation domains (HairyAct and RunAct). Using this approach, we find that Hairy and Runt efficiently target such activation domains to specific segmentation gene promoters, leading to rapid induction of transcription. Our results strongly suggest that Hairy normally acts as a promoter-bound repressor of fushi tarazu, runt and odd-skipped, and that Runt directly represses even-skipped. We also show that expressing HairyAct in early blastoderm embryos causes ectopic Sex-lethal expression and male-specific lethality, implying that the Hairy-related denominator element Deadpan represses Sex-lethal during sex determination by directly recognizing the early Sex-lethal promoter.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akazawa C., Sasai Y., Nakanishi S., Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992 Oct 25;267(30):21879–21885. [PubMed] [Google Scholar]
- Anderson K. V., Nüsslein-Volhard C. Information for the dorsal--ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984 Sep 20;311(5983):223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- Badiani P., Corbella P., Kioussis D., Marvel J., Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994 Apr 1;8(7):770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Younger-Shepherd S., Jan L. Y., Jan Y. N. deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 1992 Nov;6(11):2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- Bopp D., Bell L. R., Cline T. W., Schedl P. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 1991 Mar;5(3):403–415. doi: 10.1101/gad.5.3.403. [DOI] [PubMed] [Google Scholar]
- Brennan T. J., Chakraborty T., Olson E. N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C. V., Alonso M. C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991 Oct;10(10):2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986 Apr 11;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Carroll S. B. Zebra patterns in fly embryos: activation of stripes or repression of interstripes? Cell. 1990 Jan 12;60(1):9–16. doi: 10.1016/0092-8674(90)90711-m. [DOI] [PubMed] [Google Scholar]
- Cline T. W. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 1993 Nov;9(11):385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- Coulter D. E., Swaykus E. A., Beran-Koehn M. A., Goldberg D., Wieschaus E., Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990 Nov;9(11):3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens D. J., Greaves R., Goding C. R., O'Hare P. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 1989 Aug;8(8):2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992 May 15;256(5059):1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- Dawson S. R., Turner D. L., Weintraub H., Parkhurst S. M. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995 Dec;15(12):6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearolf C. R., Topol J., Parker C. S. Transcriptional control of Drosophila fushi tarazu zebra stripe expression. Genes Dev. 1989 Mar;3(3):384–398. doi: 10.1101/gad.3.3.384. [DOI] [PubMed] [Google Scholar]
- Delidakis C., Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C., Preiss A., Hartley D. A., Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics. 1991 Nov;129(3):803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., Gergen J. P. The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev. 1991 Dec;5(12A):2176–2187. doi: 10.1101/gad.5.12a.2176. [DOI] [PubMed] [Google Scholar]
- Frasch M., Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987 Nov;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Goodfellow M. Great value in gainsharing. Health Syst Rev. 1995 Jan-Feb;28(1):37–39. [PubMed] [Google Scholar]
- Goodrich J. A., Cutler G., Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996 Mar 22;84(6):825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- Hartley D. A., Preiss A., Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, enhancer of split, shows homology to mammalian G-protein beta subunit. Cell. 1988 Dec 2;55(5):785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- Hartmann C., Taubert H., Jäckle H., Pankratz M. J. A two-step mode of stripe formation in the Drosophila blastoderm requires interactions among primary pair rule genes. Mech Dev. 1994 Jan;45(1):3–13. doi: 10.1016/0925-4773(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A., Fisher D. E. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994 Nov 15;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987 Sep 11;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Holmgren R. Cloning sequences from the hairy gene of Drosophila. EMBO J. 1984 Mar;3(3):569–573. doi: 10.1002/j.1460-2075.1984.tb01849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper K. L., Parkhurst S. M., Ish-Horowicz D. Spatial control of hairy protein expression during embryogenesis. Development. 1989 Nov;107(3):489–504. doi: 10.1242/dev.107.3.489. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Kohyama A., Watakabe I., Inoue K., Sakamoto H., Shimura Y. Transcriptional regulation of the Sex-lethal gene by helix-loop-helix proteins. Nucleic Acids Res. 1995 Sep 11;23(17):3441–3448. doi: 10.1093/nar/23.17.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K., Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986 Mar 28;44(6):949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Pinchin S. M., Howard K. R., Ish-Horowicz D. Genetic Analysis of the Hairy Locus in DROSOPHILA MELANOGASTER. Genetics. 1985 Nov;111(3):463–486. doi: 10.1093/genetics/111.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Pinchin S. M., Ingham P. W., Gyurkovics H. G. Autocatalytic ftz activation and metameric instability induced by ectopic ftz expression. Cell. 1989 Apr 21;57(2):223–232. doi: 10.1016/0092-8674(89)90960-4. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Pinchin S. M. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987 Nov 6;51(3):405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Sasai Y., Nakanishi S., Kageyama R. Molecular characterization of HES-2, a mammalian helix-loop-helix factor structurally related to Drosophila hairy and Enhancer of split. Eur J Biochem. 1993 Aug 1;215(3):645–652. doi: 10.1111/j.1432-1033.1993.tb18075.x. [DOI] [PubMed] [Google Scholar]
- John A., Smith S. T., Jaynes J. B. Inserting the Ftz homeodomain into engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo. Development. 1995 Jun;121(6):1801–1813. doi: 10.1242/dev.121.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H., Shigesada K., Satake M., Ito Y., Miyoshi H., Ohki M., Pepling M., Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993 Oct;9(10):338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- Kania M. A., Bonner A. S., Duffy J. B., Gergen J. P. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990 Oct;4(10):1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- Klingler M., Gergen J. P. Regulation of runt transcription by Drosophila segmentation genes. Mech Dev. 1993 Sep;43(1):3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., Campos-Ortega J. A. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989 Jan;8(1):203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Schrons H., Grawe F., Campos-Ortega J. A. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992 Oct;132(2):505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Ma P. C., Rould M. A., Weintraub H., Pabo C. O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994 May 6;77(3):451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Manoukian A. S., Krause H. M. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992 Sep;6(9):1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- Manoukian A. S., Krause H. M. Control of segmental asymmetry in Drosophila embryos. Development. 1993 Jul;118(3):785–796. doi: 10.1242/dev.118.3.785. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Oellers N., Dehio M., Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994 Sep 1;244(5):465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Maruyama M., Kagoshima H., Inuzuka M., Lu J., Satake M., Shigesada K., Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako S., Hyer J., Panganiban G., Oliver I., Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994 Nov 15;8(22):2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Bopp D., Ish-Horowicz D. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell. 1990 Dec 21;63(6):1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Lipshitz H. D., Ish-Horowicz D. achaete-scute feminizing activities and Drosophila sex determination. Development. 1993 Feb;117(2):737–749. doi: 10.1242/dev.117.2.737. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Meneely P. M. Sex determination and dosage compensation: lessons from flies and worms. Science. 1994 May 13;264(5161):924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- Paroush Z., Finley R. L., Jr, Kidd T., Wainwright S. M., Ingham P. W., Brent R., Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994 Dec 2;79(5):805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Rushlow C. A., Hogan A., Pinchin S. M., Howe K. M., Lardelli M., Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989 Oct;8(10):3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992 Dec;6(12B):2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Schrons H., Knust E., Campos-Ortega J. A. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992 Oct;132(2):481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H., Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985 Jan;4(1):167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. Near-reciprocal phenotypes caused by inactivation or indiscriminate expression of the Drosophila segmentation gene ftz. Nature. 1985 Dec 19;318(6047):677–680. doi: 10.1038/318677a0. [DOI] [PubMed] [Google Scholar]
- Takebayashi K., Sasai Y., Sakai Y., Watanabe T., Nakanishi S., Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994 Feb 18;269(7):5150–5156. [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tietze K., Oellers N., Knust E. Enhancer of splitD, a dominant mutation of Drosophila, and its use in the study of functional domains of a helix-loop-helix protein. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol J., Dearolf C. R., Prakash K., Parker C. S. Synthetic oligonucleotides recreate Drosophila fushi tarazu zebra-stripe expression. Genes Dev. 1991 May;5(5):855–867. doi: 10.1101/gad.5.5.855. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Tsai C., Gergen J. P. Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development. 1994 Jun;120(6):1671–1683. doi: 10.1242/dev.120.6.1671. [DOI] [PubMed] [Google Scholar]
- Tsai C., Gergen P. Pair-rule expression of the Drosophila fushi tarazu gene: a nuclear receptor response element mediates the opposing regulatory effects of runt and hairy. Development. 1995 Feb;121(2):453–462. doi: 10.1242/dev.121.2.453. [DOI] [PubMed] [Google Scholar]
- Van Doren M., Bailey A. M., Esnayra J., Ede K., Posakony J. W. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994 Nov 15;8(22):2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Wainwright S. M., Ish-Horowicz D. Point mutations in the Drosophila hairy gene demonstrate in vivo requirements for basic, helix-loop-helix, and WRPW domains. Mol Cell Biol. 1992 Jun;12(6):2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Younger-Shepherd S., Vaessin H., Bier E., Jan L. Y., Jan Y. N. deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell. 1992 Sep 18;70(6):911–922. doi: 10.1016/0092-8674(92)90242-5. [DOI] [PubMed] [Google Scholar]