Abstract
Nosiheptide is a parent compound of thiopeptide family that exhibit potent activities against various bacterial pathogens. Its C-terminal amide formation is catalyzed by NosA, which is an unusual strategy for maturating certain thiopeptides by processing their precursor peptides featuring a serine extension. We here report the crystal structure of truncated NosA1-111 variant, revealing three key elements, including basic lysine 49 (K49), acidic glutamic acid 101 (E101) and flexible C-terminal loop NosA112-151, are crucial to the catalytic terminal amide formation in nosiheptide biosynthesis. The side-chain of residue K49 and the C-terminal loop fasten the substrate through hydrogen bonds and hydrophobic interactions. The side-chain of residue E101 enhances nucleophilic attack of H2O to the methyl imine intermediate, leading to Cα-N bond cleavage and nosiheptide maturation. The sequence alignment of NosA and its homologs NocA, PbtH, TpdK and BerI, and the enzymatic assay suggest that the mechanistic studies on NosA present an intriguing paradigm about how NosA family members function during thiopeptide biosynthesis.
Thiopeptides are a class of sulfur-rich, highly modified peptide antibiotics that are active against various drug-resistant bacterial pathogens. These antibiotics share a common ribosomally synthesized paradigm in biosynthesis, featuring conserved post-translational modifications of a precursor peptide to afford a family-characteristic framework in which a nitrogen-containing, six-membered ring is central to multiple azoles and dehydroamino acids. Many thiopeptides, including the bimacrocyclic members nosiheptide and thiostrepton (Fig. 1), possess a terminal amide moiety, formation of which, however, can proceed in completely different biosynthetic routes. In thiostrepton biosynthesis1,2, terminal amide formation involves an asparagine synthetase-like protein to incorporate an exogenous amino group arising from Gln, a precursor peptide-independent residue (where enzymes TsrB and TsrC catalyzes deesterification and amidation for thiostrepton maturation, respectively). In contrast, the amino group of the terminal amide moiety in nosiheptide is endogenous and derives from an extended Ser residue of the precursor peptide3,4. Dehydration of this residue at the early stage in the nosiheptide biosynthetic pathway generates enamide, and subsequent dealkylation requires the activity of NosA, which has recently been characterized as a new terminal amide synthase, leading to a Cα-N bond cleavage for nosiheptide maturation with release of the co-product pyruvate. NosA catalyzed reaction is apparently distinct from those of known endogenously C-terminal amide-forming proteins, which typically catalyze an oxidative cleavage of C-terminal Gly-extended peptides accompanying glyoxylate production5,6,7,8. In this study, we provide the structural basis of NosA for investigation into its enzymatic mechanism.
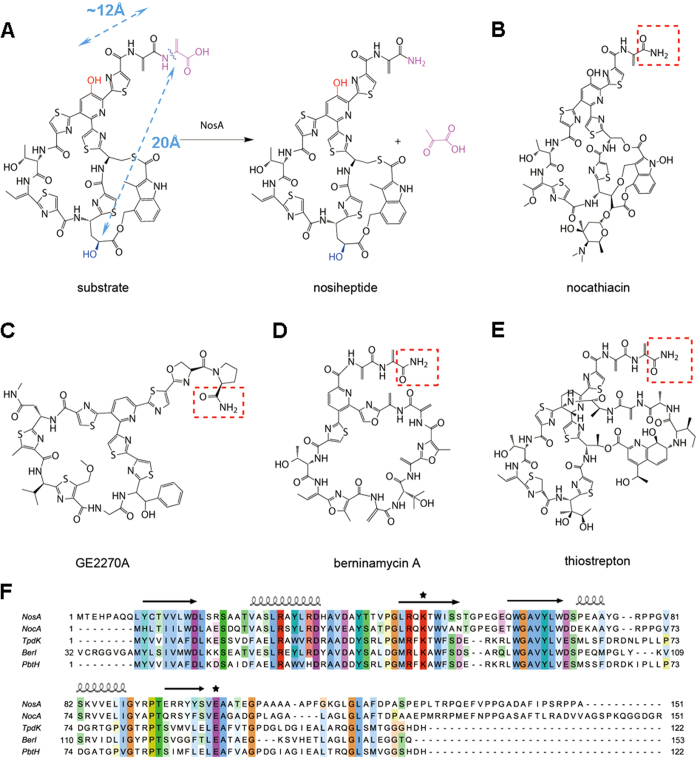
Figure 1. NosA and its homologs share a common mechanism on the post-modifications of thiopeptides.
(A) The catalytic reaction for nosiheptide maturation by NosA. The –OH groups involved in the interactions with NosA are highlighted in red and blue, respectively. The Cα-N bond cleavage site is marked in pink and a wavy line. The distances between the oxygen atoms of the –OH groups and the Cα-N bond cleavage site in the substrate were measured as 12 Å and 20 Å, indicated by blue dotted lines, respectively. (B–D) the C-terminal amide formation of the thiopeptides similar to nosiheptide, such as nocathiacin (catalyzed by NocA), (C) GE2270A (catalyzed by TpdK or PbtH). (D) berninamycin A (catalyzed by BerI). (E) For comparison, thiostrepton maturation is done through deesterification and amidation by TsrB and TsrC, respectively. In (B–E), the products of the catalytic reactions were highlighted in red dashed boxes. (F) The sequence alignments of NosA and its homologs NocA, TpdK, PbtH and BerI. The conserved residues K49 and E101 were marked with stars on the top of the sequences. On the top of the amino acid sequence, the secondary structures of NosA were displayed, where arrows indicate β-sheets, and coils represent α-helices. The stars indicate the active sites observed in this report for enzymatic reaction.
Results and Discussion
Derivation of the truncated NosA1-111 variant and its X-ray crystal structure
The full-length NosA contains 151 amino acids in total. Within ten days, it degrades as a large fragment with a molecular weight about 12 KDa (supporting information, Fig. S1). This instability is likely due to the sequence (residues 106-151) at the C-terminus, which has a potential to form a flexible loop based on the secondary structure analysis (http://bioinf.cs.ucl.ac.uk/psipred/) (supporting information, Fig. S2). Thus, we truncated NosA by cutting off the residues at its C-terminus ten by ten, and constructed the pET28a plasmids containing the genes NosA1-140 (i.e., residues 1-140), NosA1-130 (i.e., residues 1-130), NosA1-120 (i.e., residues 1-120), and NosA1-111 (i.e., residues 1-111), and carried out their overexpression and purification as we did on wild-type (WT) NosA. By running SDS_PAGE gels (supporting information, Fig. S1), we found that, except NosA1-111, other NosA variants are still unstable (Among them, NosA1-130 is too unstable to be obtained in the process of purification).
We thus overexpressed NosA1-111 variant and its Se-Met form, and purified them to homogeneity for crystallization. The purified proteins were estimated to have a purity of >95%. The crystals grew in a cubic form. The diffraction of Se-Met NosA1-111 was extended to 2.40 Å resolution. Its crystals belong to the primitive cubic space group P4132, with unit cell parameters of a = b = c = 143.3 Å. X-ray diffraction data sets of Se-Met NosA1-111 were processed using data in the resolution ranges 50.0–2.4 Å. The typical Matthews coefficient and solvent content were estimated as 3.94 Å3 Da−1 and 40.3%, respectively. The three-dimensional (3D) structure of NosA1-111 was determined using the single-wavelength anomalous-dispersion (SAD) method. The crystal parameters and data-collection statistics were summarized in Table 1.
Table 1. Summary of diffraction data and structure refinement statistics.
| Se-NosA1-111 | |
|---|---|
| Summary of diffraction data | |
| Wavelength (Å) | 0.9794 |
| Space group | P4132 |
| Cell parameters | |
| a = b = c (Å) | 143.3 |
| Resolution (Å) | 50.0-2.4 (2.59-2.40)a |
| Observed reflections | 1,483,177 |
| Unique reflections (I/σ(I) > 0) | 18,105 |
| Average redundancy | 81.9 (81.8) |
| Average I/σ(I) | 90.0 (17.6) |
| Completeness (%) | 100.0 (100.0) |
| Rmerge (%)b | 9.2 (46.4) |
| Refinement and structure model | |
| Reflections (Fo ≥ 0σ(Fo)) | |
| Working set | 17,056 |
| Test set | 915 |
| R factor/Free R factor (%)c | 17.9/21.5 |
| No. of protein atoms | 2,207 |
| No. of water atoms | 128 |
| Average B factor (Å2) | |
| All atoms | 48.4 |
| Main chain/side chain | 46.6/51.4 |
| Water | 40.3 |
| RMS deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.0 |
| Ramachandran plot (%) | |
| Most favoured regions | 96.6 |
| Allowed regions | 3.4 |
| Generously allowed regions | 0.0 |
aNumbers in parentheses represent the highest resolution shell.
bRmerge = ∑hkl∑i|Ii(hkl)i− < I(hkl) > |/∑hkl∑iIi(hkl).
cR = ∑hkl||Fo|−|Fc||/∑hkl|Fo|.
Three monomers occupy one asymmetric unit (Fig. 2A,B), two of them form a dimer. One monomer constitutes a dimer with one monomer in an adjacent asymmetric unit. Each monomer is identical to the others with an RMSD value of 0.21 Å for the backbone Cα atoms in the secondary structural regions, consisting of four anti-parallel β-sheets (β1, β2, β3, and β4), three α-helices (α1, α2 and α3) and six loops (L1, L2, L3, L4, L5 and L6), which are arranged in the order of β1- L1- α1- L2- β2- L3- β3- L4- α2- L5- α3- L6- β4 (Fig. 2C). The anti-parallel β-sheets form a semicircular hydrophobic surface, the α-helices and loops are located on the outside of the circle. The first six or fewer residues at the N-terminus are invisible in all monomers, and the residues from 35 to 42 are also invisible in the two monomers that form a dimer conformation. This dimeric structure adopts a global fold, resembling an elliptic β-barrel of 28.9 Å in height, with diameters of 15 Å and 23 Å (Fig. 2A). The α-helices and loops surround the β-barrel.
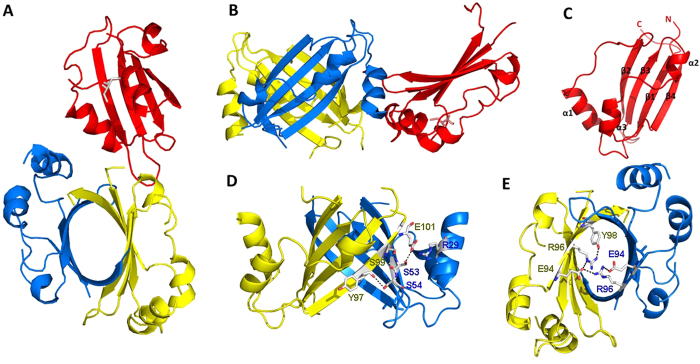
Figure 2. NosA1-111 overall fold.
(A): vertical view, (B): lateral view) Ribbon representations of a NosA trimer observed in an asymmetric unit, monomers were highlighted in red, blue and yellow, respectively. (C) monomer conformation, N-terminal and C-terminal and secondary structures were marked; (D) residues forming the hydrogen bonds in the β4 strand and the β2’ strand outside of β-barrel; (E) the salt-bridge and hydrogen-bonds formation between R96 and E94’ within the β-barrel.
The β-barrel is formed through several hydrogen bonds between the residues of the β4 strand in one monomer and the β2’ strand in another monomer (Fig. 2D), including hydrogen bonds between the E101 carboxyl and S53’ –OH group or R29’ imide group, between the S99 backbone carboxyl and the S53’ backbone nitrogen, and between the S99 backbone nitrogen and the S53’ carboxyl; and a hydrogen bond between the Y97 backbone oxygen and the S54’ side-chain –OH group. The β-barrel is also stabilized by salt bridge within the β-barrel between the side chains of the R96 and E94’ residues (Fig. 2E). Among these residues, R29 and E101 are conserved in the NosA homologs (Fig. 1), indicating that they may be important for enzyme dimerization.
The possible key sites for the catalytic reaction
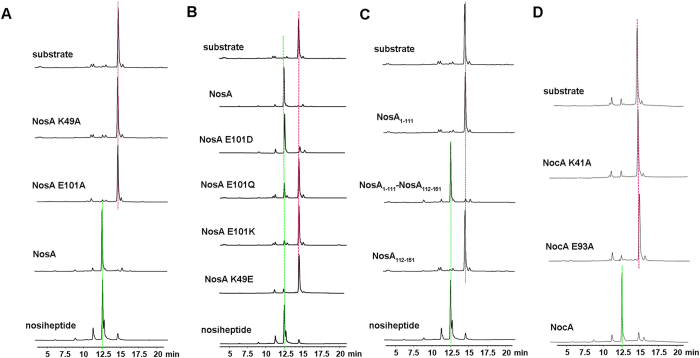
To understand how NosA interacts with its substrate, we searched NosA structural homologs in protein data bank using DALI server9. The structures with Z-score higher than 7.0 were selected. They are heme-degrading enzymes10,11,12,13,14,15 or HapK protein involved in prodigiosin biosynthesis16, demonstrating that their ligands bind to the region outside of the β-barrel (supporting information, Fig. S3). Based on these observations, we assumed that the substrate of NosA might also bind to the region out of the β-barrel. To confirm this, we performed alanine-scanning mutagenesis assay on the conserved residues that are located at the entrances of the β-barrel (such as C11 and Q48), within the β-barrel (such as R29 and S53), outside of the β-barrel (such as Q48, K49, W51 and E101), or in the C-terminal loop (such as F122, D123, P124, S126, D128, P129, R132, P133, E135, F136, P138 and P139) in the full-length NosA, respectively. Then, we measured the catalytic activities of these variants by running enzymatic assay on HPLC system (Fig. 3A and supporting information, Fig. S4). Only K49A and E101A variants abolish the catalytic activities, indicating that K49 and E101 might be key elements for catalytic reaction. As shown in Fig. 3B, when E101 was replaced by D101, Q101 or K101, and residue K49 was mutated into E49, respectively, the E101K, K49E and E101Q variants abolish or significantly lose catalytic activities, while E101D retains the catalytic activity, indicating that the charged side-chains of E101 and K49 are critical to the catalytic reaction.
Figure 3. Enzymatic assay on the full-length NosA, NocA and their variants:
(A) NosA and its K49A and E101A variants; (B) NosA and its E101D, E101Q, E101K and K49E variants; (C) NosA1-111, NosA1-111 mixed with NosA112-151 at mole ratio 1:1, and NosA112-151; (D) NocA and its K41A and E93A variants. In all cases, the substrate (top) and the product nosiheptide (bottom) were used as controls, highlighted in a dotted red line and green line, respectively.
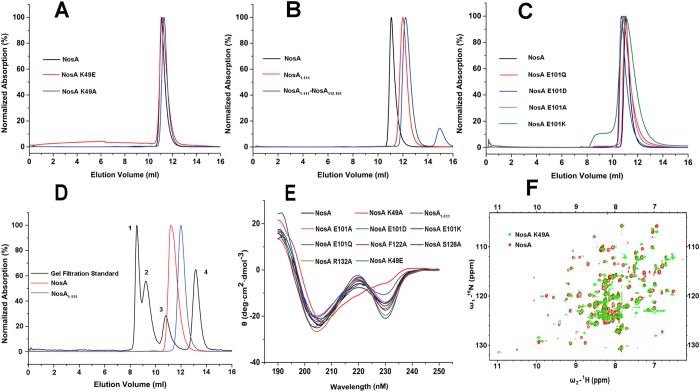
Finally, we tested the possible effects on the folding and the aggregation state of NosA by these mutations by running the size-exclusion chromatography (SEC) assay, circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopies. The results from the SEC assay suggested that the aggregation states of these variants were not affected by these mutations (Fig. 4A–C). Moreover, as shown in Fig. 4D, the retention time of the WT full-length NosA and the truncated NosA1-111 locates between gel filtration protein markers (3 and 4) with molecular weights of 44KDa and 17KDa, respectively, indicating that they are dimer in solution. Thus, the C-terminal loop is not helpful to NosA dimerization. However, the CD spectrum of the K49A variant looks much different from those of WT protein and other variants (Fig. 4E), and its 2D 1H-15N HSQC spectrum did not overlap well with that of WT NosA (Fig. 4F), indicating that the mutation from K49 to A49 might affect the folding of the protein. Thus, the loss of the catalytic activity of K49A might also result from the changes in the folding of the protein.
Figure 4. The folding and aggregation states of full-length NosA, NosA1-111 and its variants.
(A–D) The aggregation states detected by size-exclusion chromatography assay; In (D), gel filtration protein standard markers were highlighted with arabic numerals 1, 2, 3 and 4, representing thyroglobulin with a molecular weight (MW) of 670KDa, γ-globulin with a MW of 158 KDa, Ovalbumin with a MW of 44 KDa, myoglobin with MW of 17 KDa and vitamin B12 with a MW of 1.35 KDa, respectively. (E) The folding of NosA and its variants detected by circular dichroism (CD) spectroscopies respectively; (F) The folding of NosA K49A variant further confirmed by two-dimensional NMR 1H-15N HSQC spectrum of the full-length NosA K49A variant (green), overlapped with that of wild-type NosA (red). In these two NMR experiments, the concentration of the WT NosA protein and its K49A variant is about 0.2 mM in NMR buffer.
NosA may function as a dimer
It was reported that the removal of two –OH groups on the substrate (highlighted in red and in blue in Fig. 1, respectively) (by knockouting the genes nosC and nosB in nosiheptide biosynthesis responsible for these –OH groups formation, respectively) made NosA completely lose the catalytic activities17, suggesting that these two –OH groups in the substrate are important for the catalytic reaction. Interestingly, as shown in supporting information, Fig. S5, in the current crystal structure of NosA1-111, the intra-molecule distance between the oxygen atom in the side-chain of E101 and the nitrogen atom in the side-chains of K49 is approximately 26 Å, much longer than the corresponding inter-molecular distance (13.6 Å) between these two atoms. The latter is close to that (~12 Å) between the –OH group in the pyridine ring (highlighted in red, Fig. 1A) and the Cα-N bond cleavage site in the substrate (measured from the crystal structure of nosiheptide in complex with ribosomal subunit, pdb code 2ZJP, in which the distance between –OH group (highlighted in blue, Fig. 1A) and the Cα-N bond cleavage site is 20 Å)18. Thus, we suggest that NosA may function as a dimer to catalyze the maturation reaction of nosiheptide, which is consistent with the dimer conformation of the full-length NosA and its truncated NosA1-111 variant detected by SEC assay above.
NosA112-151 is crucial to the catalytic reaction
As we mentioned above, to get a stable form of NosA, we prepared several truncated NosA variants. Among them, NosA1-111 is the most stable. However, the enzymatic assay performed on the HPLC system indicates that NosA1-111 has no catalytic activity (Fig. 3C), revealing that the C-terminal loop NosA112-151 is extremely important to the catalytic reaction. To probe whether or not the catalytic efficiency of NosA1-111 can be rescued, we directly mixed NosA1-111 with NosA112-151 at mole ratio of 1:1, and performed the enzymatic assay again. The results demonstrate that the catalytic efficiency of the N-terminal NosA1-111 is partially recovered by the C-terminal NosA112-151. Further enzymatic parameter measurements suggest that the mixture of NosA1-111 and NosA112-151 has a catalytic efficiency of kcat/Km = 4.93 × 10−3 min−1 μM−1 (where the catalytic power kcat = 3.63 ± 0.7 min−1, and the Michaelis constant Km = 736.1 ± 173.7 μM, respectively), decreased by approximately 2000-fold, compared to the full-length NosA (the catalytic efficiency kcat/Km = 9.85 min−1 μM−1, where kcat = 728.1 ± 144.7 min−1, and Km = 73.9 ± 43.5 μM). This observation may be due to weak interaction between NosA1-111 and NosA112-151 (KD = 2.4 ± 1.2 mM measured by ITC binding assay, supporting information, Fig. S6). In the case of NosA1-111 mixed with the peptide NosA112-151, the Michaelis constant Km (the binding affinity of the substrate to the enzyme) is reduced by ten-fold, leading to a much weaker catalytic power (kcat is decreased by 200-fold) than that of full-length NosA, suggesting that NosA1-111 might coordinate with NosA112-151 to interact with the substrate. Therefore, by mixing with NosA112-151, the catalytic activity of NosA1-111 is only slightly recovered with a significant drop in catalytic efficiency.
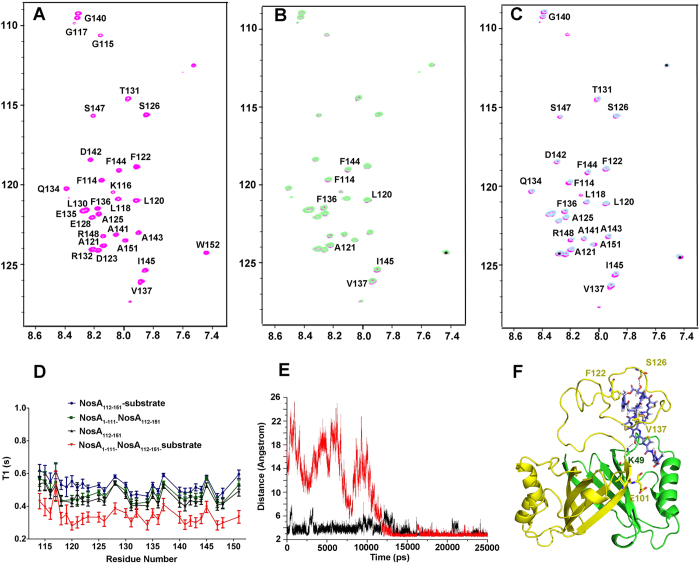
To confirm this hypothesis, we performed the following biochemical assay. The CD spectrum of NosA112-151 reveals that the NosA112-151 peptide is disordered in its free state (supporting information, Fig. S7-A). Upon mixing with the N-terminal NosA1-111, the cross-peaks of the 1H-15N spectrum acquired on NosA112-151 are still not dispersed, mainly located in the region between 8.0 ppm and 8.5 ppm, similar to the observation in 1H-15N HSQC spectrum acquired on free NosA112-151 (Fig. 5A) (both spectra overlapped very well in supporting information, Fig. S7-B). This observation suggests that the C-terminal NosA112-151 peptide is still folded as a random coil conformer upon being mixed with NosA1-111. Moreover, the cross-peaks of the 1H-15N spectra acquired on NosA112-151 did not shift (supporting information, Fig. S7-B), indicating that NosA1-111 does interact very weakly with NosA112-151, consistent with the measurement of the binding affinity of NosA1-111 to NosA112-151 by ITC assay (supporting information, Fig. S6). Moreover, Titrating the substrate into 15N-labeled NosA112-151 solution only results in slight shift of several cross-peaks in 1H-15N HSQC of NosA112-151, suggesting that individual NosA112-151 interacts with the substrate weakly (Fig. 5B). Adding the substrate into the mixture of NosA1-111 and NosA112-151 leads to obvious, but still small chemical shift changes in the 1H-15N spectra (Fig. 5C), indicating that NosA1-111 may coordinate with NosA112-151 to interact with the substrate, consistent with the measurements of the kcat and Km values above. To further confirm this conclusion, we measured the dynamic properties of the backbone atoms (relaxation time T1 and T2 and 15N-1H NOE values) of free NosA112-151, and of NoxA112-151 mixed with NosA1-111, and of NoxA112-151 mixed with the substrate, and of NoxA112-151 mixed with both NosA1-111 and the substrate, respectively. The T1 values of the backbone atoms of NosA112-151 mixed with the substrate and the N-terminal NosA1-111 are the smallest among these cases (Fig. 5D), indicating that the conformation of the NosA112-151 peptide in the ternary complex is the most rigid among these cases. This observation reveals that the flexible conformation of NosA112-151 may be fixed in the presence of NosA1-111 and the substrate.
Figure 5. NosA1-111 coordinates with NosA112-151 to interact with the substrate.
(A) 1H-15N HSQC spectra acquired on free NosA112-151, highlighted with NMR signal assignment of residues; (B) 1H-15N HSQC spectrum of NosA112-151 in complex with the substrate (in green) was overlapped with that of free NosA112-151 (in pink); the signals with chemical shifts changes were marked; (C) 1H-15N HSQC spectrum of NosA112-151 in complex with NosA1-111 and substrate (in grey), overlapped with that of free NosA112-151 (in pink); the signals with chemical shifts changes were marked; In all cases of (A–C), the concentration of NosA112-151 was about 0.2 mM in NMR buffer. (D) Relaxation time T1 measurements of backbone atoms of each residue in 15N-labeled NosA112-151 in its free state (black), in complex with NosA1-111 (green), in complex with the substrate (blue), in complex with NosA1-111 and substrate (red), respectively. (E) Two hydrogen bonds between NosA and the substrate are formed, supported by the distance changes during MD simulation trajectory. The change in the distance between nitrogen atom of the side-chain of K49 and oxygen atom in –OH group in pyridine of the substrate is shown in black, and the change in the distance between the backbone oxygen of S126 and oxygen atom in –OH group in the substrate highlighted in blue in Fig. 1 is shown in red, respectively. (F) The last snapshot of the MD simulation trajectory, where the two monomers of NosA were displayed in green and yellown ribbon modes, respectively. The substrate was displayed in cyan-sticks mode. The main residues contributing to the protein-substrate interactions, including K49 and E101 in NosA1-111 and residues in NosA1-120, were also shown in sticks mode. The two hydrogen-bonds between NosA and substrate were displayed in dotted lines.
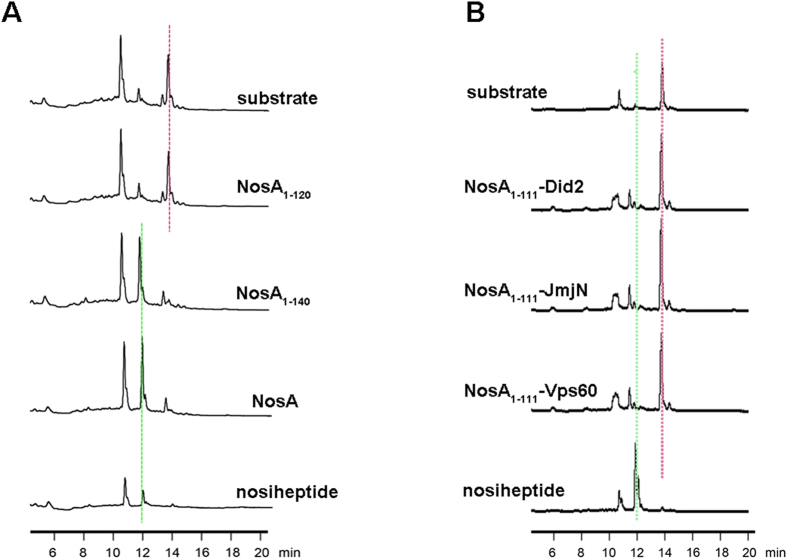
To probe whether the whole sequence of NosA112-151 affects the catalytic reaction, we measured the catalytic activities of the truncated NosA variants with different length, and found that the NosA1-140 variant maintained the catalytic activity almost similar to the full-length NosA, whereas NosA1-120 significantly lost the catalytic activity (Fig. 6A), revealing that the residues from 120 to 140 (i.e., A121FDPASPEPLTRPQEFVPPG140) of NosA112-151 are important to the catalytic reaction. To investigate whether the sequence and the coiled-coil conformation of NosA112-151 are specific to the catalytic reaction, we replaced NosA112-151 in the mixture by three randomly-selected peptides (Did2, Vps60 and JmjN) available in the lab19,20,21,22, respectively, and tested the catalytic activities of these mixtures, all displaying no catalytic activities at all (Fig. 6B). These observations suggest that the sequence and the flexible loop conformation of the NosA112-151 variant is crucial to the catalytic reaction.
Figure 6. The enzymatic assay performed on HPLC systems:
(A) from top to bottom, only substrate as a control, NosA1-120, NosA1-140 variants, full-length NosA, and the catalytic reaction product nosiheptide used as another control; (B) from top to bottom, only substrate as a control, NosA1-111 plus Did2, NosA1-111 plus JmjN from KDM5C, NosA1-111 plus Vps60, and the catalytic reaction product nosiheptide used as another control. In (A,B), the controls substrate and the product nosiheptide were indicated by pink and green dotted line, respectively.
NosA1-111 coordinates with NosA112-151 to bind the substrate
Since the substrate is not able to dissolve in solution, it’s impossible for us to get the crystals of the enzyme NosA in complex with the substrate. We also failed to get the crystals of NosA1-111 in complex with NosA112-151. Thus, to understand how the substrate interacts with NosA, ligand docking, homology modeling and molecular dynamics (MD) simulation were conducted, respectively. During MD simulation trajectory, we observed that: (1) the C-terminal NosA112-151 loop wrapped the substrate after 20–25 ns (supporting information, Fig. S8); (2) The hydrogen bond between the backbone carbonyl oxygen atom of residue S126 and the oxygen atom in the –OH group next to the carbonyl group on the substrate (highlighted in blue in Fig. 1A) was formed around 15 ns, and was maintained during the next 10 ns of simulation (Fig. 5E). We assumed that the formation of this hydrogen bond might induce the C-terminal loop to bend and wrap the substrate. The second hydrogen bond was formed between K49 side-chain in NosA and the -OH group in pyridine ring of the substrate, because the distance between them was kept less than 4 Å (Fig. 5E). These observations are supported by the fact that the catalytic power of NosA was completely abolished after removing these two hydroxyl groups in the substrate17. Moreover, the –OH group in pyridine ring is obviously more acidic than –OH group next to the carbonyl group in the substrate, which could interpret why the side-chain of residue K49 of NosA easily interacts with the –OH group in pyridine ring. (3) Residues involved in the protein-substrate interaction mainly locate at residues 120–140 on the C-terminal loop (Fig. 5F), consistent with the results from the enzymatic assay above. (4) The distance between the side-chain carboxylic oxygen atoms of E101 and the carbon atom at the Cα-N bond cleavage site on the substrate is kept larger than 5 Å, smaller than 12 Å during the MD simulation (data not shown).
The proposed mechanism for NosA catalytic amidation reaction
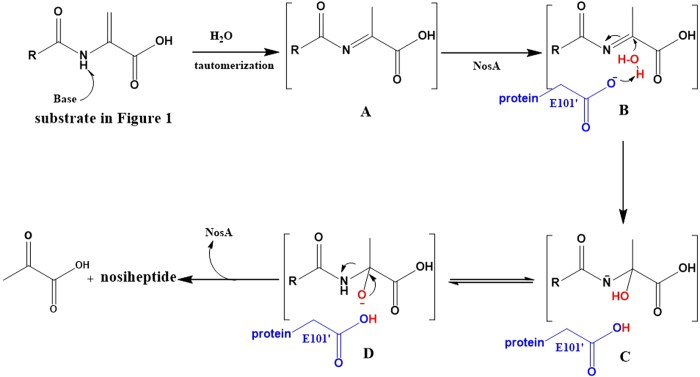
Taken all results above together, we proposed the following catalytic mechanism (Fig. 7): (1) The terminal dehydroalanine unit is tautomerized to methyl imine intermediate A in basic buffer condition, supported by the previous studies on thiostrepton synthesis23,24, where similar reaction is initiated by Et2NH; (2) The substrate is fixed into the active sites by hydrogen-bond and hydrophobic interactions between the substrate and residue K49, the C-terminus of NosA, generating intermediate B, supported by the structural and MD studies above; (3) The negatively charged side-chain of E101’ interacts with one molecule H2O, supported by the findings that several water molecules exist close to E101’ in the crystal structure; (4) The nucleophilic attack by H2O to methyl imine produces intermediates C and D, leading to the final Cα-N bond cleavage to yield nosiheptide and pyruvate.
Figure 7. The proposed mechanism of NosA to catalyze nosiheptide maturation.
NosA family members catalyze the amidation reaction through a similar way
Most importantly, it was reported that NosA homologs could also catalyze the terminal amide formation of some thiopeptides (Fig. 1). For example, NocA (64% identity to NosA) catalyzes the formation of nocathiacin25, TpdK (34% identity to NosA) and PbtH (44% identity to NosA) catalyzes the final formation step in GE2270A biosynthesis26,27, BerI (48% identity to NosA) catalyzes the final step of berninamycin A in its biosynthesis28. Among these enzymes, the residues K49 and E101 are conserved, corresponding to residues K41 and E93 in NocA. Thus, we made the NocA K41A and E93A variants, and measured their catalytic activities on the same substrate (Fig. 3D). The results indicate that either the NocA K41A or E93A variant loses catalytic activities, revealing that NosA and its family members might share a common mechanism to catalyze the terminal amidation reaction.
In conclusion, we characterized three key elements (basic lysine, acidic glutamic acid and flexible C-terminal loop) for the terminal amide formation in nosiheptide-represented thiopeptide biosynthesis, these mechanistic studies on how NosA works present an intriguing paradigm about how NosA family members function during thiopeptide biosynthesis.
Methods
The expression and purification of NosA, NocA and their variants
The ORFs of full-length NosA (151 amino acids in total), truncated NosA variants, including the truncated NosA1-111 (i.e., residues 1-111), NosA1-120 (i.e., residues 1-120), NosA1-130 (i.e., residues 1-130), NosA1-140 (i.e., residues 1-140), and NocA (151 amino acids in total) were engineered into a pET28a vector with a His6-tag using NheI and HindIII restriction sites. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene Inc.). The ORF of the C-terminal NosA112-151 (i.e., residues 112-151) was engineered into a pSMT3 vector with a SUMO tag, which can be removed using ULP1 protease. The constructs were verified by DNA sequencing, and the plasmids were transformed into Escherichia coli BL21(DE3) competent cells. The transformed cells were grown at 310 K in a Luria Broth (LB) medium containing 50 μg ml−1 kanamycin and were induced (24 h, 291 K) by the addition of 0.1 mM isopropyl-d-thio-b-D-galactopyranoside (IPTG) when the OD600 value was measured as 0.5 - 0.6. The cells were harvested and resuspended in lysis buffer (50 mM phosphate, pH 7.5, 500 mM NaCl), lysed with 10 μg ml−1 PMSF by sonication on ice. The lysates were clarified by centrifugation (30 min, 16,000 rpm), and the soluble proteins were purified by nickel-affinity chromatography (GE Healthcare) though a linear gradient of 0–500 mM imidazole in the lysis buffer. Protein fractions were collected and dialyzed twice at 277 K against the lysis buffer. To remove the His6-tag, the fusion proteins were digested with thrombin protease (ULP1 protease for NosA112-151) overnight at 4 °C, followed by running a second nickel column.
The fractions were collected and further purified by running gel-filtration chromatography on a Superdex 75 column (GE Healthcare) with a buffer containing 25 mM phosphate, pH 7.5, and 50 mM NaCl. Finally, the peak fractions containing proteins were concentrated to 50 mg ml−1 using an ultra-centrifugal filter tube (Millipore) and were used for crystallization (50 mM Tris-HCl, pH 7.5 50 mM NaCl for crystallization), NMR experiments, stability testing experiment by running SDS-PAGE gels, or kinetic experiments. The pure protein fractions were further verified by running an SDS-PAGE gel and electrospray mass spectrometry. The protein concentrations were estimated from the absorbance at 280 nm with their corresponding absorption coefficients.
The truncated NosA1-111 variant has one Met residue at its N-terminal sequence. Thus, for crystallization, the selenomethionine NosA1-111 (Se-Met NosA1-111) was successfully expressed in M9 medium using the reported methionine-pathway inhibition protocol29,30 and was purified as performed on native NosA1-111 above.
For NMR experiments, the 15N-labeled NosA112-151, the full-length NosA and its K49A variant, and the 13C- and 15N-labeled NosA112-151 were expressed in M9 medium containing 15NH4Cl and/or 13C-glucose as the nitrogen and carbon source, respectively.
NosA1-111 crystallization and its X-ray data collection
Initial crystallization trials were performed at 293 K with Crystal Screen HT and Index HT kits in 96-well plates using the sitting-drop vapor-diffusion method (Hampton Research). For refinement of the crystallization conditions, 1 μl of protein solution was mixed with an equal volume of reservoir solution and equilibrated against 0.5 ml of the reservoir solution at 293 K in 24-well plates using the sitting-drop vapor-diffusion method. Crystals of Se-Met NosA suitable for X-ray analysis were obtained under the following conditions: 0.02 M magnesium chloride hexahydrate, 0.1 M HEPES pH 6.0 - 9.0, and 22% w/v polyacrylic acid 5100.
The crystals were picked up in a nylon loop (Hampton Research) and mounted in liquid nitrogen for flash-cooling. X-ray diffraction data were collected at beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF, China) using a MAR CCD MX-225 detector. The wavelength of the radiation was 0.9794 Å, and the distance between the crystal and the detector was 400 mm. The exposure time for each frame was 1 s with a 1o oscillation, and 360 frames were collected. The data were indexed, integrated and scaled using the HKL-2000 program suite31. The structure was solved using the single-wavelength anomalous dispersion method implemented in Phenix32, which identified 3 distinct Se atoms and yielded a figure of merit of 0.22. Model building was performed using Coot33. Structure refinement was carried out using Phenix and Refmac532,34. Structure analysis was carried out using programs in CCP435. The figures were generated using Pymol (http://www.pymol.org). The statistics of the structure refinement and the quality of the final structure models are summarized in Table 1.
The enzymatic assay on HPLC system
To assay the activities of the full-length NosA, NocA and their variants, the truncated N-terminal NosA1-111 variant, the mixture of NosA1-111 with the C-terminal NosA112-151 peptide, each protein solution (including the full-length NosA, NocA or their mutants, or the mixture of NosA1-111 with NosA112-151 at mole ratio of 1:1.) was diluted to 1 mg/ml in buffer A (50 mM Glycine buffer, pH 9.0). To avoid degrading, all samples are used after purification was finished. The substrate was first dissolved in DMSO, and then diluted to 1 mg/ml by buffer A. For simple comparison, 5 μL of diluted protein solution and 5 μL of substrate solution were added into 40 μL of buffer A, respectively, and then was incubated at 303 K for 60 min. To quench the reaction, 5 μL of methanol were added. To determine whether the product nosiheptide was generated or not, HPLC analysis was performed by running C-18 reverse phase column (Agilent 1100) with a MeCN/H2O gradient mobile phase. The wavelength of nosiheptide detection was specified as 254 nm.
To investigate the importance of the sequence and the coiled-coil conformation of the C-terminal NosA112-151 in the catalytic reaction, three randomly selected peptides (Did2, Vps60 and JmjN from KDM5B) available in the lab were used to replace NosA112-151, and to mix with the N-terminal NosA1-111 variant. Both Did2 (176-204 aa, with an amino acid sequence as follows: NVPEIKAKEV NVDDEKEDKL AQRLRALRG) and Vps60 (128-186 aa, with an amino acid sequence as follows: INIDKLQDMQ DEMLDLIEQG DELQEVLAMN NNSGELDDIS DAELDAELDA LAQEDFTLP) peptides were reported in ESCRT-III system involved in multivesicular bodies (MVB) pathway21,22, with a helix conformation and a random-coiled conformation in their free states, respectively. The JmjN peptide (with an amino acid sequence as follows: ECPVFEPSWA EFRDPLGYIA KIRPIAEKSG ICKIRPPAD) locates at the N-terminal histone H3K4me3/2 demethylase KDM5B19,20, with a regular conformation. NosA1-111 was first mixed with each peptide at mole ratio 1:1, then the assay was performed as we did on the mixture of NosA1-111 and NosA112-151.
For kcat and Km measurements of the full-length NosA, a time course was carried out to determine the initial rate in 50 mM Glycine-NaOH buffer (20 μL, pH = 9.0) that contained 10 nM NosA and 200 μM substrate. The reactions were initiated by the addition of NosA into diluted substrate solution, incubated at 303 K, and then terminated by adding 50 μL of methanol into the solution at 2, 5, 10, 15, 30, 60, 120, and 240 min, respectively. The samples were subjected to the same workups and HPLC analysis as described above. The production of product (i.e., the intensity of the peak was used to quantify the nosiheptide), linear with respect to time during 0–5 min, was fitted into a linear equation to obtain the initial velocity. To determine the kinetic parameters for the conversion of substrate to product, the reactions were carried out at 303 K for 5 min in each 20 μl of the mixture that contained 10 nM NosA, 50 mM Glycine-NaOH (20 μL, pH = 9.0), and the substrate concentration varying at 50, 100, 150, 200, 250, and 500 μM, respectively.
For kcat and Km measurement of NosA1-111 mixing with NosA112-151, a time course was carried out to determine the initial rate in 50 mM Glycine-NaOH (20 μL, pH = 9.0) buffer that contained 4 μM NosA1-111, 4 μM of NosA112-151 and 200 μM substrate. The reactions were initiated by the addition of mixture of NosA1-111 and NosA112-151 into the diluted substrate solution, incubated at 303 K, and then terminated by adding 50 μL of methanol into the reaction solution at 2, 5, 10, 15, 30 min, respectively. The samples were subjected to similar HPLC analysis as described above. The production of product, linear with respect to time during 0–2 min, was fitted into a linear equation to obtain the initial velocity. To determine the kinetic parameters for the conversion of substrate to product, the reactions were carried out at 303 K for 5 min in each 20 μl of the mixture that contained 4 μM NosA1-111, 4 μM NosA112-151, 50 mM Glycine-NaOH (20 μL, pH = 9.0), and the substrate concentration varying at 50, 100, 150, 200, 250, and 500 μM, respectively.
All assays were performed in duplicate. Each conversion was analyzed by HPLC as described above. The resulting initial velocities were then fitted to the Michaelis-Menten equation using GraphPad Prism 5 to extract Km and kcat parameters.
Molecular modeling of complexation between full-length NosA with its substrate
To understand how the substrate interacts with NosA, as the first step, the substrate molecule was docked onto the crystal structure of NosA1-111. A distance-restrained docking was performed based on two observations. Firstly, the results of enzymatic assay indicate that the C-terminal NosA112-151 loop is important for catalytic reaction, which may be due to the stabilizing effect of the C-terminus on the complex structure. Due to the absence of C-terminus, it is probably difficult to generate a rational binding mode for the substrate and NosA1-111 by an unrestrained docking. Secondly, based on the information that the residues K49 and E101 play a vital role in maintaining the catalytic activity of NosA, the distances between the side-chain amino group of K49 and the oxygen atom of the –OH group in the pyridine ring of the substrate and between the side-chain carboxylic oxygen atoms of E101 and the nitrogen atom at the Cα-N bond cleavage site on the substrate were restricted within 2.5–3.5 Å during molecular docking. All ligand docking jobs were performed using the GOLD software suite36.
As the second step, a structural model of C-terminal NosA112-151 was derived through homology modeling. Our homology search found that the C-terminus of the HTLV-II matrix protein (PDB code 1JVR)37, which is also an irregular loop, had a sequence identity of 37.5% to the C-terminal NosA112-151 peptide. Thus, it was used as the template to build a model of the NosA C-terminus. The homology modeling job was completed by using the MOE software (version 2013) (Chemical Computing Group Inc., Montreal, Quebec, Canada). The resulting structure was adjusted manually to avoid steric hindrance between C-terminus and the rest part of the complex. Simultaneously, the main chain of C-terminus should stay as close as possible to the –OH group next to the carbonyl group of the substrate (Fig. 1A, highlighted in blue) according to the fact that the catalytic activity was abolished due to the deletion of this –OH group17. Finally, the adjusted structure was assembled onto the crystal structure of the N-terminal NosA1-111.
The third step was to refine the binding mode of the substrate to NosA, which was generated by molecular docking as described above, through molecular dynamics (MD) simulation. Our MD simulations were performed by using the AMBER software (version 12)38. The docking model was supplied as the initial configuration for MD simulation. The AMBER FF12SB force field39 was applied. The partial charges were calculated with the Gaussian 09 software40 at the HF/6-31G(d) level and were further processed by the RESP model41. The complex structure was soaked in a TIP3P42 water box with a margin of 10 Å at each direction. The whole system was neutralized by addition of a proper number of counterions (Na+). The system was gradually heated to 300 K over 100 ps. Then, a total of 25 ns simulation was performed at a 2-fs interval under a constant temperature of 300 K and a constant pressure of 1 atm. During simulation, all covalent bonds containing hydrogen atoms were constrained with the SHAKE algorithm43. Long-range electrostatic effects were modeled using the particle-mesh-Ewald method44 with a cutoff of 12 Å. The Langevin equilibration scheme45 was used to control and equalize the temperature. Snapshots were saved every 500 fs during simulation. The resulting MD trajectories were analyzed using the PTRAJ module and visualized with the VMD software46.
Circular dichroism spectra of NosA112-151, full-length NosA and its variants
To probe the folding of full-length NosA and its variants, and the C-terminal NosA112-151 peptide, circular dichroism (CD) experiments were performed at 298 K on a JASCO-715 spectropolarimeter (Jasco International Co., Tokyo, Japan). Data were collected at 0.1-nm intervals at a scan speed of 20 nm/min, a 1-nm bandwidth, and a 0.25-s response time from 250 to 190 nm. Circular quartz cells of 1- and 0.1-cm path lengths were used for the far-UV regions. The CD intensities are expressed as the molar residue ellipticities given in units of degrees cm2 mol−1. The concentration of protein or peptide was about 20 μm. The buffer conditions used for running the CD spectra were 50 mM sodium phosphate (pH 7.0).
NMR spectroscopy and analysis
To assign NMR resonances of the backbone atoms of C-terminal NosA112-151, the NMR sample was made containing 1.5 mM uniformly 13C/15N-labelled NosA112-151 in NMR buffer (50 mM Na2HPO4, 50 mM NaCl, 0.01% NaN3, pH 7.0 and 10% D2O). All NMR experiments for assigning the 1H, 13C and 15N backbone atoms (including 2D 15N-1H HSQC, 3D HNCA, HNCO, HN(CO)CA, HNCACB, CBCA(CO)NH) were performed at 298 K on a Varian Unity Inova 600 NMR spectrometer equipped with a triple resonances cryoprobe and pulsed field gradients.
To probe the effects on the folding of NosA by the mutation from K49 to A49, we first run circular dichroism (CD) spectra on both wild-type (WT) NosA and its K49A variant. The results from CD spectra indicate that the folding of K49A looks much different from that of WT full-length protein. To double check these changes on the folding of the K49A variant, 1H-15N HSQC spectra were acquired on WT full-length NosA and its K49A variant at 293 K in NMR buffer (50 mM Na2HPO4, 50 mM NaCl, 0.01% NaN3, pH 7.0 and 10% D2O).
To study interactions among NosA1-111, NosA112-151 and the substrate, 1H-15N HSQC spectra were acquired at 293 K on 1 mM 15N-labeled NosA112-151 in its free state, in complex with unlabeled NosA1-111 (the mole ratio of them is NosA1-111 : NosA112-151 = 1: 1.2), or in complex with unlabeled NosA1-111 and the substrate (the mole ratio of them is NosA1-111 : NosA112-151 : substrate = 1: 1.2 : 1.2) in NMR buffer (50 mM Na2HPO4, 50 mM NaCl, 0.01% NaN3, pH 7.0 and 10% D2O).
To determine the flexibility of the NosA112-151 in the presence of NosA1-111 and the substrate, the measurements of the relaxation times, T1 and T2 and 15N-1H NOEs of the backbone atoms of the NosA112-151 were performed at 293 K in NMR buffer (50 mM phosphate, 50 mM NaCl, 0.01% NaN3, pH 7.0 and 10% D2O) at 293 K for NosA112-151 on a Varian INOVA 600-MHz spectrometer. Ten different values for the relaxation delay were used for the T1 (delays 60, 80, 100, 140, 240, 360, 540 and 760 ms) and T2 (Carr-Purcell-Meiboom-Gill mixing times of 10, 30, 50, 70, 90, 110, 150, 170 and 190 ms) relaxation experiments. The T1 and T2 values were obtained by nonlinear squares fits using the program Prism 5. The 15N-1H NOE values were calculated as the ratio of the intensities of the paired 15N-1H correlation peaks from interleaved spectra acquired with and without 1H pre-saturation during a recycle time of 3 s.
All NMR spectra were processed with the NMRPipe program47 and analyzed using Sparky 3 (http://www.cgl.ucsf.edu/home/sparky/).
Size-exclusion chromatography (SEC) assay
To probe effects on the aggregation state of NosA by the mutations, the size-exclusion chromatography assay was performed on a Superdex 75 column (10/300 GL) (GE Healthcare), which was previously equilibrated with buffer B (50 mM Tris-HCl, pH 7.5, 50 mM NaCl).
Isothermal titration calorimetry (ITC) binding assay
To investigate the binding affinity of the N-terminal NosA1-111 with the C-terminal NosA112-151, the isothermal titration calorimetry (ITC) binding assay was performed. An ITC-200 microcalorimeter (GE Healthcare) was used with a buffer containing 20 mM Tris-HCl, 150 mM NaCl, pH 7.0 at 298 K. The reference titration of small molecules in the buffer was subtracted from the experimental data, and the data were fitted using the Origin 7.0 (OriginLab Corporation) software.
Additional Information
How to cite this article: Liu, S. et al. Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis. Sci. Rep. 5, 12744; doi: 10.1038/srep12744 (2015).
Supplementary Material
Acknowledgments
We thank all staff members of beamline BL17U at Shanghai Synchrotron Radiation Facility (SSRF) for their help with data collection. This work was supported by grants from the Ministry of Science and Technology of China (2011CB966300), the National Natural Science Foundation of China (21272261, 21472229 and 21275154), Science and Technology Commission of Shanghai Municipality (15ZR1449300).
Footnotes
Author Contributions C.C. conceived and designed the experiments, and wrote the manuscript; S.L. performed protein preparation, crystallization and enzymatic assay; H.G., Y.Y. and W.L. made the substrate; L. H. and R.W. conducted the molecular modeling; T.Z. and J.D. solved the crystal structure. Y.Z., N.R., W.L. and C.W. did mutation studies. W.L., R.W. and J.D. also analyzed the data and help writing manuscript. All authors reviewed the manuscript.
References
- Liao R. & Liu W. Thiostrepton maturation involving a deesterification-amidation way to process the C-terminally methylated peptide backbone. Journal of the American Chemical Society 133 (9), 2852–2855 (2011). [DOI] [PubMed] [Google Scholar]
- Liao R. et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chemistry & biology 16 (2), 141–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS chemical biology 4 (10), 855–864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. et al. NosA catalyzing carboxyl-terminal amide formation in nosiheptide maturation via an enamine dealkylation on the serine-extended precursor peptide. Journal of the American Chemical Society 132 (46), 16324–16326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A. F., Finnie M. D. & Smyth D. G. Mechanism of C-terminal amide formation by pituitary enzymes. Nature 298 (5875), 686–688 (1982). [DOI] [PubMed] [Google Scholar]
- Weinig S., Hecht H. J., Mahmud T. & Muller R. Melithiazol biosynthesis: further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chemistry & biology 10 (10), 939–952 (2003). [DOI] [PubMed] [Google Scholar]
- Kulathila R., Merkler K. A. & Merkler D. J. Enzymatic formation of C-terminal amides. Natural product reports 16 (2), 145–154 (1999). [DOI] [PubMed] [Google Scholar]
- Eipper B. A., Mains R. E. & Glembotski C. C. Identification in pituitary tissue of a peptide alpha-amidation activity that acts on glycine-extended peptides and requires molecular oxygen, copper, and ascorbic acid. Proceedings of the National Academy of Sciences of the United States of America 80 (16), 5144–5148 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. & Sander C. Protein structure comparison by alignment of distance matrices. Journal of molecular biology 233 (1), 123–138 (1993). [DOI] [PubMed] [Google Scholar]
- Lee W. C., Reniere M. L., Skaar E. P. & Murphy M. E. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. The Journal of biological chemistry 283 (45), 30957–30963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. The Journal of biological chemistry 280 (4), 2840–2846 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L. et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Molecular microbiology 75 (6), 1529–1538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim N., Iniguez A., Nguyen T. Q. & Goulding C. W. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. Journal of molecular biology 395 (3), 595–608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukpabi G., Takayama S. J., Mauk A. G. & Murphy M. E. Inactivation of the heme degrading enzyme IsdI by an active site substitution that diminishes heme ruffling. The Journal of biological chemistry 287 (41), 34179–34188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciara G. et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. The EMBO journal 22 (2), 205–215 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. J., Kim K.-J., Kim M. H. & Kang B. S. Structure insight of the role of the Hahella chejuensis HapK protein in prodigiosin biosynthesis. Proteins 70 (1), 257–262 (2007). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Multiple oxidative routes towards the maturation of nosiheptide. Chembiochem : a European journal of chemical biology 14 (13), 1544–1547 (2013). [DOI] [PubMed] [Google Scholar]
- Harms J. M. et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Molecular cell 30 (1), 26–38 (2008). [DOI] [PubMed] [Google Scholar]
- Yamane K. et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Molecular cell 25 (6), 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B. Protein & cell 5 (11), 837–850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA-ATPase regulator Vta1 in the multivesicular body pathway. The Journal of biological chemistry 287 (52), 43899–43908 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. (1)H, (1)(3)C and (1)(5)N resonance assignments of the N-terminal domain of Vta1-Vps60 peptide complex. Biomolecular NMR assignments 7 (2), 331–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou K. C. et al. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. Journal of the American Chemical Society 127 (31), 11176–11183 (2005). [DOI] [PubMed] [Google Scholar]
- Schoof S., Baumann S., Ellinger B. & Arndt H. D. A fluorescent probe for the 70 S-ribosomal GTPase-associated center. Chembiochem : a European journal of chemical biology 10 (2), 242–245 (2009). [DOI] [PubMed] [Google Scholar]
- Ding Y. et al. Moving posttranslational modifications forward to biosynthesize the glycosylated thiopeptide nocathiacin I in Nocardia sp. ATCC202099. Molecular bioSystems 6 (7), 1180–1185 (2010). [DOI] [PubMed] [Google Scholar]
- Morris R. P. et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. Journal of the American Chemical Society 131 (16), 5946–5955 (2009). [DOI] [PubMed] [Google Scholar]
- Tocchetti A. et al. Capturing linear intermediates and C-terminal variants during maturation of the thiopeptide GE2270. Chemistry & biology 20 (8), 1067–1077 (2013). [DOI] [PubMed] [Google Scholar]
- Malcolmson S. J., Young T. S., Ruby J. G., Skewes-Cox P. & Walsh C. T. The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proceedings of the National Academy of Sciences of the United States of America 110 (21), 8483–8488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar S. S. et al. Expression, purification, and crystallization of the Escherichia coli selenomethionyl beta-ketoacyl-acyl carrier protein synthase III. Biochemical and biophysical research communications 270 (1), 100–107 (2000). [DOI] [PubMed] [Google Scholar]
- Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L. & Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. Journal of molecular biology 229 (1), 105–124 (1993). [DOI] [PubMed] [Google Scholar]
- Otwinowski ZaM W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276 (Macromolecular Crystallography, part A), 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66 (Pt 2), 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D60 (Pt 12 Pt 1), 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53 (Pt 3), 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D67 (Pt 4), 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W. & Taylor R. D. Improved protein-ligand docking using GOLD. Proteins 52 (4), 609–623 (2003). [DOI] [PubMed] [Google Scholar]
- Christensen A. M., Massiah M. A., Turner B. G., Sundquist W. I. & Summers M. F. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. Journal of molecular biology 264 (5), 1117–1131 (1996). [DOI] [PubMed] [Google Scholar]
- Case D. A. D., T. A. et al.; AMBER 12. University of California: San Franciso. (2012)
- Wang J., Wolf R. M., Caldwell J. W., Kollman P. A. & Case D. A. Development and testing of a general amber force field. Journal of computational chemistry 25 (9), 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- Frisch M. J. et al. gaussian 09. Gaussian, Inc.: Wallingford CT (2009). [Google Scholar]
- Bayly C. I., Cieplak P., Cornell W. & Kollman P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. . J. Phys. Chem. 97, 10269–10280 (1993). [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- Ryckaert J. P., Ciccotti G. & Berendsen H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Phys 23 (3), 327–341 (1977). [Google Scholar]
- Darden T., York D. & Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10093 (1993). [Google Scholar]
- Loncharich R. J., Brooks B. R. & Pastor R. W. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N’-methylamide. Biopolymers 32 (5), 523–535 (1992). [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics 14 (1), 33-38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- Delaglio F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR 6 (3), 277–293 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.