Abstract
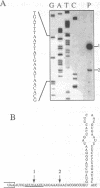
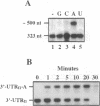
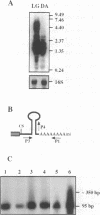
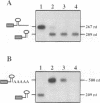
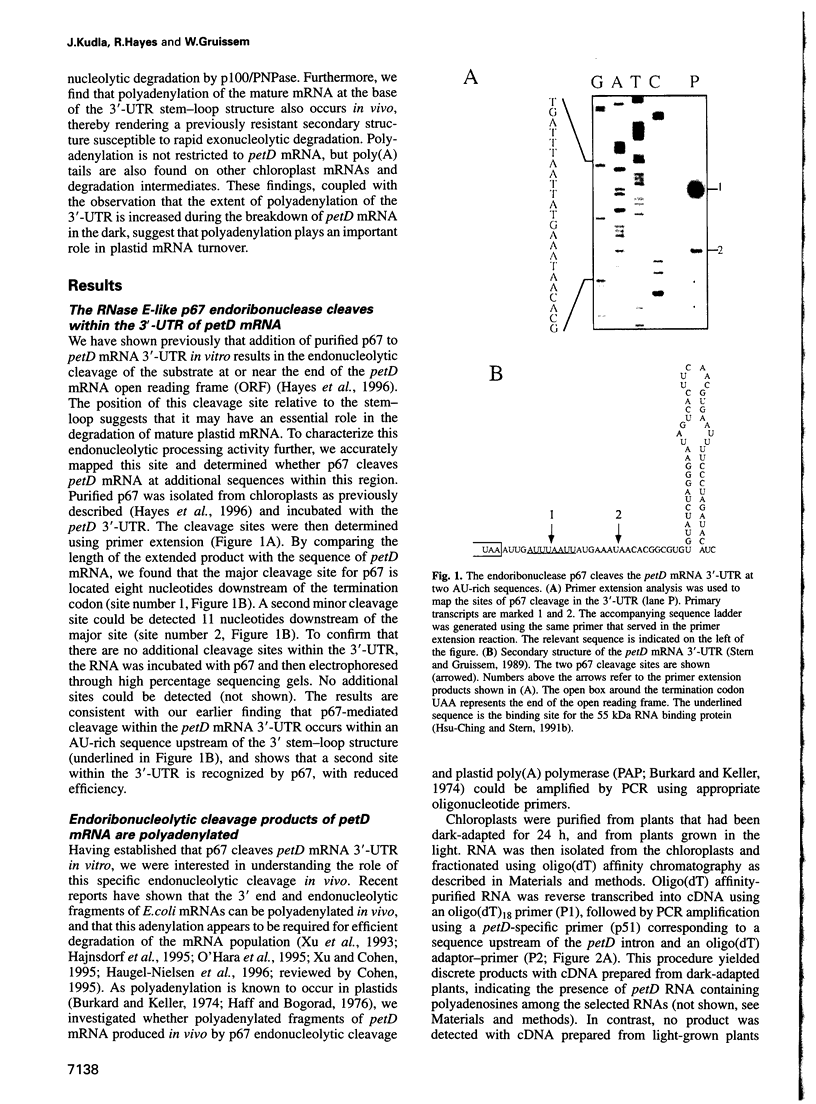
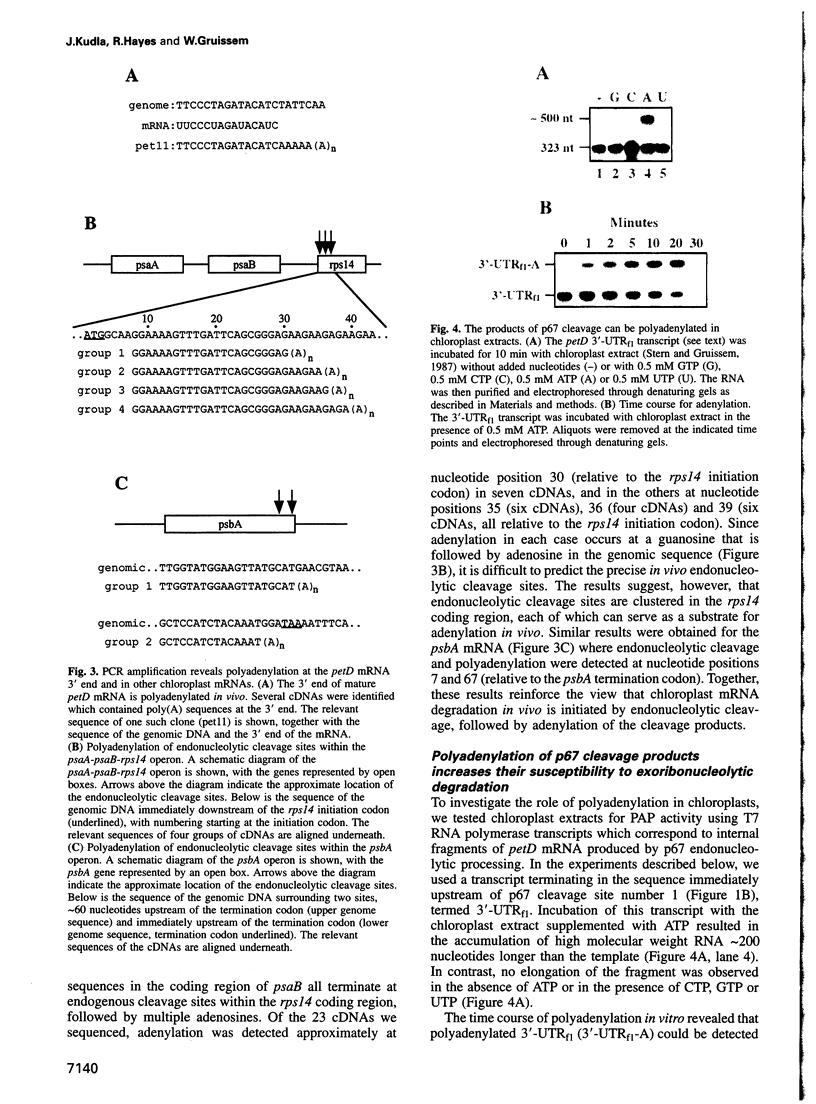
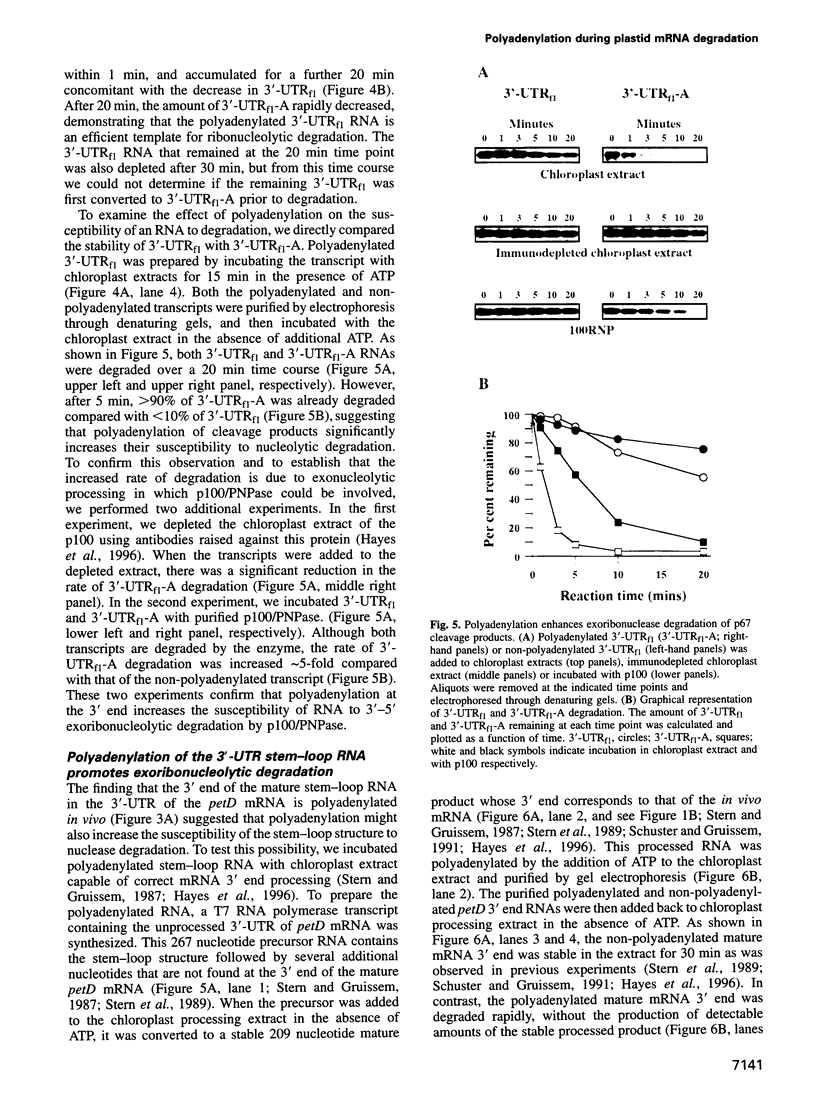
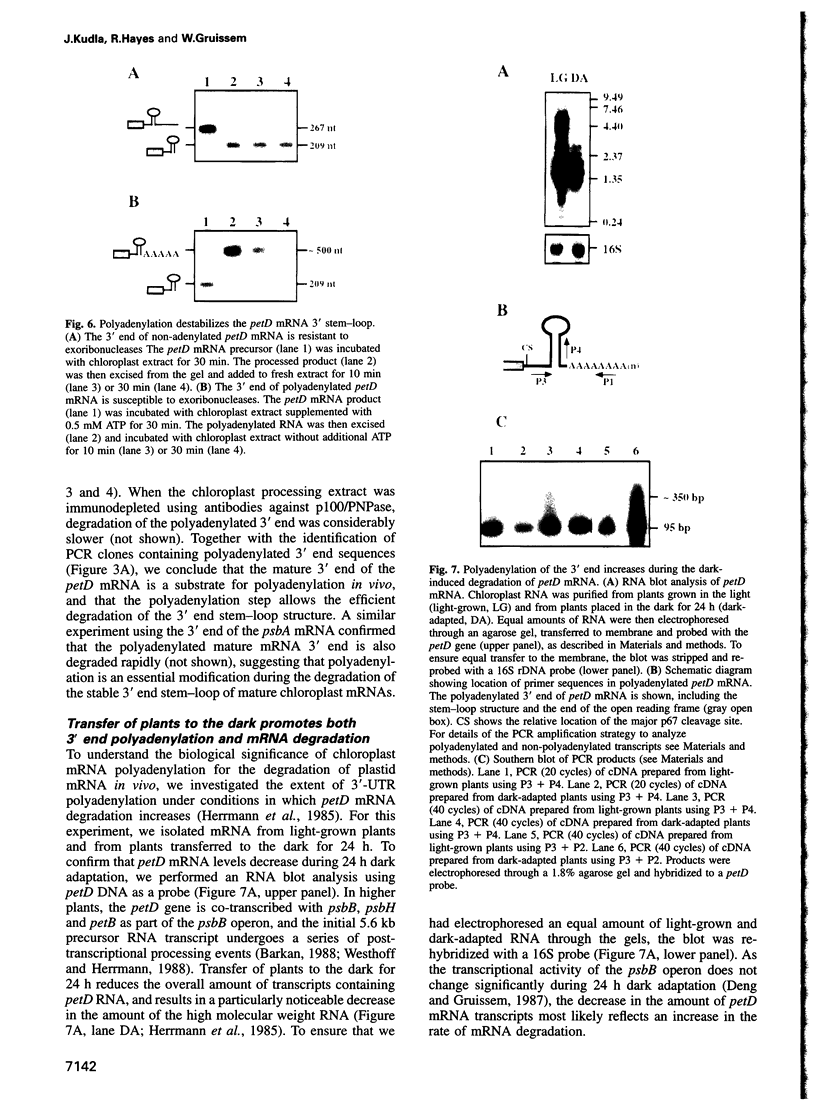
The expression of chloroplast genes is regulated by several mechanisms, one of which is the modulation of RNA stability. To understand how this regulatory step is controlled during chloroplast development, we have begun to define the mechanism of plastid mRNA degradation. We show here that the degradation petD mRNA involves endonucleolytic cleavage at specific sites upstream of the 3' stem-loop structure. The endonucleolytic petD cleavage products can be polyadenylated in vitro, and similar polyadenylated RNA products are detectable in vivo. PCR analysis of the psbA and psaA-psaB-rps14 operons revealed other polyadenylated endonucleolytic cleavage products, indicating that poly(A) addition appears to be an integral modification during chloroplast mRNA degradation. Polyadenylation promotes efficient degradation of the cleaved petD RNAs by a 3'-5' exoribonuclease. Furthermore, polyadenylation also plays an important role in the degradation of the petD mRNA 3' end. Although the 3' end stem-loop is usually resistant to nucleases, adenylation renders the secondary structure susceptible to the 3'-5' exoribonuclease. Analysis of 3' ends confirms that polyadenylation occurs in vivo, and reveals that the extent of adenylation increases during the degradation of plastid mRNA in the dark. Based on these results, we propose a novel mechanism for polyadenylation in the regulation of plastid mRNA degradation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano P., Bruni C. B., Carlomagno M. S. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94(2-3):157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988 Sep;7(9):2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C. A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995 Apr 21;81(2):179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Burkard G., Keller E. B. Poly(A) polymerase and poly(g) polymerase in wheat chloroplasts. Proc Natl Acad Sci U S A. 1974 Feb;71(2):389–393. doi: 10.1073/pnas.71.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. Poly(A) RNA in Bacillus subtilis: identification of the polyadenylylation site of flagellin mRNA. FEMS Microbiol Lett. 1993 Apr 15;108(3):281–285. doi: 10.1111/j.1574-6968.1993.tb06116.x. [DOI] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7546–7550. doi: 10.1073/pnas.89.16.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994 Mar 11;76(5):889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chen H. C., Stern D. B. Specific binding of chloroplast proteins in vitro to the 3' untranslated region of spinach chloroplast petD mRNA. Mol Cell Biol. 1991 Sep;11(9):4380–4388. doi: 10.1128/mcb.11.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. C., Stern D. B. Specific ribonuclease activities in spinach chloroplasts promote mRNA maturation and degradation. J Biol Chem. 1991 Dec 15;266(35):24205–24211. [PubMed] [Google Scholar]
- Chen Q., Adams C. C., Usack L., Yang J., Monde R. A., Stern D. B. An AU-rich element in the 3' untranslated region of the spinach chloroplast petD gene participates in sequence-specific RNA-protein complex formation. Mol Cell Biol. 1995 Apr;15(4):2010–2018. doi: 10.1128/mcb.15.4.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G. A., Mackie G. A. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J Biol Chem. 1996 Jan 12;271(2):1048–1053. doi: 10.1074/jbc.271.2.1048. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Surprises at the 3' end of prokaryotic RNA. Cell. 1995 Mar 24;80(6):829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987 May 8;49(3):379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Drager R. G., Zeidler M., Simpson C. L., Stern D. B. A chloroplast transcript lacking the 3' inverted repeat is degraded by 3'-->5' exoribonuclease activity. RNA. 1996 Jul;2(7):652–663. [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B. M., Zurawski G., Hallick R. B. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986;118:253–270. doi: 10.1016/0076-6879(86)18077-3. [DOI] [PubMed] [Google Scholar]
- Haff L. A., Bogorad L. Poly(adenylic acid)-containing RNA from plastids of maize. Biochemistry. 1976 Sep 7;15(18):4110–4115. doi: 10.1021/bi00663a030. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E., Braun F., Haugel-Nielsen J., Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugel-Nielsen J., Hajnsdorf E., Regnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996 Jun 17;15(12):3144–3152. [PMC free article] [PubMed] [Google Scholar]
- Hayes R., Kudla J., Schuster G., Gabay L., Maliga P., Gruissem W. Chloroplast mRNA 3'-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 1996 Mar 1;15(5):1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991 Sep 12;353(6340):178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Karnik P., Gopalakrishna Y., Sarkar N. Construction of a cDNA library from polyadenylated RNA of Bacillus subtilis and the determination of some 3'-terminal sequences. Gene. 1986;49(1):161–165. doi: 10.1016/0378-1119(86)90397-5. [DOI] [PubMed] [Google Scholar]
- Karnik P., Taljanidisz J., Sasvari-Szekely M., Sarkar N. 3'-terminal polyadenylate sequences of Escherichia coli tryptophan synthetase alpha-subunit messenger RNA. J Mol Biol. 1987 Jul 20;196(2):347–354. doi: 10.1016/0022-2836(87)90695-4. [DOI] [PubMed] [Google Scholar]
- Klaff P. mRNA decay in spinach chloroplasts: psbA mRNA degradation is initiated by endonucleolytic cleavages within the coding region. Nucleic Acids Res. 1995 Dec 11;23(23):4885–4892. doi: 10.1093/nar/23.23.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Wong T. T., McDowall K. J., Cohen S. N. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J Biol Chem. 1994 Apr 8;269(14):10797–10803. [PubMed] [Google Scholar]
- McDowall K. J., Kaberdin V. R., Wu S. W., Cohen S. N., Lin-Chao S. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature. 1995 Mar 16;374(6519):287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- Miczak A., Kaberdin V. R., Wei C. L., Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J. E., Klein R. R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987 Jun;6(6):1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato H., Venkatesan S., Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975 Jul 10;256(5513):144–146. doi: 10.1038/256144a0. [DOI] [PubMed] [Google Scholar]
- O'Hara E. B., Chekanova J. A., Ingle C. A., Kushner Z. R., Peters E., Kushner S. R. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe C. M., Maslesa-Galić S., Simons R. W. Decay of the IS10 antisense RNA by 3' exoribonucleases: evidence that RNase II stabilizes RNA-OUT against PNPase attack. Mol Microbiol. 1994 Sep;13(6):1133–1142. doi: 10.1111/j.1365-2958.1994.tb00504.x. [DOI] [PubMed] [Google Scholar]
- Py B., Causton H., Mudd E. A., Higgins C. F. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994 Nov;14(4):717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Py B., Higgins C. F., Krisch H. M., Carpousis A. J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996 May 9;381(6578):169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Sachs A., Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993 Nov 5;268(31):22955–22958. [PubMed] [Google Scholar]
- Sarkar N., Langley D., Paulus H. Isolation and characterization of polyadenylate-containing RNA from Bacillus brevis. Biochemistry. 1978 Aug 22;17(17):3468–3474. doi: 10.1021/bi00610a007. [DOI] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. Chloroplast mRNA 3' end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991 Jun;10(6):1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Jones H., Gruissem W. Function of plastid mRNA 3' inverted repeats. RNA stabilization and gene-specific protein binding. J Biol Chem. 1989 Nov 5;264(31):18742–18750. [PubMed] [Google Scholar]
- Stern D. B., Kindle K. L. 3'end maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol Cell Biol. 1993 Apr;13(4):2277–2285. doi: 10.1128/mcb.13.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P., Herrmann R. G. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur J Biochem. 1988 Feb 1;171(3):551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- Xu F., Cohen S. N. RNA degradation in Escherichia coli regulated by 3' adenylation and 5' phosphorylation. Nature. 1995 Mar 9;374(6518):180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- Xu F., Lin-Chao S., Cohen S. N. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]