Abstract
In eukaryotes, histone acetylation and methylation have been known to be involved in regulating diverse developmental processes and plant defense. These histone modification events are controlled by a series of histone modification gene families. To date, there is no study regarding genome-wide characterization of histone modification related genes in citrus species. Based on the two recent sequenced sweet orange genome databases, a total of 136 CsHMs (Citrus sinensis histone modification genes), including 47 CsHMTs (histone methyltransferase genes), 23 CsHDMs (histone demethylase genes), 50 CsHATs (histone acetyltransferase genes), and 16 CsHDACs (histone deacetylase genes) were identified. These genes were categorized to 11 gene families. A comprehensive analysis of these 11 gene families was performed with chromosome locations, phylogenetic comparison, gene structures, and conserved domain compositions of proteins. In order to gain an insight into the potential roles of these genes in citrus fruit development, 42 CsHMs with high mRNA abundance in fruit tissues were selected to further analyze their expression profiles at six stages of fruit development. Interestingly, a numbers of genes were expressed highly in flesh of ripening fruit and some of them showed the increasing expression levels along with the fruit development. Furthermore, we analyzed the expression patterns of all 136 CsHMs response to the infection of blue mold (Penicillium digitatum), which is the most devastating pathogen in citrus post-harvest process. The results indicated that 20 of them showed the strong alterations of their expression levels during the fruit-pathogen infection. In conclusion, this study presents a comprehensive analysis of the histone modification gene families in sweet orange and further elucidates their behaviors during the fruit development and the blue mold infection responses.
Keywords: histone modifications, Citrus sinensis, epigenetics, fruit development, blue mold
Introduction
In eukaryotes, the dynamics of chromatin structure regulate DNA accessibility and DNA-templated processes, and affect various biological processes (Ho and Crabtree, 2010). Nucleosome is the basic unit of chromatin and it compacts DNA by nearly sevenfold with ∼146 bp of DNA wrapped around a histone octamer. The histone octamer is composed by two copies of H2A, H2B, H3 and H4 histone proteins. The histone tails are modified by dynamic post-translational modifications (PTMs) including methylation/demethylation, acetylation/deacetylation, and so on (Patel and Wang, 2013). Various histone modifications, which are termed as the “histone code,” collectively build up an enriched and complicated pattern of chromatin structure and powerful function modulations (Strahl and Allis, 2000). It is well reviewed that a series of gene families involved in the establishment of histone methylation/demethylation and acetylation/deacetylation (Berr et al., 2011).
Methylation of histone lysine residues is an important epigenetic regulation mechanism which can activate or silence gene expression. It is known that histone lysine methylation modifications (except the methylation of H3K79) are catalyzed by a series of histone methyltransferases (HMTs), which were mainly encoded by a family of SET DOMAIN GROUP genes (SDGs; Feng et al., 2002). The SDG family is divided into different classes according to the sequence similarities with the suppressor of variegation 3–9 [SU(VAR)3-9], enhancer of zeste [E(z)], trithorax (TRX), and absent, small, or homeotic disks 1 (ASH1). The functions of histone lysine methylation in plant biological processes are involved in floral organ development, flowering transition, shoot and root branching, endodormancy release, carotenoid biosynthesis, hormone regulation, thigmomorphogenesis, and fungal pathogens resistance (Dong et al., 2008; Cazzonelli et al., 2009, 2014; Berr et al., 2010a,b; Sui et al., 2012; Sun et al., 2012; Kim et al., 2013a; Saito et al., 2015). Moreover, histone methylation also occurs at arginine residues and histone arginine methylation is involved in many cellular processes including transcription, RNA processing and transport, signaling, subcellular transport and so on (Pahlich et al., 2006). Histone arginine methylation is controlled by a conserved protein family named protein arginine methyltransferases (PRMTs). Plant PRMT genes are involved in the regulations of several essential developmental processes, including vegetative growth, circadian cycle, flowering process, and response to ABA and high salinity (Ahmad and Cao, 2012). On the other hand, histone methylation can be directly erased through the action of histone demethylases (HDMs). So far, two types of HDMs have been identified: lysine-specific demethylase 1 (LSD1) and jumonji C (JmjC) domain containing proteins. LSD1 is an amine oxidase, which removes mono- and di-methyl groups from H3K4 residue. Arabidopsis has four genes encoding LSD1 [LDL1, LDL2, LDL3 and FLOWERING LOCUS D (FLD)] which can regulate the flowering time with partial redundancies (Jiang et al., 2007). The other type of HDMs is JmjC-domain protein family (JMJ famlily) which has been assigned to distinct groups including JmjC-domain-only group, JHDM1/FBX/KDM2, JMJD1/JHDM2/KDM3, JMJD2/KDM4, JARID/KDM5, and JMJD3/KDM6. Studies on JMJs in plant have uncovered their important roles in chromatin regulation and plant development, including flowering time, floral organ development, female gametophyte development, BR signaling and circadian regulation (Chen et al., 2011).
Histone acetylation and deacetylation underlie a mechanism for reversibly modulating chromatin structure and transcriptional regulation (Tian et al., 2005). The homeostatic balance of histone acetylation is maintained by two types of antagonistic proteins: histone acetylases (HATs) and histone deacetylases (HDACs). So far, plant HATs have been distinctly divided into four groups including: (1) HAG group contains GCN5-, ELP3-, and HAT1-like histone acetylases; (2) HAM group featured by a MOZ-YBF2 (MYST) domain; (3) HAC group is similar to p300/CREB-binding protein (CBP) co-activator family in animals; (4) HAF group is related to mammalian TAFII250 (TATA binding protein-associated factors; Pandey et al., 2002). Genes encoding HATs have been widely reported in the regulations of developmental transitions, responses to environmental signals and integrations of stress hormone signals (Tian et al., 2005; Sheldon et al., 2006; Chen and Tian, 2007). Plant HDACs have been classified into three families including RPD3/HDA1 superfamily (HDA), Silent Information Regulator 2 (SRT) and HD2 (HDT) families (Hollender and Liu, 2008). Currently, studies have revealed key roles of plant HDACs in regulating plant vegetative and reproductive development, stress responses, gene silencing, as well as cell death and cycle (Ma et al., 2013; Wang et al., 2014). Overall, these gene families involved in histone modifications cooperatively alter the chromatin structures and performances of nucleosomes in order to specifically control gene expression. Moreover, in spite of involvements of these genes in developmental regulations, a growing body of studies has been revealed their crucial roles in abiotic stresses and plant immunity (Berr et al., 2010a; Kim et al., 2010; Luo et al., 2012).
Citrus is an important and widely grown fruit crop with richness of nutritional components such as carotenoids and vitamin C. Its fruit development and ripening process shows a single sigmoid curve including two stages of slow growth with a period of rapid growth in between (Bain, 1958). After the fruit ripening, most of citrus fruits have been proceeded to the post-harvest storage. Blue mold (Penicillium digitatum) is the most devastating pathogen in citrus fruit post-harvest process and responsible for nearly 90% of production losses during fruit post-harvest handling (Macarisin et al., 2007). Although HM genes had been investigated during the fruit development process in tomato (Aiese Cigliano et al., 2013) and grape (Aquea et al., 2011), as well as the plant–pathogen response in Arabidopsis (Alvarez et al., 2010; Berr et al., 2010a), little is known regarding the functions of HMs in citrus.
Given the critical roles of plant HMs in regulations of fruit development and pathogen responses, it is expected that they are also involved in citrus fruit development and fruit-blue mold infection. In this study, 136 CsHMs, belonging to 11 families were identified in sweet orange. Then, genomic organization, phylogenetic relationship, domain architecture, and gene structure of these genes were comprehensively analyzed. Additionally, expression profiles of CsHMs were analyzed in six stages of fruit development and four periods of blue mold infection. Such a comprehensive analysis of these CsHMs will provide fundamental to understanding their diverse roles in citrus development and be useful for future functional genomic studies on regulations of histone modifications in citrus.
Materials and Methods
Identification of CsHM Families
The HMM files containing conserved domain of each HM families (HMTs: SDG-PF00856, PRMT-PF05185; HDMs: HDMA-PF04433, JMJ-PF02373; HATs: HAG-PF00583, HAM-PF01853, HAC-PF08214, HAF-PF09247; HDACs: HDA-PF00850, SRT-PF02146) were downloaded from Pfam protein database1. These HMM files were used as a query to search the two sweet orange genome databases2 [Orange genome Annotation Project (Xu et al., 2013); Sweet Orange Genome Project 20103 (Wu et al., 2014)] using HMMER 3.0 software (HMMER 3.04) with the default parameters. In order to obtain the complete catalog of CsHMs, the output results from two genomes were combined and filtrated the redundant sequences. For CsHDTs, AtHDT1 (At3g44750), AtHDT2 (At5g22650), AtHDT3 (At5g03740), and AtHDT4 (At2g27840) from Arabidopsis thaliana were used to perform a Blastp algorithm from sweet orange genome2 and two sequences named as CsHDT1, CsHDT2 were obtained. The final ID numbers and DNA sequences of CsHMs are listed in Supplementary Table S1 and Data Sheet 1, respectively. The ID numbers with red fonts were the additional predicted genes from the second sweet orange genome (Wu et al., 2014).
Genomic Organization of CsHMs
To determine the physical location of HMs, the MapChart software (Voorrips, 2002) was applied to locate the CsHMs on sweet orange chromosomes according to their positions given in the genome database (Xu et al., 2013).
Analysis of Domain Compositions and Gene Structures
To investigate the domain compositions of CsHMs, the complete amino acid sequences of these genes were subjected to SMART website, including outlier homologs and PFAM domains. The genomic DNA sequences and corresponding CDS sequences of CsHMs were submitted to Gene Structure Display Server (GSDS5) website to visualize the gene structures.
Phylogenetic Analysis
The HM protein sequences from Arabidopsis, rice and maize were collected from ChromDB database6. Each HM family including citrus HMs was aligned with ClustalW program. The generated files were subjected to phylogenic analysis by using MEGA 5.05 program7 with Neighbor-Joining method. The phylogenic trees were constructed with the following settings: pairwise deletion for sequences analysis, poisson model for substitution, and bootstrap test of 1000 replicates for internal branch reliability. For SDG and HAG families, the conserved domain sequences of SET and AT1 identified in citrus together with the domain sequences from Arabidopsis, rice and maize were used for tree constructions, respectively.
Plant Materials and Blue Mold Infection
To analyze the expression patterns of HMs during the fruit development, fruit samples were collected from the adult plants of sweet orange (Citrus sinensis [L.] Osbeck), cultivated at the Institute of Citrus Research located in Guilin, Guangxi Province, China. Fruit samples with three independent repeats were collected from different position and orientation of six different trees. The fruit samples were continuously collected from July to December in 2011 as six fruit developmental stages, which were 90, 120, 150, 180, 210, and 240 days after flowering (daf), respectively. The peel and flesh tissues were separated from sampled fruits, and then immediately frozen in liquid nitrogen and kept at -80°C until further analysis.
Sweet orange fruits were used as the materials for the investigation of fruit-blue mold infection. Mature fruits were treated with 2% NaClO for 2 min and washed with distilled water for three times. A uniform lesion (5 mm wide, 3 mm deep) was made at the equator of the fruit using a sterile nail. An aliquot of 20 μL suspension of P. digitatum at 1 × 106 spore mL-1 was inoculated into each wound site. After the inoculation, fruits were incubated in a storage chamber with 95% relative humidity at 25°C temperature for 6, 24, and 48 h to collect the samples. An aliquot of 20 μL double distilled water was inoculated into the fruits as the control (CT). 10 mm of peel around the wound was collected and immediately frozen in liquid nitrogen and kept at -80°C for RNA extraction. Each of the inoculation experiment was performed with the three replications.
Expression Analysis of CsHMs
To investigate the expression patterns of all CsHMs in different citrus tissues, the normalized RPKM (reads per kilobase per million mapped reads) values of these genes were extracted from the dataset of the Citrus sinensis Annotation Project (CAP) and visualized by the heat maps with transformed log10 values using MeV 4.7 software (Saeed et al., 2006). In order to gain an insight of their roles in citrus fruit development, genes whose RPKM values were higher than 5.0 in fruit tissues were selected to further analyze their expression profiles during the six stages of fruit development using real-time PCR. Total RNA was extracted from the peel and flesh samples of citrus fruits according to the previous description (Liu et al., 2006). First strand cDNA was synthesized from 1.5 μg of total RNA using the ReverAid first strand cDNA synthesis KIT (Fermentas). Real-time PCR primer pairs were designed by Primer Express software (Applied Biosystems, Foster City, CA, USA) and their sequences were listed in Supplementary Table S2. The primers were tested to ensure amplification of single discrete bands with no primer-dimers. The primers were diluted in Power SYBR® Green PCR Master Mix (Applied Biosystems) and the amplification mixture volume was 10 μL per reaction. Reaction conditions were an initial incubation for 2 min at 50 and 95°C for 1 min, and then followed by 40 cycles of 95°C/15 s and 60°C/1 min. Reactions were run on a 7900 HT Fast Real-Time PCR System with 384-Well Block Module (Applied Biosystems). The β-actin gene was used as an endogenous control and comparative Ct method (2-ΔΔCt) was adopted to calculate the expression data (Liu et al., 2007). The expression levels of 90 daf flesh or peel were used as the calibrator for the relative expression analysis. Expression analysis of all CsHMs response to blue mold infection was performed with real-time PCR. The expression levels of control were used as the calibrator for the analysis. The heat maps and hierarchical clustering of gene expression data were visualized in MeV 4.7 software. Genes with fold change (log2 value) higher than 1.0 or lower than -1.0 were selected and their expression profiles were shown in Supplementary Figure S2. SPSS software was applied to the statistical analysis of these data in the present study.
Results
Identification of HMs in Sweet Orange Genome
In this study, a total of 136 CsHMs were identified in sweet orange genome, including 47 histone methyltransferase genes (CsHMTs), 23 histone demethylase genes (CsHDMs), 50 histone acetylase genes (CsHATs), and 16 histone deacetylase genes (CsHDACs). The 47 CsHMTs had 40 CsSDGs and 7 CsPRMTs in sweet orange (Table 1). The number of CsSDG family was closed to that in Arabidopsis (41 members), rice (37; ChromDB database), tomato (43; Aiese Cigliano et al., 2013), and grape (33; Aquea et al., 2011). The 23 CsHDMs were composed by three citrus HDMA histone demethylase genes (CsHDMAs) and 20 JMJ genes (CsJMJs). The number of CsJMJs was also similar to that in Arabidopsis (21 members), rice (20; ChromDB database) and tomato (20; Aiese Cigliano et al., 2013). The 50 identified CsHATs were classified to 45 CsHAGs, one CsHAM, two CsHACs, and two CsHAFs in sweet orange. Much more HAG members were obtained in sweet orange genome compared with Arabidopsis (three members), rice (3) and maize (4). Only one MYST histone acetyltransferase gene (CsHAM1) encoded a 449 amino acid protein was identified in sweet orange. Two CsHACs belonging to the HAC group were identified and two TAFII250-like genes named as CsHAF1 and CsHAF2 were obtained in sweet orange. In addition, the 16 CsHDACs have nine CsHDAs, five CsSRTs, and two CsHDTs (Table 1). All of gene IDs was listed in Supplementary Table S1.
Table 1.
Gene numbers of each histone modification families in sweet orange.
| Types | Family | Gene numbers |
|---|---|---|
| HMTs (Histone methylation) | SDGs | 40 |
| PRMTs | 7 | |
| HDMs (Histone demethylation) | HDMAs | 3 |
| JMJs | 20 | |
| HATs (Histone acetylation) | HAGs | 45 |
| HAMs | 1 | |
| HACs | 2 | |
| HAFs | 2 | |
| HDACs (Histone deacetylation) | HDAs | 9 |
| SRTs | 5 | |
| HDTs | 2 |
Chromosomal Distribution of CsHMs
CsHMTs/CsHDMs
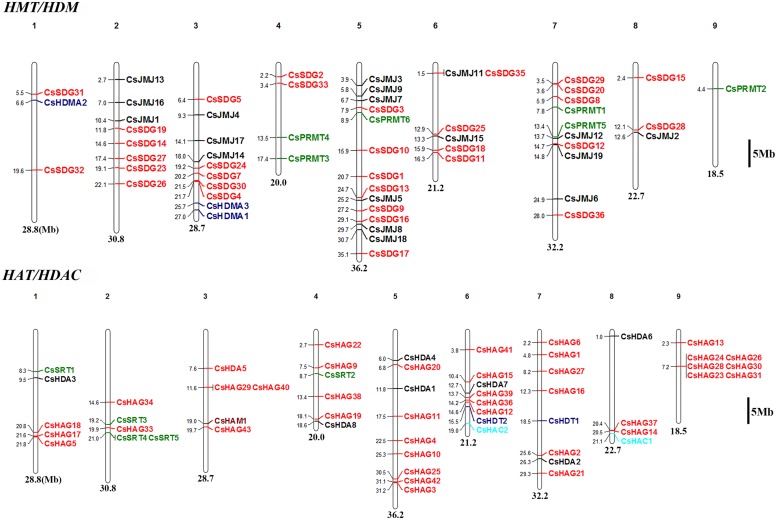
The chromosomal locations of CsHMs were demonstrated on sweet orange chromosome available at CAP8. The members of CsSDG family were widely distributed in eight chromosomes with no distribution in the ninth chromosome (Figure 1). The largest number of CsSDGs was located on chromosome 5 (seven CsSDGs). However, eight genes including CsSDG6, 21, 22, 34, 37, 38, 39, and 40 were not determined because the physical map of sweet orange was incomplete. CsPRMTs were distributed at chromosomes 4, 5, 7, and 9 (Figure 1). As regard CsHDMAs, CsHDMA2 was located in chromosome, while CsHDMA1 and CsHDMA3 were closely located in chromosome 3, suggesting the occurrence of tandem duplication. CsJMJs were widely distributed at chromosomes 2, 3, 5, 6, 7, and 8 and six of CsJMJs were located in chromosome 5.
FIGURE 1.
Chromosomal localization of CsHM genes.
CsHATs/CsHDACs
As shown in Figure 1, lots of CsHAGs displayed the close locations from each other. This might imply that the tandem duplication events occurred in this gene family. The citrus GCN5 (CsHAG25) and ELP3 gene (CsHAG20) were located in chromosome 5, while citrus HAT1 gene (CsHAG38) was in chromosome 4. Members of CsHDAs were distributed at chromosomes 1, 4, 5, 6, 7, and 8. Five CsSRTs named as CsSRT1-5 were located at chromosomes 1, 2, and 4. Two CsHDTs named as CsHDT1 and CsHDT2 were located at chromosomes 7 and 6, respectively.
Phylogenetic Analysis, Conserved Domains, and Exon/Intron Organizations of CsHMs
To explore the phylogenetic relationships among CsHM proteins and group them within the established classes, the predicted amino acid sequences of each HM family from various species were aligned and phylogenetic trees were further constructed. Furthermore, the gene structures of all CsHMs and the domain compositions of their coding proteins were analyzed.
CsHMTs
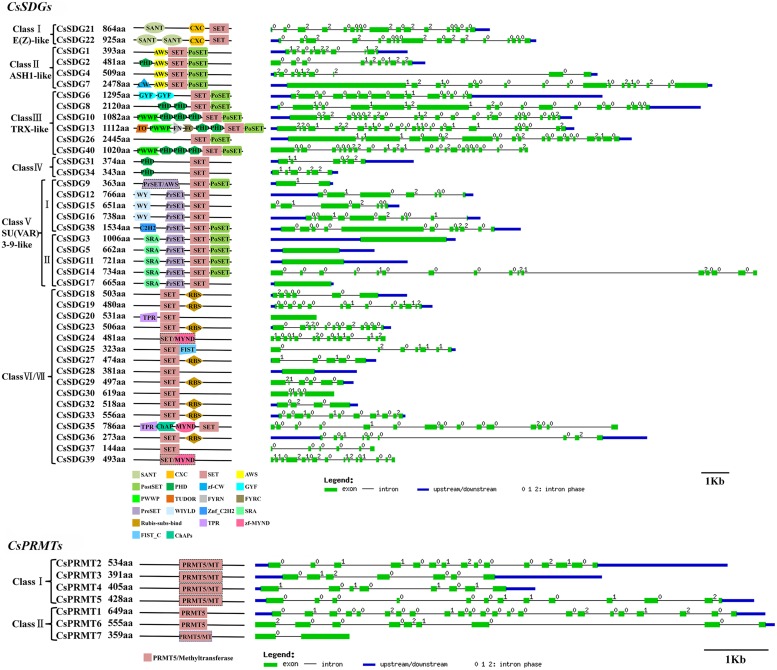
All of the 40 CsSDGs were divided into seven classes according to the classification criteria of SDG family in Arabidopsis (Springer et al., 2003; Figure 2). In detail, CsSDG21 and 22, which were clustered with three AtSDGs, belonged to class I [E(z)-like]. CsSDG21 and 22 commonly had conserved SANT, CXC and SET domain and CsSDG22 had an additional SANT domain (Figure 4). Class II was comprised of four ASH1-like CsSDGs, clustered with five Arabidopsis and five rice ASH1-like proteins. CsSDGs in this class contained the conserved SET, Post-SET and AWS domains, while CsSDG2 and CsSDG7 had an additional PHD and CW domain, respectively. The TRX (TRITHORAX) family (class III) included six CsSDGs which was featured by highly conserved SET and Post-SET domains. CsSDG31 and 34 with a SET and N-terminus PHD domain belonged to class IV. Ten CsSDGs homologous to SU(VAR)3–9 group belonged to class V and were further divided into two main clades. Each of subgroups had five CsSDG members. Three members of Subgroup I contained WIYLD domain and Subgroup II was featured by a conserved SRA domain at the N-terminus. Sixteen CsSDGs clustered within class VI and class VII and eight of them contained Rubis-subs-bind (RBS) domain. As regards CsPRMTs, seven predicted proteins characterized by PRMT5 domain (Figure 4) were categorized to two classes (Supplementary Figure S1) according to the previous study (Aiese Cigliano et al., 2013). CsPRMT2 to 5 proteins were clustered to class I, and CsPRMT1, 6, 7 proteins belonged to class II.
FIGURE 2.
Phylogenetic analysis of SDG family between Citrus Sinensis and Arabidopsis. The tree was constructed by the Neighbor-Joining method with MEGA program 5.05 using the conserved SET-domain region.
FIGURE 4.
Domain composition and gene structure of sweet orange SDGs and PRMTs. Exon/intron structures of these genes were placed on the right side of the domain composition. Exon(s) and intron(s) were represented by green boxes and black lines, respectively. The blue box represented UTR region of gene upstream and/or downstream.
CsHDMs
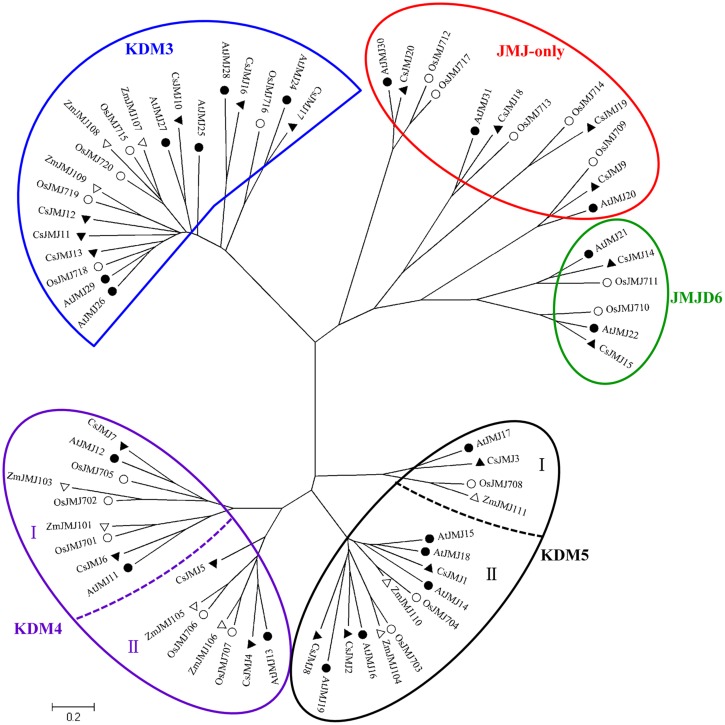
The Phylogenetic tree of HDMAs was clustered to two main clades (Supplementary Figure S1) and all of three CsHDMAs were characterized by conserved N-terminal SWIRM (PF04433) domain and C-terminal Amino_oxidase (PF01593) domain. JMJ family was grouped into five classes based on sequence similarities, including JMJ-only (class I), KDM3 (class II), KDM4 (class III), KDM5 (class IV), and JMJD6 (class V) groups (Figure 3; Lu et al., 2008). JMJ-only class included four citrus members (CsJMJ9 and CsJMJ18-20) which only had JmjC domain and were not clustered to other groups. However, amino acid analysis of Arabidopsis and rice JMJ-only members indicated that they could be active demethylases (Lu et al., 2008). KDM3 class had six citrus members (CsJMJ10-13, CsJMJ16, and CsJMJ17), featured by a JmjC domain at the C-terminal with Ring finger domains (SM000184) ahead of it (Figure 5). The CsJMJs of KDM4 class fell into two main subclasses corresponding to the domain composition. Subclass I was characterized by four tandem repeats of ZnF_C2H2 domain (SM000355), while subclass II contained a zf-C5HC2 domain (PF02928) at the C-terminal. KDM5 group was also divided into two main subclasses which contain one (CsJMJ3) and three citrus members (CsJMJ1, 2, and 8), respectively. Additionally, the class of JMJD6 had two citrus members which had JmjC and N-terminal FBOX domain.
FIGURE 3.
Phylogenetic tree for Citrus Sinensis, Arabidopsis, Oryza sativa, and Zea mays JMJ family. The tree was constructed by the Neighbor-Joining method with MEGA program 5.05.
FIGURE 5.
Domain composition and gene structure of sweet orange JMJs and HDMAs. Exon/intron structures of these genes were placed on the right side of the domain composition. Exon(s) and intron(s) were represented by green boxes and black lines, respectively. The blue box represented UTR region of gene upstream and/or downstream.
CsHATs
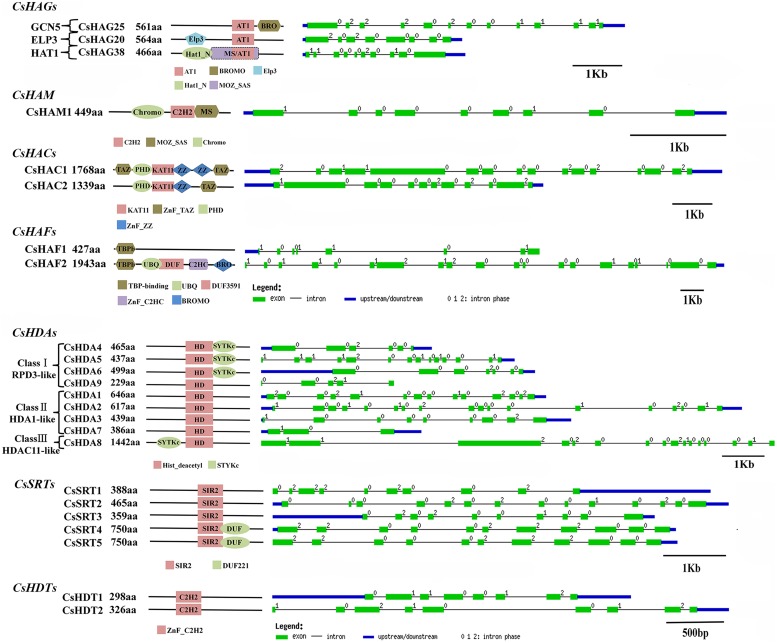
The Phylogenetic tree and domain composition demonstrated that CsHAG25, 20, and 38 were belonged to GCN5, ELP3, and HAT1 class respectively (Supplementary Figure S1). Figure 6 only presented these three proteins domain compositions, while the other members only contained the AT1 conserved domain. In detail, CsHAG25 carried a C-terminal Bromo domain (PF00439) which recognized acetylated lysine residues. CsHAG20 had an N-terminal Elp3 domain, which was conserved in AtHAG3, OsHAG703 and two maize ELP3 proteins. CsHAG38 was a GNAT/MYST-Like (GML; Aiese Cigliano et al., 2013) member featured by N-terminal Hat1_N (PF10394) and an additional MOZ_SAS (PF01853) domain (Figure 6). As regard CsHAM1, it composed by Chromo (PF00385), C2H2 (PF00096), and MOZ_SAS (PF01853) domain which was the typical domain architecture of HAM family. The two CsHACs presented the similar domain compositions, while CsHAC1 had an additional ZnF_TAZ and a ZnF_ZZ domain. Although the two citrus TAFII250-like genes CsHAF1 and CsHAF2 showed high similarity with each other, CsHAF1 only had the TBPb domain and lacked other conserved domains (Figure 6).
FIGURE 6.
Domain composition and gene structure of sweet orange CsHAGs, CsHAM, CsHACs, CsHAFs, CsHDAs, CsSRTs, and CsHDTs. Exon/intron structures of these genes are placed on the right side of the domain composition. Exon(s) and intron(s) were represented by green boxes and black lines, respectively. The blue box represented UTR region of gene upstream and/or downstream.
CsHDACs
As shown in Supplementary Figure S1, CsHDAs were divided into three classes according to the previous study in Arabidopsis, rice and maize (Alinsug et al., 2009). All of them contained conserved Hist_deacetyl domain (PF00850) and an additional STYKc (SM00221) domain was presented in CsHDA4, 5, 6, and 8. Five of CsSRTs are characterized by an SIR2 domain (PF02146) and CsSRT4 and 5 had an additional DUF domain (PF02714) at the C-terminal. In addition, the phylogenetic tree of HDTs showed that CsHDT1 was close to AtHDT1 and AtHDT2, while CsHDT2 was most closely related to AtHDT3 (Supplementary Figure S1).
Expression Patterns of CsHMs in Different Tissues
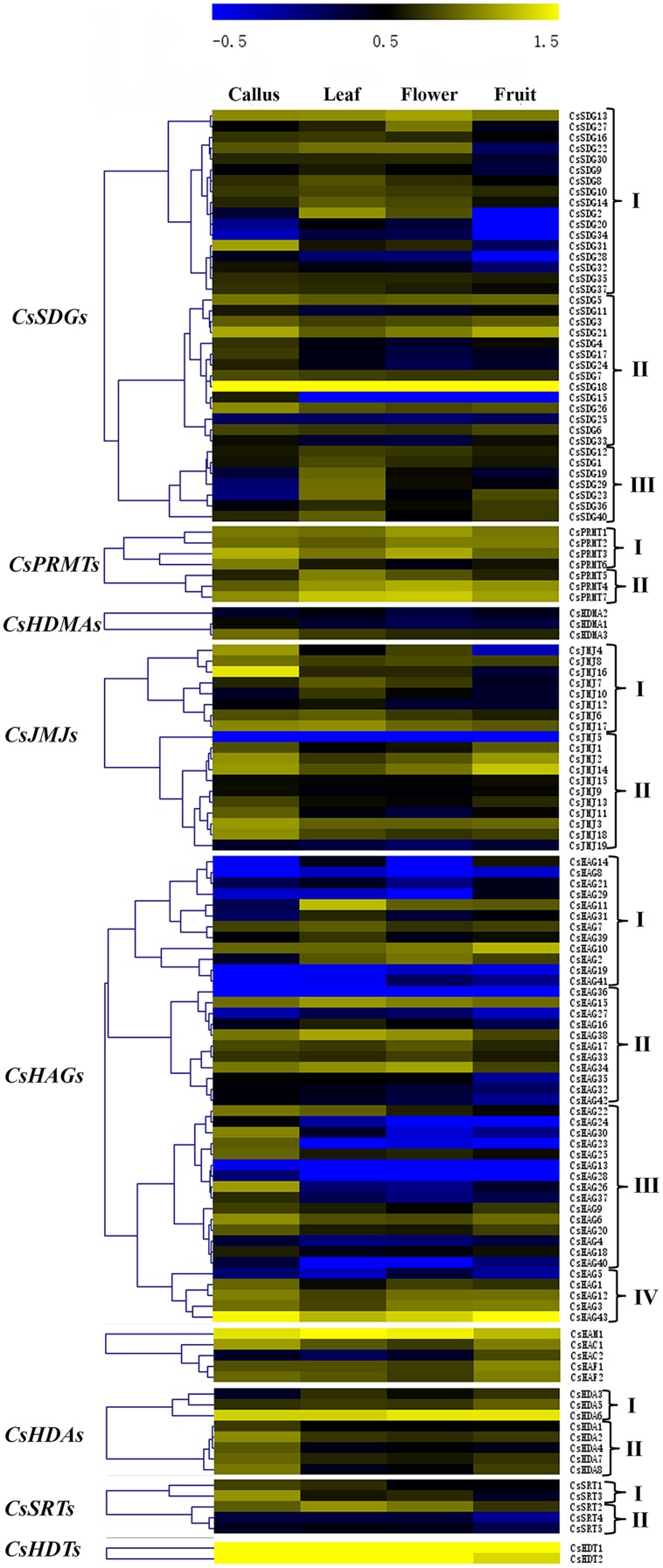
The expression patterns of all CsHMs in different tissues (callus, leaf, flower, and fruit) revealed by RNA-seq data of the CAP (Xu et al., 2013) were demonstrated in Figure 7. Hierarchical cluster analysis was performed based on the expression data of each CsHM gene family corresponding to the four different tissues. According to the hierarchical clustering results, we classified genes of each family to different expression pattern groups (I–IV). As shown in Figure 7, the group I genes of CsSDGs showed a low expression level in fruit tissues, while the members of group III were expressed highly in leaf. The members of CsPRMTs presented a high expression level among these four tissues. For CsJMJs, genes in group I were expressed lowly in fruit, while group II genes showed a relative high expression level in callus and fruit. The CsHAGs mainly grouped to four expression patterns and the high expression in callus was prevalent among the group III members. Moreover, the genes belonged to CsHDAs group I expressed highly in fruit and the group II members showed high expression in callus. Above all, the various expression profiles of CsHMs among the four tissues indicated that these genes might take part in different biological processes in sweet orange.
FIGURE 7.
Hierarchical clustering of each CsHM family. The expression data were extracted from the Citrus sinensis Annotation project (CAP). The normalized RPKM values were transformed to log10 values and visualized with the heat map using MeV 4.7 software. The members of each family were clustered to different gene expression groups (I–IV) based on the hierarchical analysis with Pearson correlation.
Dynamic Expression Patterns of CsHMs in Different Fruit Developmental Stages
In order to gain insights into the biological roles of CsHMs in citrus fruit development, genes with RPKM values higher than 5.0 in fruit tissues according to RNA-seq data were selected to further analyze their expression profiles during the six fruit developmental stages using real-time PCR. The real-time PCR primers of each CsHMs were listed in Supplementary Table S2.
CsHMTs/CsHDMs
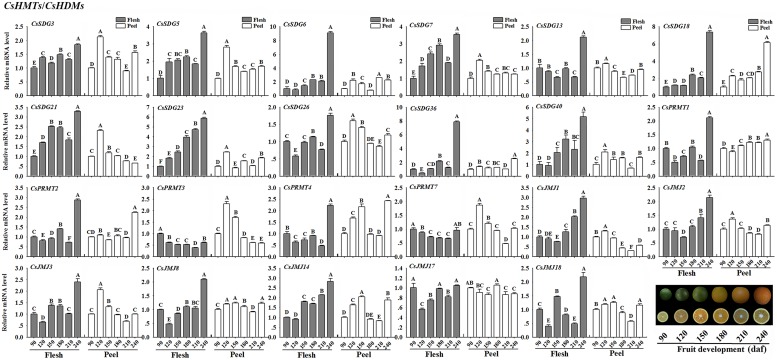
As shown in Figure 8, most of selected CsSDGs were expressed highly in flesh of citrus fruit at the mature stage (240 daf). Notably, the increasing expression levels of CsSDG6, 7, 18, 23, and 40 in flesh were strongly correlated with the fruit development process (Figure 8). For CsPRMTs, the mRNA abundance of CsPRMT1, 2, and 4 were expressed highly in flesh at the 240 daf stage. As regard CsJMJs, all members of KDM5 class (CsJMJ1, 2, 3, and 8) showed the increasing expression patterns in flesh along with the fruit development. Additionally, the expression levels of CsJMJ14 also increased during fruit development in flesh. However, the expression profiles of these selected genes showed more complicated in peel during the fruit development.
FIGURE 8.
Expression profiles of selected CsHMTs and CsHDMs (RPKM > 5 in fruit tissue based on RNA-seq data) in peel and flesh during six fruit developmental stages (90–240 daf-days after flowering) using real-time PCR. Data were mean ± SD of three separate measurements. Capital letters indicated significant differences at P < 0.01.
CsHATs/CsHDACs
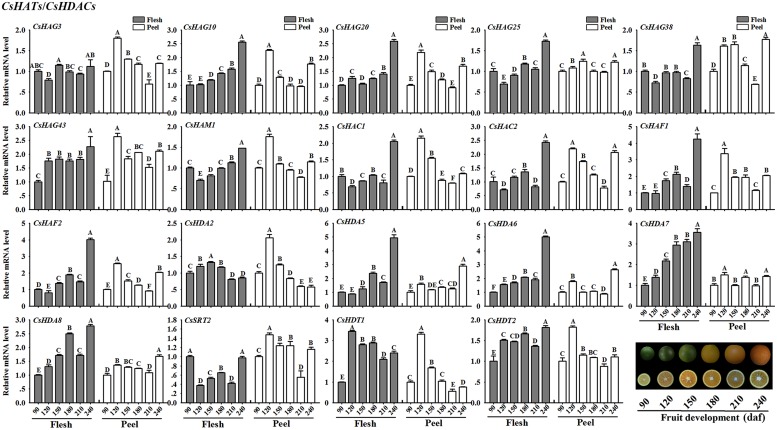
Figure 9 showed the expression profiles of the selected CsHATs and CsHDACs genes. In flesh, CsHAG10 and 20, CsHAM1, CsHAF1 and 2 presented an increasing expression level along with the fruit development. Additionally, two CsHACs showed a high expression level at the mature stage (240 daf) of fruit. As regard CsHDAs, the expression profiles of CsHDA5, 6, 7, and 8 presented an increasing trend during the fruit development. Notably, the expression level of CsHDA7 showed the strong positive correlation with the citrus fruit development. In peel, the selected CsHATs genes showed a similar expression pattern with high expression levels at 120 daf stage of fruit development.
FIGURE 9.
Expression profiles of selected CsHATs and CsHDACs (RPKM > 5 in fruit tissue based on RNA-seq data) in peel and flesh during six fruit developmental stages (90–240 daf-days after flowering) using real-time PCR. Data were mean ± SD of three separate measurements. Capital letters indicated significant differences at P < 0.01.
Expression Profiles of CsHMs Response to Blue Mold Infection
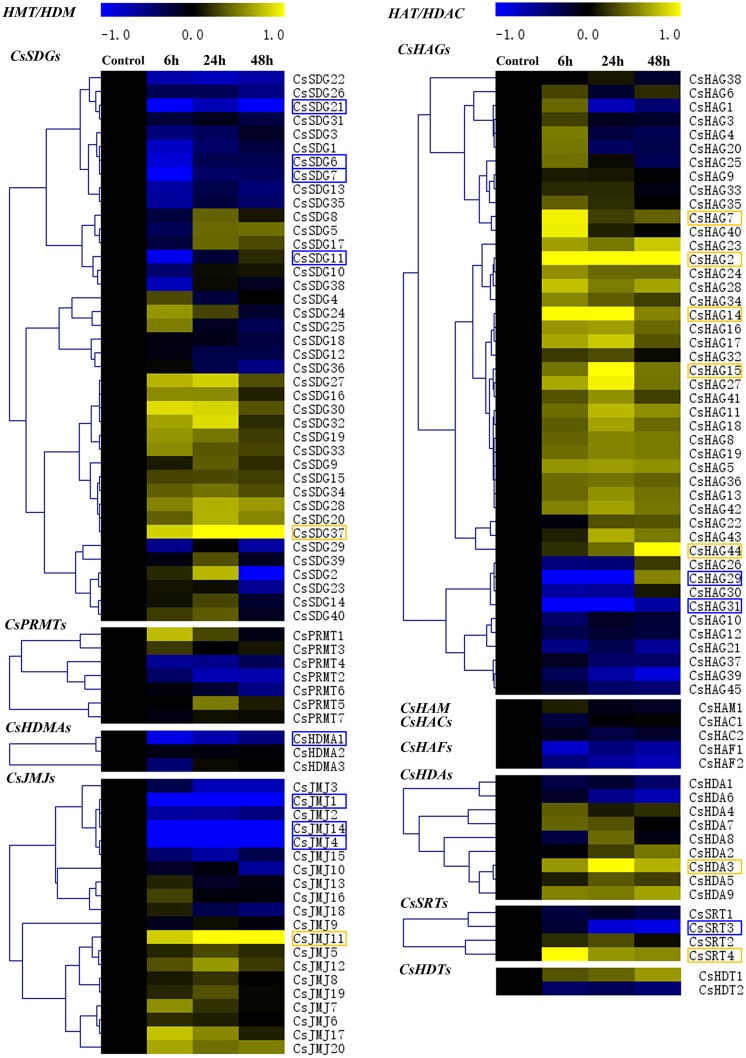
A growing body of studies had been revealed the crucial roles of HMs in various abiotic stresses and plant immunity. Blue mold is considered as the most devastating pathogen in citrus fruit post-harvest process and causes lots of rotting losses. In order to determine the CsHMs responding to fruit-blue mold infection, the expression profiles of all CsHMs were detected among the four periods of infection using real-time PCR. The results were visualized by the heat maps and hierarchical clustering was further analyzed in each CsHM family (Figure 10). A numbers of CsHMs were up (yellow color) or down (blue color) regulated by the blue mold infection. According to these data, 20 genes including five CsSDGs, one CsHDMA, four CsJMJs, seven CsHAGs, one CsHDA, and two CsSRTs were selected to present their expression patterns in Supplementary Figure S2 for their expression fold change higher than 2.0 compared with the control. The expression levels of CsSDG6, 7, and 11 were inhibited at 6 h after infection (hai) and recovered at 24 and 48 hai. On the contrast, CsSDG37 was up-regulated by the infection. Three CsJMJs (CsJMJ1, 4, and 14) were strongly down-regulated, while CsJMJ11 were induced by the infection. Five CsHAGs including CsHAG2, 7, 14, 15, and 44 were up-regulated, while the expression levels of CsHAG29 and 31 were strongly inhibited at 6 and 24 hai and then recovered at 48 hai. For CsHDAs, CsHDA3 was selected out and showed the increased expression levels under the infection. Additionally, two CsSRTs (CsSRT3 and 4) presented different responses to infection. CsSRT1 was down-regulated, while CsSRT4 was highly up-regulated at the 6 h and then slightly induced at the 24 and 48 h by the infection.
FIGURE 10.
Expression profiles of all CsHMs in response to blue mold (Penicillium digitatum) infection of citrus fruit at different periods (Control, 6, 24, and 48 h) using real-time PCR. Transcripts were normalized to Actin gene expression and the expression level of control was used as the calibrator for relative expression analysis. Hierarchical clustering of each CsHM family was performed using MeV 4.7 software. Genes with log2 value higher than 1.0 (up-regulated) or lower than -1.0 (down-regulated) were marked with yellow box or blue box, respectively.
Discussion
Chromatin based gene regulation affects various processes such as root growth, flowering time, floral organogenesis, gametophyte, and embryo development as well as plant response to pathogens and environmental changes (Alvarez et al., 2010; Deal and Henikoff, 2011; Gu et al., 2014; Kim et al., 2015). A series of gene families have been proved to be involved in establishment of histone modifications which can determine chromatin state to regulate biological processes (Berr et al., 2011). Here, genes involved in histone methylation/demethylation and acetylation/deacetylation have been genome-widely characterized in sweet orange based on the sequenced genome (Xu et al., 2013; Wu et al., 2014). Eleven gene families (SDGs, PRMTs; HDMAs, JMJs; HAGs, HAM, HACs, HAFs; HDAs, SRTs, and HDTs) containing 136 CsHMs were identified in sweet orange genome. The gene numbers of these families in sweet orange are close to the Arabidopsis. For example, CsSDGs and CsJMJs have 40 and 20 members respectively (Table 1) and the corresponding AtSDG and AtJMJ families contain 41 and 21 members (ChromDB database). However, much more HAG genes (45 members) were identified in sweet orange genome compared with in Arabidopsis (three members), rice (3) and maize (4; ChromDB database). But if we used the AT1 domain as a query to apply the Blast program, 33 HAG members were obtained in Arabidopsis (Aiese Cigliano et al., 2013), which was close to the number of CsHAGs. Moreover, 26 predicted HAG proteins were identified in tomato genome based on the similar searching methods (Aiese Cigliano et al., 2013).
Recently, an increasing number of HMs have been identified and unraveled their pivotal roles in regulations of essential processes (Berr et al., 2011). The involvements of HMs in citrus fruit development have not yet been described. However, numbers of tomato HMs had been characterized their potential roles in tomato ripening process (Aiese Cigliano et al., 2013). A study on grape SDG family has revealed that several VvSDGs were increasing their expression levels during the grape berry development (Aquea et al., 2011). In our study, we also revealed that a number of CsHMs showed the increasing expression patterns during the citrus fruit development (Figures 8 and 9). Regarding histone methylation in plants, SDG family controls the methylation of histone lysine residues, which are involved in various biological processes such as flowering transition, hormone regulation, and carotenoid biosynthesis (Cazzonelli et al., 2009; Sun et al., 2012; Kim et al., 2013a). Generally, histone H3K9 and H3K27 methylation are two repressive marks, whereas H3K4 and H3K36 methylation activates gene expression (Berger, 2007). A number of SDGs has been characterized their catalytic functions in Arabidopsis and rice (Liu et al., 2010). AtSDG9, 23, 31, 33 and OsSDG714, belonging to Class V (Figure 2), are responsible for H3K9 methylation, while AtSDG1, 5, 10 (Class I), 15 and 34 (Class IV) catalyze H3K27 methylation. For the activation marks, AtSDG27, 30 (Class III) and 4 (Class II) act on the H3K4 residues and AtSDG4, 8 and 26 (Class II) catalyze the H3K36 methylation. Although the enzymatic activity and specificity of citrus SDGs are not known, genetic data suggest that they may catalyze the same lysine residues and act the similar repression/activation functions with Arabidopsis. During citrus fruit development, CsSDG7 showed the increasing expression levels and CsSDG13 presented the high expression level at the mature stage (Figure 8). Moreover, CsSDG7 and CsSDG13 are homologous to AtSDG8 and AtSDG27 which catalyze the activation marks H3K36 and H3K4 methylation respectively, indicating that CsSDG7 and CsSDG13 could have similar functions and activate genes expression during the fruit development. Furthermore, AtSDG8 encoding a HMT can affect the carotenoid biosynthesis via regulating the H3K4 tri/di-methylation on CRTISO (CAROTENOID ISOMERASE) which controls the carotenoid isomerization in carotenoid biosynthetic pathway (Cazzonelli et al., 2009). Citrus fruits accumulated nearly 115 kinds of carotenoids and the total carotenoids content increased rapidly during the fruit development (Rouseff et al., 1996; Kato et al., 2004). Previous study revealed that the ‘Anliu’ sweet orange used in this study showed a rapid increase of total carotenoids in flesh after the green stage (150 daf), which was attributed to the increased accumulation of β-cryptoxanthin and violaxanthin (from an undetectable level to 2.28 μg/g and from 0.99 to 4.63 μg/g, respectively; Liu et al., 2007). In this study, CsSDG7, being homolog to AtSDG8 (Figure 2), showed the increasing expression levels in flesh during the fruit development process (Figure 8). This implied that CsSDG7 could also be involved in citrus fruit carotenoid accumulations during the fruit development. Additional expression analysis of CsCRTISO gene during the citrus fruit development also showed that the CsCRTISO was expressed highly in flesh at the mature stage of fruit development (Supplementary Figure S3). For histone arginine methylation, atprmt4a and atprmt4b double mutant, atprmt5/skb1, and atprmt10 display the late flowering phenotype by increasing the FLC expression in Arabidopsis, indicating these AtPRMTs are required in the Arabidopsis flowering transition process (Pei et al., 2007; Niu et al., 2008; Schmitz et al., 2008). In our study, CsPRMT1 and CsPRMT2, being homolog to AtPRMT5 and AtPRMT4 respectively, showed the high expression levels in flesh of fruits at 240 daf stage. Based on the similarity and expression profiles, we expected that these two CsPRMTs could also be required in citrus fruit ripening process. Regarding histone demethylases, Arabidopsis JMJ14 catalyzes histone demethylation at H3K4 residues and represses flowering (Lu et al., 2010). A recent study has revealed that two novel NAC transcription factors NAC050 and NAC052 interacted with the FYRC domain of AtJMJ14 to regulate gene expression and flowering time (Ning et al., 2015). AtJMJ15 is an H3K4me3 demethylase and AtJMJ15 overexpression resulted in an obvious early flowering phenotype (Yang et al., 2012). CsJMJ1 with a FYRC domain (Figure 5) is clustered with AtJMJ14 and AtJMJ15 (Figure 3), suggesting CsJMJ1 is a predicted H3K4 demethylase with repression of gene expression. Moreover, the expression levels of CsJMJ1 and the other members of KDM5 class (CsJMJ2, 3, 8) presented an increasing trend during the citrus fruit development (Figure 8) and we expect that these genes could be functional during citrus fruit development.
For histone acetylation/deacetylation, one HAF gene (SlHAF1) was identified in tomato genome and it has the strongest expression in tomato fruit at 10 days after breaking, suggesting an important role in tomato maturation (Aiese Cigliano et al., 2013). Similarly, two identified CsHAFs (CsHAF1 and 2) also showed a high expression level in flesh at the mature stage (240 daf) of citrus fruit development (Figure 9), implying that they could have the similar functions with SlHAF1 in tomato. The best studied HDACs in Arabidopsis belonged to RPD3 (Class I), including AtHDA6, 19, 7, 9 and pseudogenes AtHDA10 and 17 (Supplementary Figure S1). AtHDA19 was a member of AP2-TPL-HDA19 repressor complex which negatively regulated multiple floral organ identity genes in Arabidopsis (Krogan et al., 2012). AtHDA9 repressed Arabidopsis flowering by removing H3K9Ac and H3K27Ac on the flowering promoting gene AGAMOUS-LIKE19 (AGL19; Kim et al., 2013b). AtHDA9 and AtHDA6 worked redundantly in the repression of embryonic properties (Tanaka et al., 2008) and AtHDA7 was required for female gametophyte development and embryogenesis (Cigliano et al., 2013). Moreover, a study on tomato revealed five SlHDAs (SlHDA1, 3, 5, 6, and 7) could have the potential roles in fruit development and ripening process by their high expression levels in tomato fruit development (Aiese Cigliano et al., 2013). In this study, CsHDA5 and 6, being homolog to AtHDA9 and 19 respectively, also showed the increasing expression levels in flesh during the citrus fruit development (Figure 9), suggesting that these two genes could also have deacetylation functions with the repression on genes involved in fruit development.
A few studies have shown that the HM genes play vital roles in plant immunity (Alvarez et al., 2010). In Arabidopsis, HMT gene SDG8 was confirmed to be crucial in plant defense against fungal pathogens by regulating genes within JA (jasmonic acid) and/or ethylene signaling pathway (Berr et al., 2010a). ATX1 (AtSDG27), a TRX (TRITHORAX) member involved in H3K4 trimethylation, activated WRKY70 and SA-sensitive genes and reinforced basal resistance to Pseudomonas syringae (Alvarez-Venegas et al., 2007). In our study, CsSDG7 and CsSDG13, being homologous to AtSDG8 and AtSDG27 which catalyzed the H3K36 and H3K4 methylation respectively (both of them activated gene expression), were down-regulated under the blue mold infection (Figure 10). For HDAs, AtHDA6 was involved in regulating tolerance to necrotrophic fungi by repressing the JA and ethylene signaling in Arabidopsis (Zhu et al., 2011). AtHDA19 activated the resistance against Alternaria brassicicola and was also involved in JA and ethylene signaling of pathogen response in Arabidopsis (Zhou et al., 2005; Choi et al., 2012). In citrus, CsHDA4, being homologous to AtHDA6, was up-regulated responding to blue mold infection, while CsHDA6, clustering with AtHDA19, was suppressed (Figure 10). Overall, 20 genes including five CsSDGs, one CsHDMA, four CsJMJs, seven CsHAGs, one CsHDA, and two CsSRTs exhibited the strong alterations of their expression levels under the infection (Supplementary Figure S2). The expression change of these genes implied that they could be involved in fruit-blue mold infection process.
Conclusion
This study provided the first insight into the CsHMs in citrus and their expression patterns during the citrus fruit development as well as response to fruit-blue mold infection. These CsHM genes were further characterized from the perspectives of genomic organization, phylogenetic relationship, domain composition, and gene structure. Additional expression analysis of these genes was measured in six different fruit developmental stages and four periods of blue mold infection. From the results, we obtained a numbers of genes with the increasing expression profiles during the fruit development and 20 strongly blue mold responsive genes. The comprehensive characterizations of CsHMs presented in our study will be useful for future research to unravel the mechanisms of histone modification regulations in citrus.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yizhong He, from the Key Laboratory of Horticultural Plant Biology of Ministry of Education, Huazhong Agricultural University, for providing the peel samples of blue mold infection. We thank Prof. Zuoxiong Liu, from the College of Foreign Languages of Huazhong Agricultural University, for reading the manuscript. We also thank Dr. Chengquan Yang, from the Key Laboratory of Horticultural Plant Biology of Ministry of Education, for reading the manuscript. This project was supported by the Ministry of Science and Technology of China (2011CB100600) and the National Natural Science Foundation of China (31330066 and 31221062).
Footnotes
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00607
List of the histone modification genes (CsHMs) in sweet orange.
Real-time PCR primers of sweet orange HMs.
References
- Ahmad A., Cao X. (2012). Plant PRMTs broaden the scope of arginine methylation. J. Genet. Genomics 39 195–208. 10.1016/j.jgg.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Aiese Cigliano R., Sanseverino W., Cremona G., Ercolano M. R., Conicella C., Consiglio F. M. (2013). Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14:57 10.1186/1471-2164-14-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alinsug M. V., Yu C. W., Wu K. (2009). Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 9:37 10.1186/1471-2229-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M. E., Nota F., Cambiagno D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11 563–576. 10.1111/J.1364-3703.2010.00621.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Al Abdallat A., Guo M., Alfano J. R., Avramova Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2 106–113. 10.4161/epi.2.2.4404 [DOI] [PubMed] [Google Scholar]
- Aquea F., Vega A., Timmermann T., Poupin M. J., Arce-Johnson P. (2011). Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep. 30 1087–1097. 10.1007/s00299-011-1015-0 [DOI] [PubMed] [Google Scholar]
- Bain J. M. (1958). Morphological, anatomical, and physiological changes in the developing fruit of the Valencia orange, Citrus sinensis (L) Osbeck. Aust. J. Bot. 6 1–23. 10.1071/BT9580001 [DOI] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. 10.1038/Nature05915 [DOI] [PubMed] [Google Scholar]
- Berr A., Mccallum E. J., Alioua A., Heintz D., Heitz T., Shen W. H. (2010a). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154 1403–1414. 10.1104/pp.110.161497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Mccallum E. J., Menard R., Meyer D., Fuchs J., Dong A. W., et al. (2010b). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22 3232–3248. 10.1105/tpc.110.079962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Shafiq S., Shen W. H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809 567–576. 10.1016/j.bbagrm.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C. I., Cuttriss A. J., Cossetto S. B., Pye W., Crisp P., Whelan J., et al. (2009). Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21 39–53. 10.1105/tpc.108.063131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C. I., Nisar N., Roberts A. C., Murray K. D., Borevitz J. O., Pogson B. J. (2014). A chromatin modifying enzyme, SDG8, is involved in morphological, gene expression, and epigenetic responses to mechanical stimulation. Front. Plant Sci. 5:533 10.3389/fpls.2014.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu Y., Zhou D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809 421–426. 10.1016/j.bbagrm.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., Tian L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769 295–307. 10.1016/j.bbaexp.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. M., Song H. R., Han S. K., Han M., Kim C. Y., Park J., et al. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 71 135–146. 10.1111/j.1365-313X.2012.04977.x [DOI] [PubMed] [Google Scholar]
- Cigliano R. A., Cremona G., Paparo R., Termolino P., Perrella G., Gutzat R., et al. (2013). Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol. 163 431–440. 10.1104/pp.113.221713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R. B., Henikoff S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14 116–122. 10.1016/j.pbi.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G. F., Ma D. P., Li J. X. (2008). The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 373 659–664. 10.1016/j.bbrc.2008.06.096 [DOI] [PubMed] [Google Scholar]
- Feng Q., Wang H. B., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., et al. (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12 1052–1058. 10.1016/S0960-9822(02)00901-6 [DOI] [PubMed] [Google Scholar]
- Gu X. F., Xu T. D., He Y. H. (2014). A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant 7 977–988. 10.1093/Mp/Ssu035 [DOI] [PubMed] [Google Scholar]
- Ho L., Crabtree G. R. (2010). Chromatin remodelling during development. Nature 463 474–484. 10.1038/Nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C., Liu Z. C. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50 875–885. 10.1111/j.1744-7909.2008.00704.x [DOI] [PubMed] [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R. M. (2007). Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19 2975–2987. 10.1105/tpc.107.052373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Ikoma Y., Matsumoto H., Sugiura M., Hyodo H., Yano M. (2004). Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 134 824–837. 10.1104/pp.103.031104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Sasaki T., Ueda M., Sako K., Seki M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114 10.3389/Fpls.2015.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., To T. K., Nishioka T., Seki M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33 604–611. 10.1111/j.1365-3040.2009.02076.x [DOI] [PubMed] [Google Scholar]
- Kim S., Lee J., Yang J. Y., Jung C., Chua N. H. (2013a). Arabidopsis histone methyltransferase SET DOMAIN GROUP2 is required for regulation of various hormone responsive genes. J. Plant Biol. 56 39–48. 10.1007/s12374-012-0320-7 [DOI] [Google Scholar]
- Kim W., Latrasse D., Servet C., Zhou D. X. (2013b). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432 394–398. 10.1016/j.bbrc.2012.11.102 [DOI] [PubMed] [Google Scholar]
- Krogan N. T., Hogan K., Long J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139 4180–4190. 10.1242/Dev.085407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61 395–420. 10.1146/annurev.arplant.043008.091939 [DOI] [PubMed] [Google Scholar]
- Liu Q., Xu J., Liu Y., Zhao X., Deng X., Guo L., et al. (2007). A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 58 4161–4171. 10.1093/Jxb/Erm273 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu Q., Tao N. (2006). Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck). J. Huazhong Agri. Uni. 25 300–304. 10.13300/j.cnki.hnlkxb.2006.03.020 [DOI] [Google Scholar]
- Lu F., Cui X., Zhang S., Liu C., Cao X. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 20 387–390. 10.1038/cr.2010.27 [DOI] [PubMed] [Google Scholar]
- Lu F., Li G., Cui X., Liu C., Wang X. J., Cao X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50 886–896. 10.1111/j.1744-7909.2008.00692.x [DOI] [PubMed] [Google Scholar]
- Luo M., Liu X. C., Singh P., Cui Y. H., Zimmerli L., Wu K. Q. (2012). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819 129–136. 10.1016/j.bbagrm.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Ma X., Lv S., Zhang C., Yang C. (2013). Histone deacetylases and their functions in plants. Plant Cell Rep. 32 465–478. 10.1007/s00299-013-1393-6 [DOI] [PubMed] [Google Scholar]
- Macarisin D., Cohen L., Eick A., Rafael G., Belausov E., Wisniewski M., et al. (2007). Penicillium digitatum suppresses production of hydrogen peroxide in host tissue during infection of citrus fruit. Phytopathology 97 1491–1500. 10.1094/PHYTO-97-11-1491 [DOI] [PubMed] [Google Scholar]
- Ning Y.-Q., Ma Z.-Y., Huang H.-W., Mo H., Zhao T.-T., Li L., et al. (2015). Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 43 1469–1484. 10.1093/nar/gku1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Zhang Y., Pei Y., Liu C., Cao X. (2008). Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 148 490–503. 10.1104/pp.108.124727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S., Zakaryan R. P., Gehring H. (2006). Protein arginine methylation: cellular functions and methods of analysis. Biochim. Biophys. Acta 1764 1890–1903. 10.1016/j.bbapap.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C. A., Selinger D. A., Pikaard C. S., Richards E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30 5036–5055. 10.1093/nar/gkf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Wang Z. (2013). Readout of epigenetic modifications. Annu. Rev. Biochem. 82 81–118. 10.1146/annurev-biochem-072711-165700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Niu L., Lu F., Liu C., Zhai J., Kong X., et al. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144 1913–1923. 10.1104/pp.107.099531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouseff R., Raley L., Hofsommer H.-J. (1996). Application of diode array detection with a C-30 reversed phase column for the separation and identification of saponified orange juice Carotenoids. J. Agric. Food Chem. 44 2176–2181. 10.1021/jf950631q [DOI] [Google Scholar]
- Saeed A. I., Hagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., et al. (2006). TM4 microarray software suite. Methods Enzymol. 411 134–193. 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- Saito T., Bai S., Imai T., Ito A., Nakajima I., Moriguchi T. (2015). Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38 1157–1166. 10.1111/pce.12469 [DOI] [PubMed] [Google Scholar]
- Schmitz R. J., Sung S., Amasino R. M. (2008). Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 411–416. 10.1073/pnas.0710423104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Finnegan E. J., Dennis E. S., Peacock W. J. (2006). Quantitative effects of vernalization on FLC and SOC1 expression. Plant J. 45 871–883. 10.1111/j.1365-313X.2006.02652.x [DOI] [PubMed] [Google Scholar]
- Springer N. M., Napoli C. A., Selinger D. A., Pandey R., Cone K. C., Chandler V. L., et al. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132 907–925. 10.1104/pp.102.013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Sui P. F., Jin J., Ye S., Mu C., Gao J., Feng H. Y., et al. (2012). H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 70 340–347. 10.1111/j.1365-313X.2011.04873.x [DOI] [PubMed] [Google Scholar]
- Sun C., Fang J., Zhao T., Xu B., Zhang F., Liu L., et al. (2012). The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24 3235–3247. 10.1105/tpc.112.101436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146 149–161. 10.1104/pp.107.111674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M. P., Wang J. J., Wei N. E., Jiang H., Doerge R. W., et al. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169 337–345. 10.1534/genetics.104.033142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Wang Z., Cao H., Chen F., Liu Y. (2014). The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 84C, 125–133. 10.1016/j.plaphy.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Wu G. A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32 656–662. 10.1038/nbt.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Chen L. L., Ruan X., Chen D., Zhu A., Chen C., et al. (2013). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45 59–66. 10.1038/ng.2472 [DOI] [PubMed] [Google Scholar]
- Yang H. C., Mo H. X., Fan D., Cao Y., Cui S. J., Ma L. G. (2012). Overexpression of a histone H3K4 demethylase, JMJ 15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 31 1297–1308. 10.1007/s00299-012-1249-5 [DOI] [PubMed] [Google Scholar]
- Zhou C. H., Zhang L., Duan J., Miki B., Wu K. Q. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17 1196–1204. 10.1105/tpc.104.028514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., An F., Feng Y., Li P., Xue L., Mu A., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 12539–12544. 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the histone modification genes (CsHMs) in sweet orange.
Real-time PCR primers of sweet orange HMs.