Abstract
We recently showed that primer tRNA3Lys, human immunodeficiency virus type 1 (HIV-1) RNA and HIV-1 reverse transcriptase (RT) form a specific complex of initiation of reverse transcription that can be functionally distinguished from the elongation complex, which can be obtained by substituting an 18mer oligodeoxyribonucleotide (ODN) for the natural primer (Isel et al., 1996). Here, we compared the binding properties and the single and multiple turnover kinetics of HIV-1 RT in the initiation and elongation complexes. Even though the equilibrium dissociation constants of HIV-1 RT are not very different for the two complexes, RT dissociates approximately 200-fold faster from the initiation complex. Furthermore, nucleotide incorporation by the pre-formed primer-template-RT complexes is reduced by a approximately 50-fold factor during initiation of reverse transcription, compared with elongation. As a consequence, processivity of HIV-1 RT in the initiation complex is close to unity, while it increases by four orders of magnitude during elongation, as expected for a replication enzyme. This processivity change is reminiscent of the transition from initiation to elongation of transcription. Furthermore, our results indicate that the post-transcriptional modifications of tRNA3Lys play a role similar to that of the sigma factor in transcription by the Escherichia coli RNA polymerase: they favour the formation of the specific initiation complex but do not affect the polymerization rate of the bound enzyme.
Full text
PDF
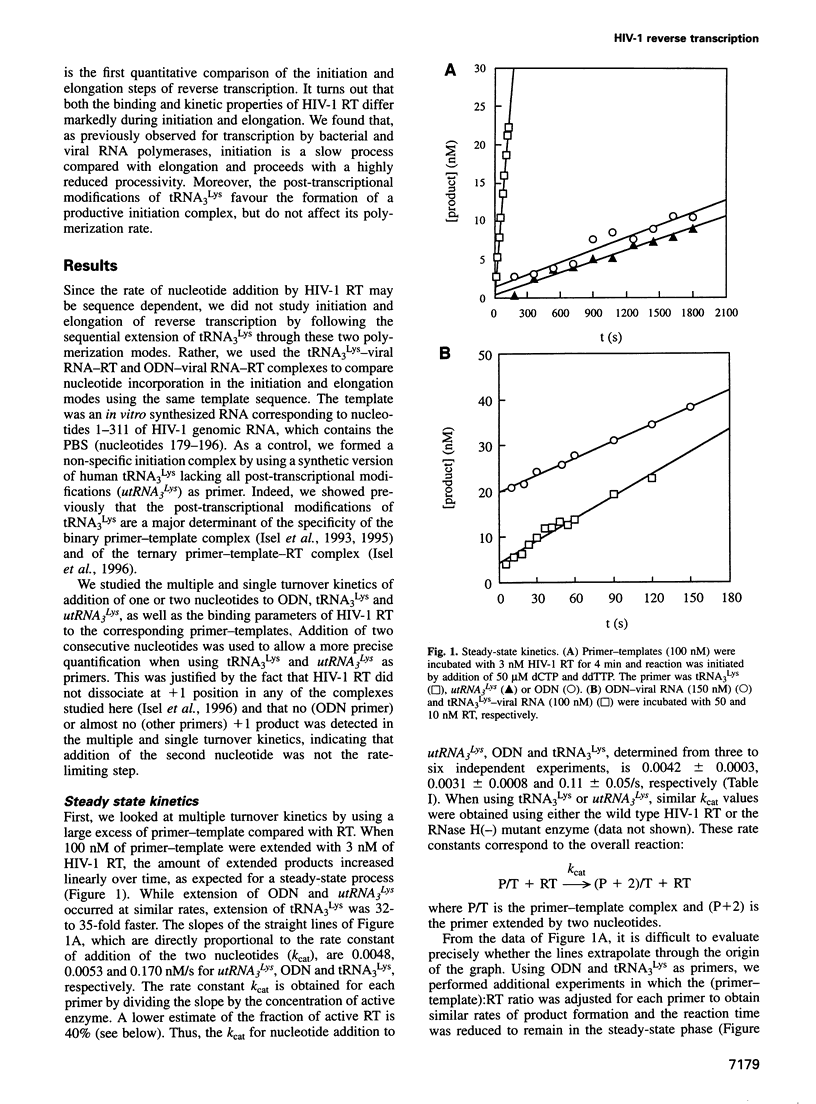

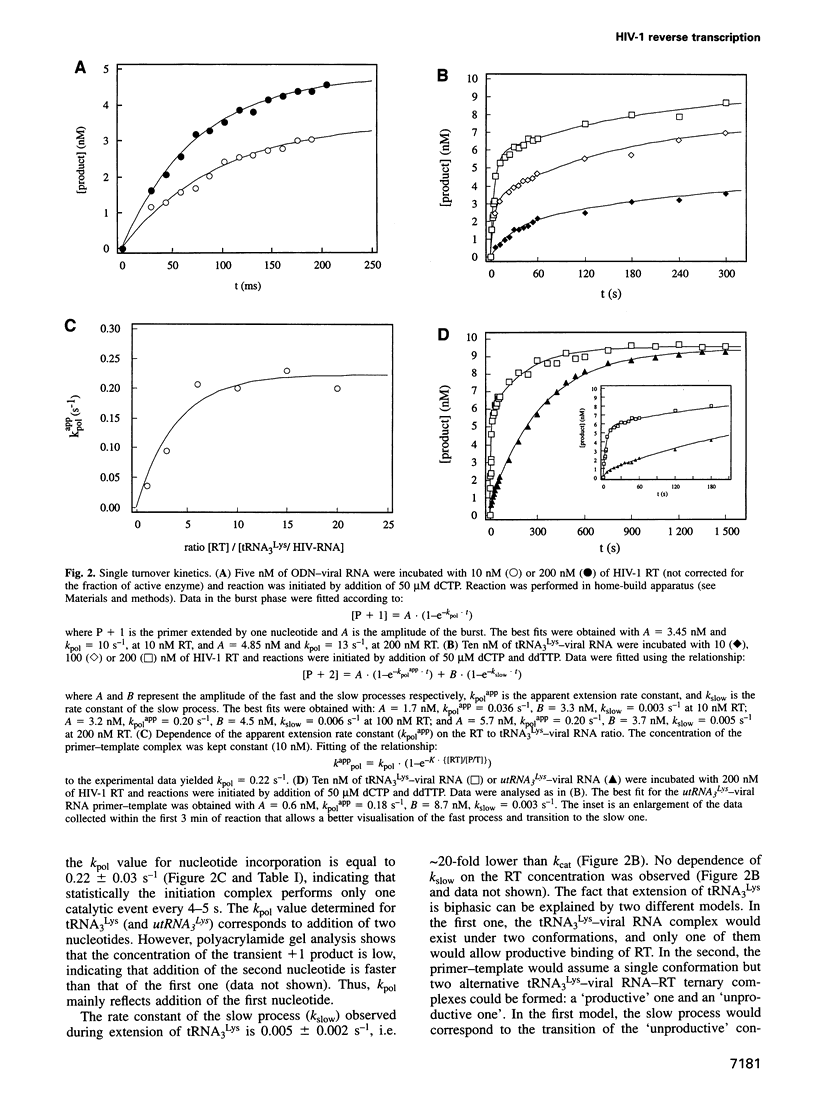

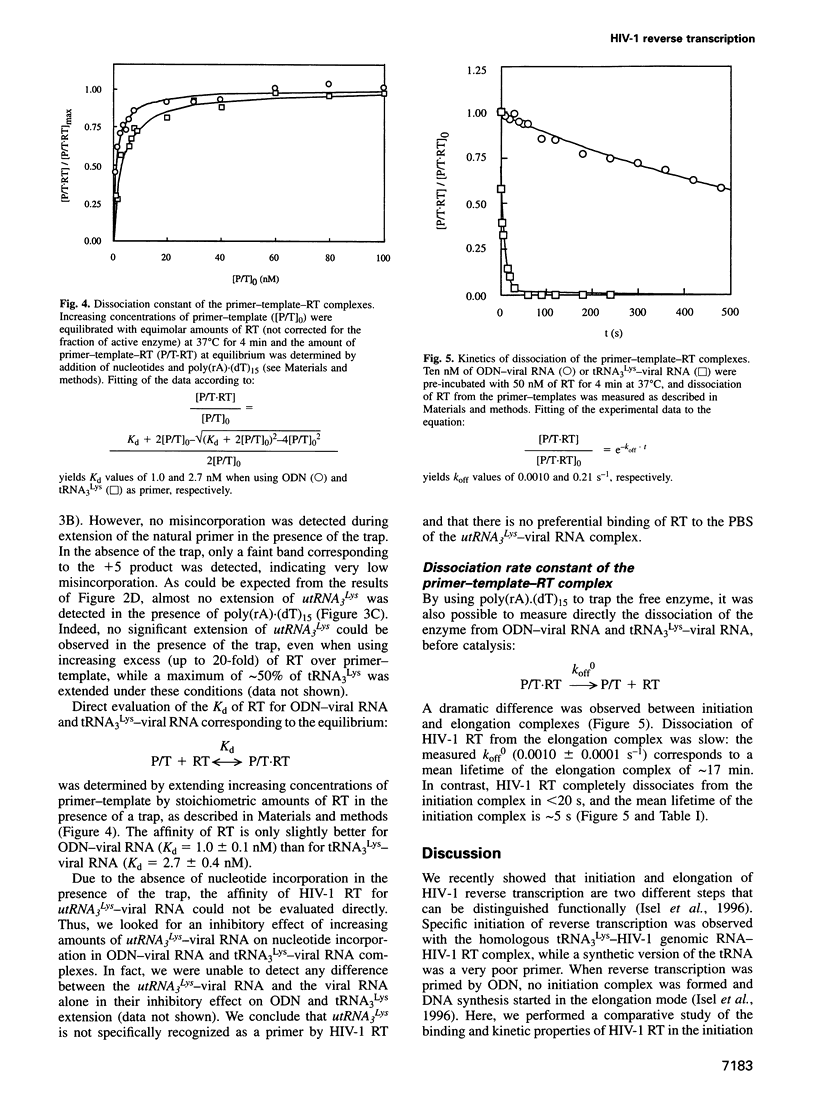




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiyar A., Cobrinik D., Ge Z., Kung H. J., Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol. 1992 Apr;66(4):2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A., Ge Z., Leis J. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J Virol. 1994 Feb;68(2):611–618. doi: 10.1128/jvi.68.2.611-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E., Jacobo-Molina A., Nanni R. G., Williams R. L., Lu X., Ding J., Clark A. D., Jr, Zhang A., Ferris A. L., Clark P. Structure of HIV-1 reverse transcriptase/DNA complex at 7 A resolution showing active site locations. Nature. 1992 May 7;357(6373):85–89. doi: 10.1038/357085a0. [DOI] [PubMed] [Google Scholar]
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Beard W. A., Wilson S. H. Kinetic analysis of template.primer interactions with recombinant forms of HIV-1 reverse transcriptase. Biochemistry. 1993 Sep 21;32(37):9745–9753. doi: 10.1021/bi00088a029. [DOI] [PubMed] [Google Scholar]
- DeStefano J. J., Bambara R. A., Fay P. J. Parameters that influence the binding of human immunodeficiency virus reverse transcriptase to nucleic acid structures. Biochemistry. 1993 Jul 13;32(27):6908–6915. doi: 10.1021/bi00078a014. [DOI] [PubMed] [Google Scholar]
- Friant S., Heyman T., Wilhelm M. L., Wilhelm F. X. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res. 1996 Feb 1;24(3):441–449. doi: 10.1093/nar/24.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff J., Pouyet J., Kern D., Dirheimer G. A quenched-flow apparatus which allows the measurement of the kinetics of a reaction in one stroke. J Biochem Biophys Methods. 1984 Jul;9(3):201–213. doi: 10.1016/0165-022x(84)90025-3. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Hsieh J. C., Zinnen S., Modrich P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J Biol Chem. 1993 Nov 25;268(33):24607–24613. [PubMed] [Google Scholar]
- Isel C., Ehresmann C., Keith G., Ehresmann B., Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer). J Mol Biol. 1995 Mar 24;247(2):236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- Isel C., Lanchy J. M., Le Grice S. F., Ehresmann C., Ehresmann B., Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996 Feb 15;15(4):917–924. [PMC free article] [PubMed] [Google Scholar]
- Isel C., Marquet R., Keith G., Ehresmann C., Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993 Dec 5;268(34):25269–25272. [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaju M., Beard W. A., Wilson S. H. Human immunodeficiency virus type 1 reverse transcriptase. 3'-Azidodeoxythymidine 5'-triphosphate inhibition indicates two-step binding for template-primer. J Biol Chem. 1995 Apr 28;270(17):9740–9747. doi: 10.1074/jbc.270.17.9740. [DOI] [PubMed] [Google Scholar]
- Kati W. M., Johnson K. A., Jerva L. F., Anderson K. S. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992 Dec 25;267(36):25988–25997. [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990 Jan 26;187(2):307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- Marquet R., Baudin F., Gabus C., Darlix J. L., Mougel M., Ehresmann C., Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991 May 11;19(9):2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R., Isel C., Ehresmann C., Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77(1-2):113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Muller D. K., Coleman J. E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988 May 31;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Patel P. H., Jacobo-Molina A., Ding J., Tantillo C., Clark A. D., Jr, Raag R., Nanni R. G., Hughes S. H., Arnold E. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995 Apr 25;34(16):5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- Patel S. S., Wong I., Johnson K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991 Jan 15;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Reardon J. E. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J Biol Chem. 1993 Apr 25;268(12):8743–8751. [PubMed] [Google Scholar]
- Reardon J. E. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry. 1992 May 12;31(18):4473–4479. doi: 10.1021/bi00133a013. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Divita G., Goody R. S. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci U S A. 1995 Aug 15;92(17):8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto T., Hsu M. Y., Inouye S., Inouye M. Reverse transcriptases from bacterial retrons require specific secondary structures at the 5'-end of the template for the cDNA priming reaction. J Biol Chem. 1993 Feb 5;268(4):2684–2692. [PubMed] [Google Scholar]
- Spence R. A., Kati W. M., Anderson K. S., Johnson K. A. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995 Feb 17;267(5200):988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970 Jun 27;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- Wakefield J. K., Kang S. M., Morrow C. D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNA(His). J Virol. 1996 Feb;70(2):966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992 Nov 13;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Wang H., Lambowitz A. M. The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell. 1993 Dec 17;75(6):1071–1081. doi: 10.1016/0092-8674(93)90317-j. [DOI] [PubMed] [Google Scholar]
- Wilhelm M., Wilhelm F. X., Keith G., Agoutin B., Heyman T. Yeast Ty1 retrotransposon: the minus-strand primer binding site and a cis-acting domain of the Ty1 RNA are both important for packaging of primer tRNA inside virus-like particles. Nucleic Acids Res. 1994 Nov 11;22(22):4560–4565. doi: 10.1093/nar/22.22.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I., Patel S. S., Johnson K. A. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991 Jan 15;30(2):526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- Zimmerly S., Guo H., Perlman P. S., Lambowitz A. M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995 Aug 25;82(4):545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]




