Abstract
Members of the double-stranded RNA-specific ribonuclease III (RNase III) family were shown to affect cell division and chromosome segregation, presumably through an RNA interference-dependent mechanism. Here, we show that in Saccharomyces cerevisiae, where the RNA interference machinery is not conserved, an orthologue of RNase III (Rnt1p) is required for progression of the cell cycle and nuclear division. The deletion of Rnt1p delayed cells in both G1 and G2/M phases of the cell cycle. Nuclear division and positioning at the bud neck were also impaired in Δrnt1 cells. The cell cycle defects were restored by the expression of catalytically inactive Rnt1p, indicating that RNA cleavage is not essential for cell cycle progression. Rnt1p was found to exit from the nucleolus to the nucleoplasm in the G2/M phase, and perturbation of its localization pattern delayed the progression of cell division. A single mutation in the Rnt1p N-terminal domain prevented its accumulation in the nucleoplasm and slowed exit from mitosis without any detectable effects on RNA processing. Together, the data reveal a new role for a class II RNase III in the cell cycle and suggest that at least some members of the RNase III family possess catalysis-independent functions.
INTRODUCTION
Double-stranded RNA (dsRNA)-specific ribonucleases play an important role in the processing and degradation of many RNAs essential for cell growth (Nicholson, 1999; Lamontagne et al., 2001; Hutvagner and Zamore, 2002). All known dsRNA-specific endoribonucleases belong to the RNase III family and are identified by the presence of two features: first, a nuclease domain (NUCD) containing a signature motif (RLEFLGD) (Blaszczyk et al., 2001); and second, a dsRNA-binding domain (dsRBD) exhibiting the αβββα structure (Kharrat et al., 1995) characteristic of dsRNA-binding proteins (Bass, 1995; Saunders and Barber, 2003). Members of this family are found in all kingdoms, with the exception of the archaebacteria where their functions are carried out by the bulge-helix-bulge nuclease (Durovic and Dennis, 1994). The RNase III family is divided into four classes based on protein structure (Lamontagne et al., 2001). Class I includes bacterial enzymes that possess a single N-terminal NUCD and a C-terminal dsRBD. Class II enzymes are identified by the presence of a highly variable N-terminal extension and include fungal RNase III (Abou Elela et al., 1996; Rotondo and Frendewey, 1996). Class III enzymes exhibit two NUCDs and include both plant and vertebrate enzymes (Filippov et al., 2000). Class IV includes the RNA interference (RNAi) enzyme Dicer (Knight and Bass, 2001; Provost et al., 2002a; Zhang et al., 2002), which possesses an N-terminal helicase domain. Most eukaryotic cells, with the exception of the yeast Saccharomyces cerevisiae contain two RNase III isoforms: one from class II or III, and one from class IV. Class II and III enzymes are expected to accumulate in the nucleolus and participate in processing rRNA (Abou Elela et al., 1996; Zhou et al., 1999; Spasov et al., 2002), whereas class IV enzymes are found in the endoplasmic reticulum and induce gene silencing (Provost et al., 2002a).
The most studied class II enzyme is the S. cerevisiae RNase III (Rnt1p). Like bacterial RNase III (Nicholson, 1999), Rnt1p is not essential, but its deletion causes severe growth defects and temperature sensitivity (Abou Elela and Ares, 1998). Depletion of Rnt1p blocks the maturation of the 25S preribosomal RNA (pre-rRNA) 3′ end (Abou Elela et al., 1996) and slows the formation of the 18S rRNA 5′ end (Kufel et al., 1999). Rnt1p also processes many of the small nucleolar RNAs (snoRNAs) required for rRNA maturation (Chanfreau et al., 1997; Abou Elela and Ares, 1998; Seipelt et al., 1999; van Hoof et al., 2000) and initiates the 3′ end processing of all Pol II-transcribed small nuclear RNAs (snRNAs). Recently, Rnt1p was shown to cleave within the intron sequence of two pre-mRNAs coding for ribosomal proteins, and it was suggested that this cleavage targets unspliced RNAs for degradation (Danin-Kreiselman et al., 2003). Rnt1p probably enters the nucleolus to cleave the rRNA, but it is not clear where snRNA and mRNA cleavages take place.
The first eukaryotic RNase III (PacI) was isolated from the fission yeast Schizosaccharomyces pombe as a suppressor of uncontrolled meiosis, and its overexpression was shown to inhibit mating and sporulation (Iino et al., 1991). Recently, other orthologues of RNase III have been implicated in cell development (Grishok et al., 2001; Knight and Bass, 2001; Banerjee and Slack, 2002) and chromosome segregation, suggesting that these enzymes may play a role in regulating cell division (Provost et al., 2002b; Volpe et al., 2002; Hall et al., 2003). However, with the exception of PacI, which is a class II RNase III, most of the enzymes implicated in cell division are Dicer-like class IV enzymes and their effects are mediated by RNAi. Indeed, in S. pombe, where the RNAi machinery is conserved, Dicer, and not PacI, is required for heterochromatin formation and chromosome segregation (Provost et al., 2002b; Volpe et al., 2002; Hall et al., 2003). In S. cerevisiae, it is not clear whether Rnt1p contributes to cell division or chromosome segregation because the only RNase III found is a class II enzyme. If S. cerevisiae uses a mechanism analogous to that of S. pombe to regulate cell division and chromosome segregation, it does so most likely through an RNAi-independent pathway and would likely involve Rnt1p.
Here, we show that the deletion of Rnt1p perturbs the cell cycle and impairs chromosome segregation. The cell cycle defects were largely restored by the expression of catalytically impaired Rnt1p, indicating that RNA cleavage is not essential for cell cycle progression. Rnt1p was found to accumulate in either the nucleoplasm or the nucleolus in a cell cycle-dependent manner. A single mutation in the Rnt1p N-terminal domain altered the protein localization pattern and slowed the exit from mitosis without any detectable effects on RNA processing. Together, the data reveal a new role for a class II RNase III in the cell cycle and suggest that Rnt1p exits the nucleolus to cleave its nuclear RNA substrates.
MATERIALS AND METHODS
Strains and Plasmids
All yeast strains described in Table 1 were grown and manipulated using standard procedures (Rose et al., 1990; Guthrie and Fink, 1991). The effects of Rnt1p depletion were studied using strains W303-1A (Thomas and Rothstein, 1989) and Δrnt1 (Chanfreau et al., 1998a). Strain RWY12 (Dionne and Wellinger, 1996) carrying a deletion in TLC1 was used as a senescence control. Strain MLY30, which was used for cell synchronization, is a derivative of BY4705 (Brachmann et al., 1998) carrying a deletion in BAR1 that improves the response to α-factor (a kind gift from R. Wellinger, University of Sherbrooke, PQ, Canada). Strains 1724 and 1730 were used for the Mcd1p and Esp1p experiments (Visintin et al., 1999). The repression of rRNA synthesis was performed using strains NOY260 and NOY265 (Wittekind et al., 1988). Yeast EGY48 was used for the nuclear localization assay (Estojak et al., 1995). All clonings were performed in either Escherichia coli DH5αF' (Invitrogen, Burlington, Ontario, Canada) or E. coli M15 (pREP4) (QIAGEN, Valencia, CA) for protein overexpression.
Table 1.
S. cerevisiae strains
| Name | Genotype | Source |
|---|---|---|
| W303-1A | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 can1-100 | Thomas and Rothstein, 1989 |
| Δrnt1 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 can1-100 rnt1Δ::TRP1 | Chanfreau et al., 1998a |
| RWY12 | MATaura3-52 lys2-801 ade2-101 his3-Δ200 trp1-Δ1 leu2-Δ1 DIA5-1 tlc1::LEU2 | Dionne and Wellinger, 1996 |
| MLY30 | MATaade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 bar1Δ::HIS3 | Personal Communication |
| 1724 | MATabar1 mcd1 trp1 ura3 CDC14-3HA | Visintin et al., 1999 |
| 1730 | MATaleu2-3,112 his3-11,15 trp1-1 ura3 | Visintin et al., 1999 |
| NOY260 | MAT α RPA190 trp1-Δ1 his4-Δ401 leu2-3,112 ura3-52 canr | Wittekind et al., 1988 |
| NOY265 | MAT α rpa190-3 trp1-Δ1 his4-Δ401 leu2-3,112 ura3-52 canr | Wittekind et al., 1988 |
| EGY48 | MAT α his3 trp1 ura3 LexAop(x6)-LEU2 | Estojak et al., 1995 |
Rnt1p was expressed in vivo by using the yeast vectors pRS315 (Sikorski and Hieter, 1989), pNS (Ueki et al., 1998), or pGAD and pGBDU (James et al., 1996). All constructions generated from these vectors and used in this study are listed in Table 2. A description of the cloning involved is presented as supplemental material 7.
Table 2.
Plasmid constructs used in this study
| Name | Description | Source |
|---|---|---|
| pRS315/GFP | GFP under RNT1 promoter control in pRS315 | This work |
| pRS315/GFP/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/ΔNT2 | RNT1 coding sequence (192-471 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/NT2 | RNT1 coding sequence (1-191 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/ds1 | RNT1 coding sequence (344-471 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/ds4 | RNT1 coding sequence (428-471 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-22st | RNT1 coding sequence (1-22 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-82st | RNT1 coding sequence (1-82 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-451ds | RNT1 coding sequence (451-471 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-K45/I | RNT1 coding sequence (1-471 aa) variant with K45I mutation cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/NT2-K45/I | RNT1 coding sequence (1-191 aa) variant with K45I mutation cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/ds1-463st | RNT1 coding sequence (344-463 aa) cloned in frame in pRS315/GFP | This work |
| pRS315/GFP/RNT1-D247/R | RNT1 coding sequence (1-471 aa) variant with D247R mutation cloned in frame in pRS315/GFP | This work |
| pGBDU-C3 | James et al. (1996) | |
| BD/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pGBDU-C3 | Lamontagne et al. (2000) |
| BD/RNT1-D247/R | RNT1 coding sequence (1-471 aa) variant with D247R mutation cloned in frame in pGBDU-C3 | This work |
| AD/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pGAD | This work |
| PNS | Ueki et al. (1998) | |
| pNS/NLS | Ueki et al. (1998) | |
| pNS/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pNS | This work |
| pNS/ΔNT2 | RNT1 coding sequence (192-471 aa) cloned in frame in pNS | This work |
| pNS/NT2 | RNT1 coding sequence (1-191 aa) cloned in frame in pNS | This work |
| pNS/ds1 | RNT1 coding sequence (344-471 aa) cloned in frame in pNS | This work |
| pQE31 | ||
| pQE/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pQE31 | Lamontagne et al. (2000) |
| pQE/RNT1-K45/I | RNT1 coding sequence (1-471 aa) variant with K45I mutation cloned in frame in pQE31 | This work |
| pQE/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pQE31 | This work |
| pQE/RNT1-D247/R | RNT1 coding sequence (1-471 aa) variant with D247R mutation cloned in frame in pQE31 | This work |
Colony-forming Assay
The viability of the different strains was analyzed >200 generations. The number of generations corresponds to the number of restreakings of a single colony (estimated to grow for 20 generations on a plate) from one plate to another. For each generation, one colony was resuspended in 1 ml of sterile water, the number of cells counted (using a hemacytometer), and 103 cells spread on a plate. For each mutant strain, the number of colonies that regrew on the plate was compared with that for its isogenic wild-type strain. Each experiment was performed in triplicate.
Live/Dead Cell Assay
Δrnt1 cells transformed with either pRS315/green fluorescent protein (GFP) or pRS315/GFP/RNT1 were grown in SCD-Leu medium to either log or stationary phase at 26°C. The cells (2 × 107) were then harvested and resuspended in DH buffer (1 ml, 10 mM Na-HEPES, pH 7.2, 2% dextrose), FUN-1 (1 ml, 5 μM in DH buffer; Molecular Probes, Eugene, OR) was then added, and the cells incubated at 26°C for 30 min with 1 ml of 5 μM FUN-1. The cells were then spotted on a slide and visualized with the 488- and 568-nm laser lines of an Olympus IX-70 microscope equipped with the Fluoview confocal upgrade (Carsen Group, Markham, Ontario, Canada). The transformation of the green dye to a red product revealed metabolically active cells.
Fluorescence-activated Cell Sorting (FACS) Procedures
Cells (107) were fixed overnight at 4°C by the addition of 4 volumes of 95% ethanol, washed twice with phosphate-buffered saline (PBS, 1 ml) containing 2.7 mM KCl, 0.8 mM Na2HPO4, 1.5 mM KH2PO4, 150 mM NaCl, 60 μM NaN3 at pH 7.3, and resuspended in PBS (500 μl) containing 1 mg/ml RNaseA (Roche Diagnostics, Laval, PQ, Canada). After overnight incubation at 37°C, the cells were harvested and resuspended in propidium iodide (500 μl, 50 μg/ml; Sigma Diagnostics Canada, Mississauga, Ontario, Canada) in PBS and incubated for 3 h at 4°C in the dark. The cells were again harvested and finally resuspended in propidium iodide (5 μg/ml) in PBS and diluted to 5 × 106 cell/ml in this solution. After sonication, cells were analyzed on FACScan (BD Biosciences Immunocytometry Systems, San Jose, CA).
Yeast Cell Cycle Synchronization
Yeasts MLY30 were synchronized in G1 by the addition of α-factor (Bioshop, Burlington, ON, Canada) to a final concentration of 500 ng/ml, followed by incubation at 30°C for 6 h. The cells were released from arrest by harvesting and resuspending the culture in prewarmed YEPD containing 200 μg/ml pronase (Roche Diagnostics). Strain 1730 was arrested in G1 by incubating the culture for 1.5 h at 26°C with 4 μg/ml α-factor. For the release, the cells were harvested and resuspended in fresh media containing 200 μg/ml pronase preincubated at either the permissive (26°C) or the restrictive temperature (37°C). Yeast 1724 was arrested in G2/M by addition of nocodazole (15 μg/ml; Sigma Diagnostics Canada) to cells in logarithmic phase and incubated for 3 h.
Time-Lapse Confocal Microscopy
W303-1A and Δrnt1 cells transformed with plasmid pTS417 (Carminati and Stearns, 1997) expressing GFP-Tub3p under the control of the GAL1,10 promoter were grown to early logarithmic phase in SC-URA containing 2% galactose and 0.5% dextrose and mounted for live cell imaging as described previously (Waddle et al., 1996; Carminati and Stearns, 1999). Cells were pictured using an inverted Olympus IX-70 microscope equipped with the Fluoview confocal upgrade in a room set at 23°C.
Immunolocalization
Yeast cells were prepared for immunofluorescence as described previously (Tremblay et al., 2002). The primary antibodies used in this study were mouse monoclonal anti-3-phosphoglycerate kinase (PGK) (Molecular Probes) at a dilution of 1:100; rabbit polyclonal anti-Rnt1p at a dilution of 1:1000 (Tremblay et al., 2002); mouse monoclonal B15 anti-Nop1p at a dilution of 1:10,000 (Aris and Blobel, 1988); and rabbit polyclonal anti-Nhp2p at a dilution of 1:1000 (Henras et al., 1998). Oregon green 488-conjugated goat antibodies against mouse or rabbit IgG, and Texas Red X-conjugated goat anti-rabbit secondary antibodies (Molecular Probes) were used at 1:1000 dilutions for detection. DNA was stained with 4′6′-diamidino-2-phenylindole (DAPI, 1 μg/ml; Sigma Diagnostics Canada).
Nuclear Localization Assay
Yeast EGY48 transformed with the different pNS and pNS/NLS constructs were grown in SCD-His medium to an OD600 of 0.8. Serial dilutions of the cultures (10-fold increments) were spotted on SCD-His or SCD-His-Leu plates and then grown at 30°C for 3 d.
In Vitro RNA Cleavage
Cleavage reactions were performed as described previously (Lamontagne and Abou Elela, 2001) using recombinant Rnt1p (0.2 pmol) and 5′ end-labeled U5 snRNA 3′ end (3 fmol) (Lamontagne et al., 2000) in 20 μl of reaction buffer (Lamontagne and Abou Elela, 2001) containing 150 mM KCl and incubated for 20 min at 30°C.
Northern Blot Analysis
Northern blot analysis was performed with total RNA (15 μg) run on a 6% denaturing polyacrylamide gel as described previously (Abou Elela and Ares, 1998) using an oligonucleotide against snR43 (Chanfreau et al., 1998b) as probe.
RESULTS
Deletion of RNT1 Impairs Cell Division
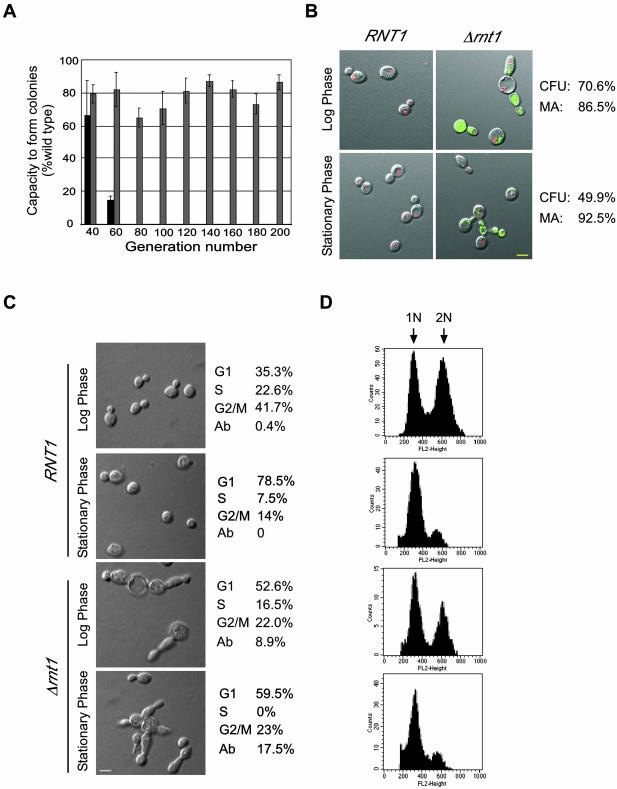
The deletion of RNT1 allows cells to grow slowly at 26°C but not at higher temperatures (Lamontagne et al., 2000). In rich media Δrnt1 cells grow 2 to 3 times slower than wild-type cells and stop growing at a lower density than wild-type cells (Abou Elela and Ares, 1998; Lamontagne et al., 2000). This phenotype could be caused by slow metabolism, cell death, or senescence. To determine the nature of the growth defect caused by the deletion of RNT1, we compared the viability of Δrnt1 cells to that of wild-type cells and to that of cells carrying a deletion in TLC1 (RWY12). TLC1 is a component of the telomerase ribonucleoprotein (RNP) complex required for sustained cell growth, and it was included as a control for rapid senescence. As shown in Figure 1A, compared with wild type, Δtlc1 lost 85% of its capacity to form colonies at 60 generations and stopped growing after 60. However, Δrnt1 cells continued to grow even after 200 generations, but their capacity to form colonies was consistently reduced by 20-30% compared with that of wild-type cells. We conclude that RNT1 deletion does not cause senescence but instead impairs colony formation.
Figure 1.
Deletion of RNT1 impairs colony formation and cell division. (A) Capacity of Δtlc1 (dark gray) and Δrnt1 (light gray) cells to form colonies over different generations is shown as a percentage of that obtained with wild-type cells (set to 100%). (B) Assay for cell vitality of Δrnt1 cells transformed with pRS315/GFP or pRS315/GFP/RNT1 and grown to either log or stationary phase. Transformation of the green FUN-1 dye to red demonstrates an active metabolism. The yellow colored bar represents 5 μm. The pictures shown depict cells after 30 min of incubation with the FUN-1 dye. The percentage of the cells that are metabolically active (MA) is shown on the right. Colony-forming units (CFU) of Δrnt1 cells under the same growth conditions are indicated on the right. (C) Phenotypic examination of Δrnt1 in comparison to wild-type cells from cultures growth to either log or stationary phases. The number of cells in each phase of the cell cycle was identified by observing the bud size. Abnormally large cells, those with very long bodies or buds and those with multiple buds were scored as abnormal (Ab). The yellow colored bar on micrographs represents 5 μm. (D) Cell analysis of Δrnt1 and wild-type by using flow cytometry. Cells were grown and analyzed as described in MATERIALS AND METHODS. Histograms of fluorescence intensity versus cell number are shown. 1N represents unreplicated DNA, whereas 2N represents replicated DNA.
To determine whether Δrnt1 cells fail to form colonies due to a high rate of cell death, or to defects in cell division, we directly examined the viability of Δrnt1 cells by monitoring the metabolism of the degradable dye FUN-1. The FUN-1 dye accumulates in vacuoles as defined red pigments in metabolically active cells, but looks like a diffused green stain in dead cells. As shown in Figure 1B, the FUN-1 dye became red after 30 min in all wild-type cells (RNT1), whereas most Δrnt1 cells remained green. The FUN-1 dye slowly turned red in most Δrnt1 cells grown either to log or stationary phases as the incubation time increased, leaving only 10% of the cells green at the end of the experiment (our unpublished data). This indicates that most of the Δrnt1 cells are not dead but convert the Fun-1 dye slower than wild-type cells. This is consistent with the known slow growth rate of Δrnt1 cells (Abou Elela and Ares, 1998) and defects in ribosome biogenesis (Abou Elela et al., 1996). Colony-formation assays conducted using the cell cultures tested for metabolic activity showed that only 70% of Δrnt1 cells in log phase and 50% of Δrnt1 cells in stationary phase form colonies compared with wild-type cells (Figure 1B). This indicates that despite a constant rate of mortality, Δrnt1 cells' capacity to form colonies dramatically decreases as the cell culture ages, suggesting that Rnt1p is required for efficient cell division. Observation of cell morphology (Figure 1C) revealed a 17% increase in the number of unbudded cells (G1) in Δrnt1 cell culture as compared with a wild-type culture at the same cell density. In addition, ∼9% of Δrnt1 cells in a culture grown to log phase were abnormal and exhibited multiple elongated buds. The number of cells with an aberrant morphology increased as the culture aged, reaching ∼18% in a culture in stationary phase. Fewer cells in S and more cells in the G2/M phases of the cell cycle were observed in Δrnt1 cell culture grown to stationary phase compared with that of wild-type cells. Analysis of the DNA content by using FACS confirmed the increased number of cells in G1 of the Δrnt1 strain (Figure 1D). In addition, comparison of DAPI-stained nuclei of wild-type and Δrnt1 cells (our unpublished data) revealed that many dividing Δrnt1 cells have either empty buds, or buds with two nuclei, indicating that Rnt1p deletion causes nuclear missegregation. We conclude that the deletion of RNT1 slows cell metabolism and hinders the cell cycle progression.
RNT1 Deletion Impairs Nuclei Positioning and Slows Mictrotubule Movement in Dividing Cells
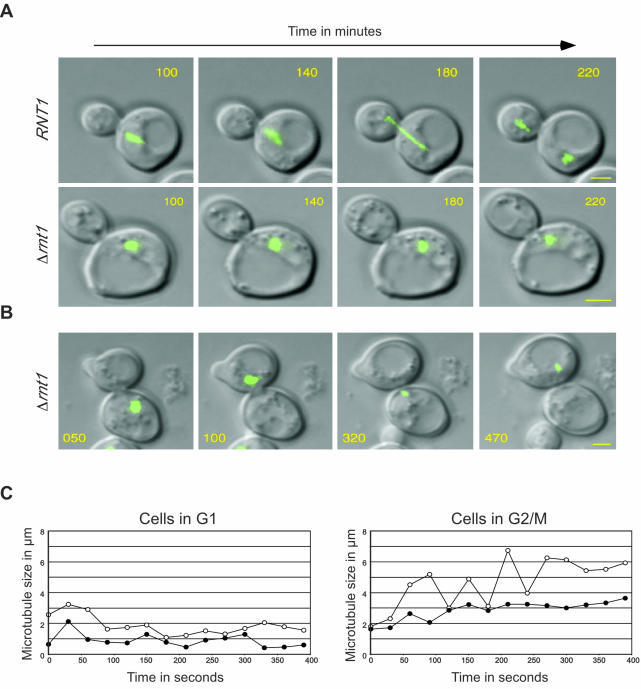
Previous studies showed that the deletion of the Rnt1p orthologue Dicer in S. pombe results in elongated cells, multiple and fragmented nuclei, and chromosome segregation defects (Provost et al., 2002b; Hall et al., 2003). To determine whether the cell division defects observed upon the deletion of RNT1 are also mediated by segregation or microtubule anomalies, we monitored microtubule movement during cell division in Δrnt1 cells. We used time-lapsed confocal microscopy to follow a complete cycle of cell division. Photos from multiple focal points of both wild-type cells and Δrnt1 cells expressing GFP-Tub3p (which allows the visualization of the microtubule complex in living cells) were taken every 10 min and merged into single two-dimensional pictures (see supplementary materials 1 and 2). Photos of wild-type and Δrnt1 cells taken in the time period needed for wild-type cells to pass from S phase to the end of cytokinesis are shown in Figure 2A. In this time frame, wild-type nuclei moved in long steps and directed manner from one end of the mother cell to the bud neck (Figure 2A, top), leading to a successful division after 240 min (supplementary material 1). In contrast, the nuclei of Δrnt1 cells moved in random short steps toward and away from the bud neck (Figure 2A, bottom), and the cells took 440 min to pass from the S phase to the end of division (supplementary material 2). This means that Δrnt1 cells take at least twice as long as wild-type cells to complete one cycle of cell division, clearly explaining why Δrnt1 cells take more time to reach the same optical density as wild-type cells. Strikingly, the hesitant movement of the microtubules and slow positioning of the nuclei in the bud were observed in all Δrnt1 cells. Consistent with this observation, the nuclei were correctly positioned near the bud neck in only 50% of Δrnt1 cells in G2/M phase of the cell cycle compared with that of wild-type cells (Table 3). We conclude that Rnt1p is required for nuclei positioning at the bud neck and hence for efficient cell division.
Figure 2.
Deletion of RNT1 affects nuclear positioning, nuclear division, and microtubule movement. Pictures were taken every 10 min, and representative pictures are shown (see videos in supplementary material 1-3). The times from the beginning of the acquisitions are indicated in minutes on each photo. The microtubule complex revealed by GFP-Tub3p is shown in green. The yellow bars represent 2 μm. (A) Comparison between the division speed and microtubule movement of RNT1 and Δrnt1 cells starting from late S phase. (B) Δrnt1 cells displaying movement of the microtubule bundle from the mother to daughter cell without elongation and division of the spindle. (C) Representative plot of microtubule size over time for RNT1 (open circles) and Δrnt1 cells (black circles) in G1 (left), or G2/M (right). Microtubule length was calculated using stacks of three images taken every 30 s.
Table 3.
Effects of Rnt1p mutants on nuclear positioning and microtubules
| Mutant expressed | % of Nuclei near the bud necka | % of Cells with telophase spindleb |
|---|---|---|
| GFP-Rnt1p | 100 | 10 ± 1.0 |
| GFP | 55.2 | 2.2 ± 0.5 |
| GFP-Rnt1p-K45/I | 99 | 5 ± 1.0 |
| GFP-Rnt1p-D247/R | 98 | 9 ± 1.0 |
Percentage nuclei near the bud neck of cells in G2/M phase of the cell cycle.
Percentage of fully elongated spindles of cells grown to log phase as observed by immunostaining.
In addition to the uncoordinated nuclei movement and the slow cell division shown in Figure 2A, we found that several cell divisions resulted in either of the daughter or the parent cell lacking a nucleus (our unpublished data). This phenotype is very similar to that observed with cells carrying mutations affecting spindle assembly (Spang et al., 1995, 1996; Geissler et al., 1996). To observe the process underlying this defect, we monitored microtubule movement during the division of cells resulting in the production of enucleated cells (see supplementary material 3). As shown in Figure 2B, in these cells the microtubule complexes were compact and did not elongate or divide, even at the end of cell division. The compacted complex was observed to shuttle back and forth through the bud neck several times during the 7 h of the experiment. This phenomenon is probably caused by the inability of the microtubules to form a spindle attached to both extremities of the daughter and mother cells.
The defects observed in the division of Δrnt1 cells greatly resemble the phenotypes caused by the deletions of genes involved in nuclei positioning (Adames and Cooper, 2000). Because most of these genes also affect microtubule movement, we examined the microtubule shrinking and elongating in Δrnt1 cells and compared it with that of wild-type cells. Pictures were taken every 30 s, and the length of microtubules was determined in cells in either the G1 or G2/M phases of the cell cycle. As shown in Figure 2C, in the G1 phase of the cell cycle, the microtubule movements in RNT1 and Δrnt1 cells are similar. In contrast, Δrnt1 cells in G2/M showed severe defects in the rate of microtubules elongation and shrinkage. Moreover, the elongated microtubules in Δrnt1 cells were much shorter than those of wild-type cells. Clearly, the nuclear positioning defect in Δrnt1 cells is due, at least in part, to the inefficient growth of the microtubules to the neck.
Rnt1p Catalytic Activity Is Not Required for Cell Division
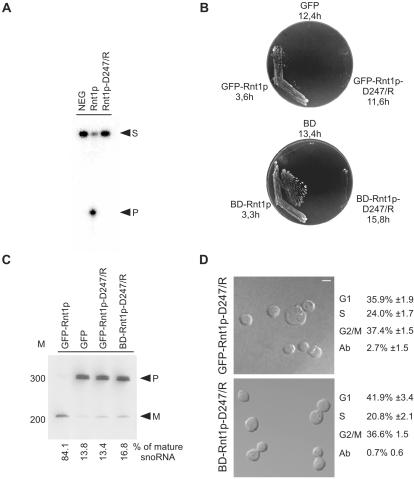
Rnt1p is a dsRNA-specific endoribonuclease that could influence cell division by degrading the mRNAs of related proteins. To test this possibility, we generated a mutation in Rnt1p that blocks RNA cleavage but allows binding to the RNA and interaction with other cellular proteins. As shown in Figure 3A, a mutation in the catalytic domain that changes the conserved aspartic acid in position 247 to arginine (Rnt1p-D247/R) blocks RNA cleavage in vitro. Like Rnt1p, Rnt1p-D247/R bound to RNA efficiently in the absence of Mg2+ (our unpublished data), indicating that the cleavage defect is specific and was not caused by general perturbation of the enzyme's structure. We tested the impact of this mutation on Rnt1p activity in vivo by expressing the mutated protein either fused to GFP under the control of the Rnt1p promoter (pRS315/GFP/RNT1-D247/R), or fused to simian virus 40 nuclear localization signal under the control of the ADH1 promoter (BD/RNT1-D247/R), in Δrnt1 cells. As shown in Figure 2B, the expression of Rnt1p in Δrnt1 cells from either the Rnt1p promoter (GFP/RNT1) or the ADH1 promoter (BD/RNT1) complemented the RNT1 deletion, thereby allowing the cells to grow at 37°C. In contrast, cells expressing either GFP-Rnt1p-D247/R or BD-Rnt1p-D247/R failed to grow at 37°C and grew as slowly at 26°C as Δrnt1 cells. The expression level of Rnt1p-D247/R was similar to that of Rnt1p (our unpublished data), indicating that the failure of this mutant to complement cell growth was not due to expression anomalies. This result confirms that Rnt1p catalytic activity is required for normal cell growth. In addition, the processing of known Rnt1p substrates, like the snoRNA snR43 and the rRNA, were shown by Northern blot analysis, to be also blocked in cells expressing Rnt1p-D247/R (Figure 3C; our unpublished data). Together, these data show that Rnt1p catalytic activity is essential for RNA processing and cell growth at 37°C.
Figure 3.
Rnt1p catalytic activity is not required for cell division. A catalytically inactive mutant of Rnt1p (D247/R) was created by changing aspartic acid 247 to arginine. (A) In vitro cleavage assay of Rnt1p-D247/R. Mutant and wild-type recombinant proteins were tested for their ability to cleave, a 5′ end-labeled model of the Rnt1p natural substrate found at the 3′ end of U5 snRNA (16). NEG indicates RNA incubated under cleavage conditions in the absence of Rnt1p. Arrowheads indicate the position of the substrate (S) and the 5′ cleaved product (P). (B) Rnt1p catalytic activity is required for growth at 37°C. Δrnt1 cells expressing either GFP, or GFP fused with either Rnt1p or Rnt1p-D247/R (top), and cells expressing either BD or BD fused with either Rnt1p or Rnt1p-D247/R (bottom), were compared for growth on plates at 37°C. The doubling times of cells growing in liquid SCD media at 26°C are indicated under the name of each plasmid. (C) Northern blot analysis of RNA processing in vivo. RNA was extracted from the different yeast strains grown at 26°C, fractionated using denaturing PAGE, and transferred to nylon membrane. A probe specific to a known Rnt1p substrate (snoRNA, snR43) was used to visualize both mature (M) and premature RNAs (P). The bands were quantified using Instant Imager, and the ratios of mature to total snR43 were calculated (shown at bottom). The position of the 100-base pair DNA molecular weight marker is indicated on the left. (D) Phenotypic examination of cells expressing catalytically impaired Rnt1p. The phenotypes of Δrnt1 cells expressing either GFP-Rnt1p-D247/R, or Rnt1p in fusion with a heterologous nuclear localization signal (BDRnt1p-D247/R), were examined using confocal microscopy with the white colored bar representing 5 μm. Cells in different stages of the cell cycle were identified using bud size, and the cell number of an average of three independent experiments are indicated on the right. The cell cycle pattern of both cultures was also confirmed by FACS analysis (our unpublished data).
Surprisingly, microscopic examination showed that unlike Δrnt1 cells, cells expressing Rnt1p-D247/R do not accumulate in the G1 phase of the cell cycle and display only a 2-3% level of budding anomalies (Figure 3D). Further examination of the microtubules and FACS profiles indicate that all defects related to cell division were complemented by the expression of Rnt1p-D247/R (our unpublished data). Thus, the role of Rnt1p in the regulation of cell division is not mediated by RNA cleavage nor caused by a general disturbance of RNA metabolism.
Rnt1p Nucleolar Localization Is Cell Cycle Dependent
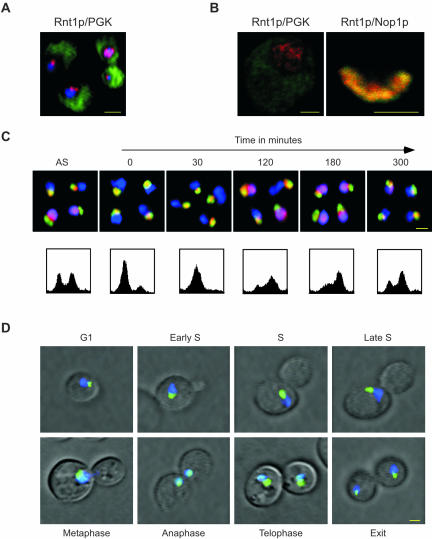
Because Rnt1p catalytic activity seems not to be required for cell cycle progression, we wanted to know whether Rnt1p cellular distribution contributes to its function in the cell cycle. First, we examined the cellular distribution of Rnt1p and compared it with that of known cytoplasmic and nucleolar proteins. Yeast cells were triple stained using DAPI to identify the nucleus, antibodies against PGK to highlight the cytoplasm, and antibodies against Rnt1p. As shown in Figure 4A, Rnt1p-specific stain (red) either partially or completely overlapped with that of DAPI (blue), but not with PGK (green). This indicates that Rnt1p is not cytoplasmic and accumulates in the nucleus. Careful examination of Rnt1p subnuclear localization by using confocal microscopy and either anti-PGK or the nucleolar marker Nop1p showed that Rnt1p accumulates in the nucleolus of some cells and in the nucleoplasm of others (Figure 4B). To determine the effect of the cell cycle on Rnt1p subnuclear localization, we synchronized a culture of yeast cells in the G1 phase of the cell cycle by using α-factor, and then followed Rnt1p's localization in the released culture (Figure 4C). In asynchronized cell cultures, Rnt1p was nucleolar in ∼60% of the cells and nuclear in the other 40%, consistent with the data shown in Figure 4, A and B. In contrast, Rnt1p was nucleolar in all cells arrested in G1 (Figure 4C, 0 min). After release, Rnt1p remained nucleolar through the S phase (30 min after release), whereas an increasing number of cells showed nuclear staining as the culture entered the G2/M phase (120 min). After 180 min, when most cells were in G2/M phase of the cell cycle, Rnt1p was nuclear in most cells. From that point on, Rnt1p gradually returned to the nucleolus as the G1 peak of the FACS profile reappeared. In all stages of the cell cycle no release of Nop1p (Figure 4C), nor of other nucleolar proteins such as Nhp2p and Gar1p (our unpublished data), into the nucleoplasm was observed, indicating that the observed changes in Rnt1p localization are not a general effect of the cell cycle on the nucleolus. This result shows that Rnt1p's localization changes from the nucleolus to the nucleus in synchrony with the cell cycle. To ensure that the observed release of Rnt1p from the nucleus occurs in living cells and is not an artifact caused by fixation or the method of synchronization we used a GFP-labeled version of Rnt1p to observe the Rnt1p subnuclear localization in living cells. Dividing Δrnt1 cells expressing the GFP-Rnt1p fusion protein were stained with DAPI, and the subnuclear localization of Rnt1p was determined in cells at different stages of the cell cycle (Figure 4D). As observed in fixed cells, Rnt1p was nucleolar in G1 and remained so until the end of the S phase of the cell cycle. When the nucleus started to penetrate the bud neck, Rnt1p was released from the nucleolus, becoming completely nuclear when cells entered anaphase. After this point Rnt1p began to return to the nucleolus and became entirely nucleolar once cytokinesis was completed. The same program of Rnt1p localization was observed in a time-lapse study of single cells undergoing cell division (see supplementary material 4). Together, the results indicate that Rnt1p specifically enters the nucleoplasm in the G2/M phase of the cell cycle during which nuclei positioning and microtubule movement into the bud takes place. This suggests that Rnt1p is needed in the nucleus at a specific stage of the cell cycle and may explain why deletion of Rnt1p perturbs nuclear positioning and cell division. We conclude that Rnt1p subnuclear localization is regulated in a cell cycle-dependent manner.
Figure 4.
Rnt1p shuttles between the nucleolus and the nucleoplasm during cell division. (A) In situ immunofluorescence analysis using the epifluorescence of wild-type cells immunostained with antibodies against Rnt1p (red) and PGK (green). The DAPI-stained DNA (blue) indicates the position of the nucleus. The yellow colored bar represents 2 μm. (B) In situ immunofluorescence analysis using confocal microscopy of wild-type cells double-stained for either Rnt1p in red and PGK in green (left), or Rnt1p in red and Nop1p in green (right). The yellow colored bar represents 1 μm. (C) In situ immunofluorescence of Rnt1p by using epifluorescence after the release of cells synchronized by α-factor in G1. Cells are shown before synchronization (AS), after synchronization (0) and at different times after release from α-factor (indicated in minutes). Rnt1p is labeled in red, Nop1p in green and DNA in blue. The lower panels show the FACS profiles of the cell culture at the time points indicated on top of the upper panel. (D) Determining the localization pattern of the GFP-Rnt1p fusion protein in living cells. Δrnt1 cells expressing the GFP-Rnt1p fusion (green) were incubated in SCD growth media with the DNA stain DAPI (blue) and observed using epifluorescence. Areas of overlap between Rnt1p and the DNA appear in light blue. Yellow bar, 2 μm. The cell cycle phase is indicated for each picture.
To determine whether Rnt1p release from the nucleolus is induced by sister-chromatid separation or cell division, we monitored the localization of Rnt1p in cells carrying mutations in separin (Esp1p) (Ciosk et al., 1998) and cohesin (Mcd1p) (Guacci et al., 1997; Michaelis et al., 1997). Cells carrying a temperature sensitive mutation in ESP1 (esp1-1) exit mitosis and divide without nuclear division. Therefore, these cells allow us to determine whether Rnt1p requires nuclear division to exit from the nucleolus. As shown in Figure 5A, Rnt1p exits to the nucleus in esp1-1 dividing synchronized cultures at both permissive and restrictive temperatures, indicating that nuclear division is not required for Rnt1p nuclear localization. Consistently, Rnt1p localization is not affected in Mcd1p-defective cells that undergo premature sister-chromatids separation when drug arrested at metaphase at the restrictive temperature (Figure 5B). We conclude that cell division and not sister-chromatid separation induces Rnt1p release from the nucleolus.
Figure 5.
Rnt1p release from the nucleolus is cell division dependent. (A) Cells carrying a temperature sensitive allele of separin (esp1-1) were grown to log phase and then synchronized in G1 with α-factor. The synchronized cells were released either at the permissive (26°C) or the restrictive (37°C) temperatures and fixed once they reached the G2/M phase of the cell cycle. Antibodies against Rnt1p (red) were used to determine its localization pattern. The position of the nucleolus was highlighted with antibodies against the nucleolar protein Nop1p (green). DNA stained with DAPI is indicated in blue. Yellow bar, 2 μm. The graph shown on the right indicates the percentage of cells in which Rnt1p is nucleolar (No) or nucleoplasmic (Nu). The data shown represent the results of three independent experiments. (B) Cells carrying a temperature sensitive allele of cohesin (mcd1-1) were arrested in G2/M with nocodazole and then shifted to either the permissive (26°C) or the restrictive (37°C) temperature. Rnt1p was visualized and the number of cells with different localization pattern was quantified as described in A. (C) Wild-type cells (RPA190), or cells expressing a temperature-sensitive component of RNA polymerase I (rpa190-3), were either grown at the permissive temperature (23°C) or shifted to the restrictive temperature (37°C). Samples were taken at different intervals and the localization pattern of Rnt1p and two other nucleolar proteins (Nop1p and Nhp2) were visualized using immunofluorescence. The graphs shown on the left indicate the percentages of budded cells (black box) and of cells where Rnt1p is either nucleolar (open circle) or nucleoplasmic (black circle) as a function of time. The graph shown represents the average of three experiments with average error rates of ± 2.4%. (D) Photos of rpa190-3 cells grown at 37°C for either 1 or 4 h are shown on the right. The cells shown in the left panel were stained with antibodies against Nop1p (green) and Rnt1p (red), whereas those in the right panel were stained with antibodies against Nop1p (green) and Nhp2p (red). In both cases the DNA was colored by DAPI (blue). Yellow bar, 2 μm.
Because Rnt1p is required for pre-rRNA processing, we wanted to determine whether Rnt1p retention in the nucleolus requires actively transcribed rRNA. If Rnt1p nucleolar localization is mediated by binding to unprocessed prerRNA, then it should be immediately released from the nucleolus upon the cessation of rRNA transcription, and before the release of any other nucleolar proteins, and subsequent destruction of the nucleolus. We tested this hypothesis using a strain carrying a temperature-sensitive mutation in the largest subunit of Pol I (rpa190-3), which allows rapid shutdown of rRNA transcription upon shifting to the restrictive temperature (Wittekind et al., 1988). As shown in Figure 5, C and D, the number of budded cells decreased upon shifting either the wild-type strain RPA190 or the mutated strain rpa190-3 to 37°C. However, although the number of cells exhibiting Rnt1p in the nucleolus decreases in RPA190, the number of rpa190-3 cells with nuclear Rnt1p dramatically increases. Thus, Rnt1p localization in the nucleolus is linked to rRNA transcription. To ensure that the release of Rnt1p from the nucleolus does not simply reflect the destruction of the nucleolus in the absence of rRNA, we examined the localization of several known nucleolar proteins. In contrast to Rnt1p, which became mostly nuclear 1 h after shifting rpa190-3 cells to 37°C, nucleolar proteins such as Nop1p and Nhp2p remained nucleolar even after shifting rpa190-3 cells to 37°C for 4 h. It is important to note that unlike higher eukaryotes, yeast nuclei remain intact throughout the cell cycle and rRNA transcription is not completely arrested during the cell cycle (Sogin et al., 1974). However, subtle changes in the rate of rDNA transcription that could signal Rnt1p release from the nucleus cannot be excluded at this moment. We conclude that Rnt1p nucleolar localization requires actively transcribed RNA and suggest that Rnt1p is anchored to the rRNA processing machinery through binding to its RNA substrate.
Rnt1p Nuclear Localization Is Mediated by Two Distinct Signals
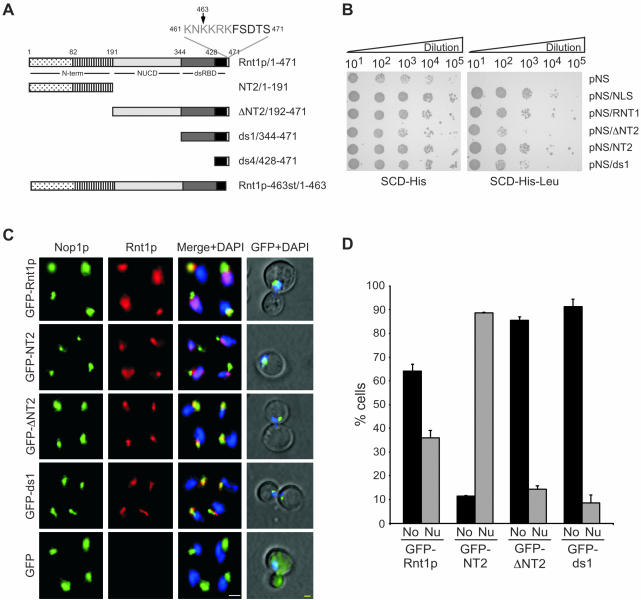
To understand how the cycle-dependent localization of Rnt1p is achieved, we needed to determine the amino acid residues required for Rnt1p localization. The Rnt1p domains required for the protein's nuclear localization were first identified using a nuclear transportation trap assay (Ueki et al., 1998). In this assay, different fragments of Rnt1p (Figure 6A) were expressed fused to the LexA DNA-binding domain, the Gal4 transactivation domain, and a strong nuclear export signal (Ueki et al., 1998). Fusion of the nuclear export signal to a nuclear localization signal (NLS) transports the LexA DNA-binding domain-Gal4 transactivation domain into the nucleus, resulting in the expression of a marker LEU2 gene and allowing the cells to grow in media lacking leucine. As shown in Figure 6B, all fusion proteins allowed the cells to grow in media containing leucine, but only those with a control NLS or expressing either the N-terminal domain or the dsRBD of Rnt1p grew on media lacking leucine. This indicates that both the N- and C-terminal domains of Rnt1p may enter the nucleus independently and suggests that Rnt1p contains two different NLSs. This possibility was further verified by monitoring the localization pattern of different fragments of Rnt1p fused to GFP. As expected, the GFP by itself accumulated in the cytoplasm, whereas GFP-Rnt1p accumulated both in the nucleus and in the nucleolus, depending on which phase the cell cycle is in (Figure 6, C and D). Surprisingly, both the N-terminal fusion protein (GFP-NT2) and the dsRBD fusions (GFP-ΔNT2 and GFP-ds1) exhibited two distinct localization patterns. GFPNT2 accumulated in the nucleoplasm, whereas GFP-ΔNT2 and GFP-ds1 accumulated exclusively in the nucleolus (Figure 6, C and D). The same results were obtained using both fixed and living cells (Figure 6C). Examination of cells transformed with different fragments of Rnt1p indicates that none of Rnt1p domains could independently rescue Δrnt1 defects in the cell cycle (our unpublished data). We conclude that both the N-terminal domain and dsRBD are required for Rnt1p subnuclear localization but that these domains cannot independently support the Rnt1p functions required for the normal progression of the cell cycle.
Figure 6.
Rnt1p possesses two distinct nuclear localization signals. (A) Schematic representation of the different Rnt1p domains and truncations used in this study. Rnt1p nuclease domain and dsRBD are depicted as light and dark gray boxes, respectively. The C-terminal extension harboring the “KNKKRK” NLS is shown in white, and its sequence is indicated on top. The first 82 residues of the N-terminal domain required for Rnt1p localization in the nucleoplasm are indicated as a dotted box. The rest of the N-terminal domain is indicated as a hatched box. The amino acid positions corresponding to the beginning and end of each domain are stated beside the name of each fragment. (B) The different domains of Rnt1p were expressed in EGY48 cells by using the nuclear localization trap vectors (62) and tested for the presence of a nuclear localization signal. The transformed cells were grown on media lacking either histidine (left), or histidine and leucine (top). All transformed cells could grow on SCD-His, whereas only those harboring a nuclear localization signal could grow on SCD-His-Leu. Each construct contains a nuclear export signal, whereas the positive control (pNS/NLS) contains both export and import nuclear signals. The names of the different vectors are indicated on the right, and the cell dilution factors are indicated on top. (C) Immunofluorescence assisted visualization of the different domains of Rnt1p. GFP fusions with different Rnt1p fragments were expressed from the RNT1 natural promoter in Δrnt1 cells and the proteins were either visualized by the immunostaining of fixed cells (the three columns on the left) or by direct epifluorescence of living cells (the last column on the right). In the case of immunostained cells the nucleolar protein Nop1p is shown in green, Rnt1p in red and DNA in blue. The different stains are shown either independently (first two columns on the left) or merged (third column). Rnt1p was visualized in living cells as described in Figure 4D and is shown in green, whereas DNA is colored blue. Yellow bars, 2 μm. (D) Graphical representation of the nucleolar (No) and nucleoplasmic (Nu) localizations of the different truncations of Rnt1p used in B. The data shown represent the results of three independent experiments and margin of errors is indicated on the chart.
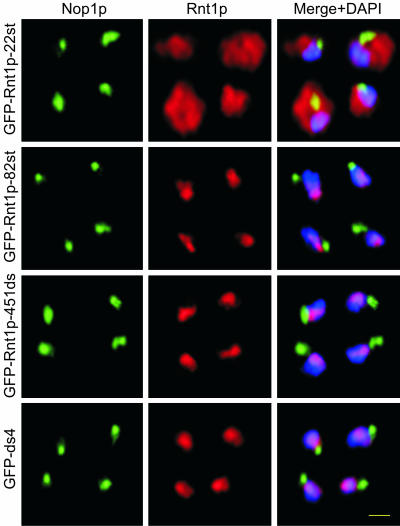
In general, protein localization in yeast is mediated by NLSs rich in basic amino acids (Jans et al., 2000). Therefore, to identify any potential Rnt1p NLS, we searched both the N- and C-terminal amino acids sequences of the protein for clusters of positively charged residues. No conserved NLS was found in the N-terminal domain, but we noticed that the first 82 residues of the N terminus, and especially the first 22 residues, were rich in lysine. In contrast, a typical KNKKRK nuclear localization signal motif was found near the C terminus in positions 461-466 (Figure 6A). To determine whether either or both fragments could promote nuclear localization, we expressed them in fusion with GFP and followed their localization by using either immunofluorescence microscopy (Figure 7) or by monitoring GFP fluorescence in living cells (our unpublished data). GFP fused to the N-terminal 22 amino acids alone (GFP-Rnt1p-22st) was largely cytoplasmic, whereas GFP fused with the first 82 amino acids (GFP-Rnt1p-82st) was nuclear. On the other hand, either the C-terminal 43 amino acids, or the last 11 amino acids, of Rnt1p were sufficient to transport the GFP to the nucleus. This indicates that the observed KNKKRK sequence functions as a nuclear localization signal. However, none of these sequences resulted in the accumulation of the protein in the nucleolus (Figure 7) as GFP-ds1 did. Consistently, Rnt1p carrying either deletions in the dsRBD, or mutations that destroy the dsRBD structure, also fail to accumulate in the nucleolus (our unpublished data). Together, these data indicate that Rnt1p nuclear localization could be independently mediated via a canonical NLS at the C terminus and through a less defined sequence at the N-terminal domain. The data also show that the accumulation of Rnt1p in the nucleolus requires the presence of a functional dsRBD and suggest that Rnt1p is retained in the nucleus through interaction with its RNA substrates.
Figure 7.
Immunofluorescence of cells expressing GFP fused to the Rnt1p N-terminal 22 amino acids (GFP-Rnt1p-22st), N-terminal 82 amino acids (GFP-Rnt1p-82st), C-terminal 43 amino acids (GFP-ds4), or the last 22 amino acids at the C terminus (GFP-Rnt1p-451ds). Rnt1p is shown in red, Nop1p in green, and DNA in blue. Colocalization of Rnt1p and Nop1p is in yellow and colocalization of Rnt1p with DNA is in purple. Yellow bar, 2 μm.
Rnt1p Release into the Nucleoplasm Is Required for Exit from the G2/M Phase of the Cell Cycle
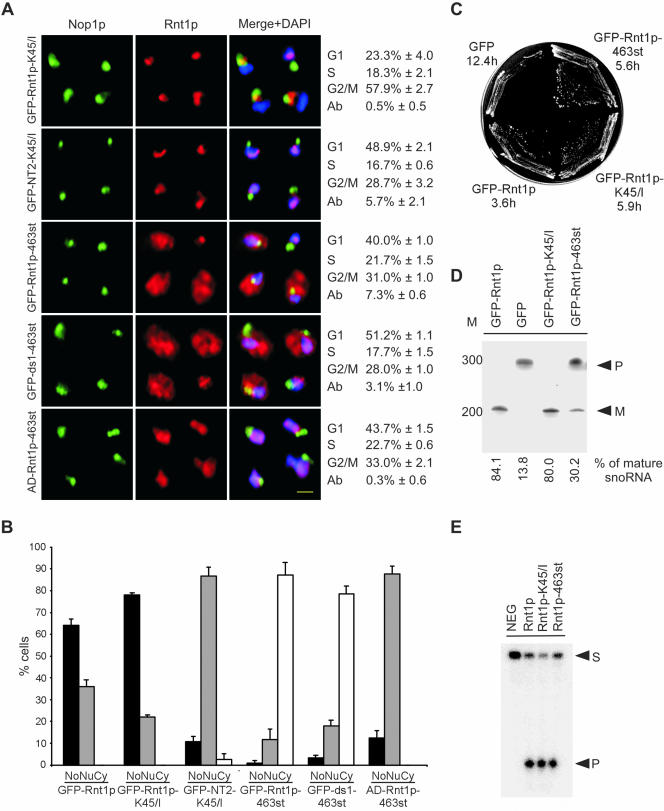
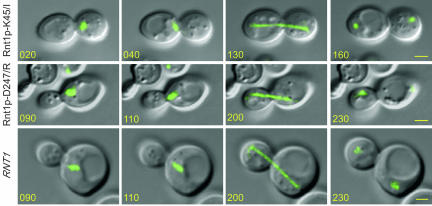
To further understand the impact of Rnt1p localization on the cell cycle, we searched for mutations that block the rhythmic localization of Rnt1p and render it either constitutively nuclear or exclusively nucleolar. GFP-Rnt1p-463st, which carries a stop codon at position 463 eliminating the last eight amino acids of Rnt1p, was detected in all cellular compartments, including the cytoplasm. This localization pattern was also displayed when the 463st mutation was introduced into an independently expressed dsRBD. This mutation partially rescued growth of Δrnt1 cells and reduced the number of cells accumulating in the G1 phase of the cell cycle, but it did not eliminate the aberrant budding characteristic of the Δrnt1 cells (Figure 8, A-C). Rnt1p-dependent RNA processing in vivo was partially restored in GFP-Rnt1p-463st (Figure 8D), but RNA cleavage in vitro was not affected (Figure 8E). This indicates that the mutant 463st does not affect Rnt1p catalytic activity but instead affects its nucleolar localization and thus impairs snoRNA processing. However, the general distribution of GFP-Rnt1p-463st, and its associated defects in RNA processing, prevent clear assessment of the impact of Rnt1p nuclear localization on the cell cycle. To address this, we screened a library of Rnt1p mutants produced by random mutagenesis of the N-terminal region to identify mutations that specifically block Rnt1p nuclear localization without affecting RNA processing. One mutation that changes the lysine at position 45 to isoleucine (K45/I) rendered Rnt1p nucleolar, even in cells at the G2/M phase of the cell cycle (Figure 8, A and B). Truncated Rnt1p N-terminal domain carrying the K45/I mutation constitutively accumulated in the nucleoplasm, suggesting that this mutation does not block the N terminus-dependent nuclear import pathway. The expression of GFP-Rnt1p-K45/I in Δrnt1 cells partially complemented Rnt1p functions, allowing cells to grow at both 26 and 37°C, albeit at a slower rate than GFP-Rnt1p (Figure 8C). Analysis of Rnt1p dependent RNA processing in cells expressing GFP-Rnt1p-K45/I showed no effects on RNA maturation. This indicates that a single mutation in the N-terminal domain could slow cell growth without affecting the classical function of Rnt1p in RNA processing. Consistently, recombinant Rnt1p-K45/I efficiently and specifically cleaved known Rnt1p substrates in vitro (Figure 8E). Surprisingly, cells carrying GFP-Rnt1p-K45/I were delayed at the G2/M phase of cell cycle, indicating that Rnt1p presence in the nucleus is important for the end of mitosis. Consistently, the number of cells in the G2/M phase of the cell cycle with telophase spindles decreased dramatically in cultures of Δrnt1 and GFP-Rnt1p-K45/I cells (Table 3). In addition, the nuclei of cells expressing GFP-Rnt1p-K45/I but not the catalytically impaired Rnt1p (Rnt1p-D247/R) shuttled back and forth through the bud neck (Figure 9 and supplementary material 5 and 6) as previously noted with Δrnt1 cells (Figure 2B). This clearly shows that the defect in nuclear division is caused by the absence of Rnt1p from the nucleoplasm and not due to anomalies in RNA or rRNA processing. Thus, a single mutation in the Rnt1p N terminus slowed cell growth and impaired cell progress without affecting Rnt1p-dependent RNA processing. These data suggest that the presence of Rnt1p in the nucleoplasm is required for exit from the G2/M phase of the cell cycle.
Figure 8.
Rnt1p release into the nucleoplasm is required for exit from the G2/M phase of the cell cycle. (A) Immunofluorescence of Δrnt1 cells expressing different Rnt1p mutations fused to GFP. Rnt1p lacking the last eight amino acids was also expressed in fusion with a heterologous nuclear localization signal (AD-Rnt1p-463st). Rnt1p is shown in red, Nop1p in green, and DNA in blue. Yellow bar, 2 μm. The percentage of cells in each phase of the cell cycle is indicated on the right of each mutant. The percentage of abnormal cells (Ab) is also indicated. The numbers are the average of three independent experiments. (B) Graphical representation of the percentage of cells where Rnt1p was found in the nucleolus (No), nucleoplasm (Nu), or the cytoplasm (Cy). (C) Cells lacking RNT1 were transformed with plasmids expressing either GFP-Rnt1p-463st or GFP-Rnt1p-K45/I, and then were tested for growth at 26°C. The corresponding doubling time (in hours) in liquid media at 26°C for each construct is indicated below the name of each fragment. (D) Northern blot analysis by using a probe against snR43 was performed as described in Figure 3C. The positions of premature (P) and mature (M) snR43 are indicated on the right, whereas the position of the DNA molecular weight marker is indicated on the left. The percentage of mature snR43 in each strain is indicated below. (E) In vitro cleavage assay of the different Rnt1p mutants. The reaction was carried using a model Rnt1p substrate as described in Figure 3A. The positions of the substrate (S) and the product (P) are indicated on the right.
Figure 9.
Mutations in Rnt1p affect division and microtubule movement. Photos of cells expressing Rnt1p-K45/I, Rnt1p-D247/R, or Rnt1p were taken every 10 min, and representative pictures are shown (see videos in supplementary material videos 1, 5, and 6). The times from the beginning of the acquisitions are indicated in minutes on each photo. The microtubule complex revealed by GFP-Tub3p is shown in green. Yellow bars, 2 μm.
DISCUSSION
In fission yeast, orthologues of RNase III have been implicated in cell division and chromosome segregation, presumably through their role in RNA interference (Provost et al., 2002b; Hall et al., 2003). Here, we show in baker's yeast, where the components of the RNA interference machinery do not exist, that Rnt1p is required for nuclear positioning, nuclear division, and normal progression of the cell cycle. Surprisingly, we found that Rnt1p activities in the cell cycle do not involve RNA processing and degradation, but instead require the presence of Rnt1p in the nucleoplasm. Rnt1p was localized to the nucleolus in all phases of the cell cycle except the G2/M phase where it became nucleoplasmic. This localization pattern is similar to that observed with human RNase III (Wu et al., 2000). The Rnt1p subnuclear localization pattern was found to be mediated by two distinct localization signals: a canonical importin-dependent signal required for both nuclear and nucleolar localization, and a less defined signal at the N terminus needed for the accumulation of Rnt1p in the nucleoplasm. Point mutations that specifically block the accumulation of an otherwise functional Rnt1p in the nucleoplasm slowed initiation of mitosis. Together, the results presented in this study suggest a new noncatalytic role of yeast RNase III that ensures cell cycle progression and nuclear division.
Novel Functions of Rnt1p Unrelated to RNA Processing
Like other eukaryotic RNase IIIs (Nicholson, 1999; Wu et al., 2000), Rnt1p is required for the processing of rRNA and other noncoding RNAs, including snRNAs and snoRNAs (Lamontagne et al., 2001). In addition, the deletion of RNT1 causes severe growth defects and temperature sensitivity (Abou Elela and Ares, 1998) the cause of which remains unclear. In this study, we carefully examined the growth and cell division of Δrnt1 cells and identified new phenotypes unrelated to RNA processing. The results suggest that the deletion of RNT1 has two distinct phenotypes: the first is slow growth stemming from defects in RNA processing and the second is a processing-independent defect in cell division. The slow growth phenotype was shown to result from slow metabolism (Figure 1), consistent with defects in ribosome biogenesis. Indeed, cells expressing Rnt1p, but lacking its processing signal at the end of the 25S rRNA, displayed defects in ribosome biogenesis and cell growth (Allmang and Tollervey, 1998) similar to those observed upon the deletion of RNT1. Consistently, a single mutation in the Rnt1p catalytic domain that impairs RNA cleavage blocked rRNA processing and slowed cell growth (Figure 3) to the same extent as the complete deletion of RNT1 (Abou Elela et al., 1996; Abou Elela and Ares, 1998; Kufel et al., 1999). In contrast, the defect in cell division was not observed in cells expressing catalytically impaired Rnt1p (Figure 3), indicating that the observed effects on the cell cycle are not caused by a general perturbation of RNA processing. This is further supported by the failure of mutations in known Rnt1p processing signals, including that at the end of rRNA, to reproduce the cell cycle defects observed upon the deletion of Rnt1p (Abou Elela and Ares, 1998; Allmang and Tollervey, 1998). The data clearly indicate that Rnt1p has new cellular functions unrelated to RNA processing. It is not clear at this moment whether the noncatalytic function of Rnt1p is shared by other RNase IIIs. It was previously shown that bacterial RNase III stimulates the synthesis of lambda cIII protein by binding to, and not by cleaving, the cIII mRNA, suggesting that bacterial RNase III may have cleavage independent functions (Altuvia et al., 1987). On the other hand, eukaryotic orthologues of RNase III have been shown to affect chromosome segregation (Provost et al., 2002b; Hall et al., 2003), cell development (Knight and Bass, 2001), mating (Iino et al., 1991; Jacobsen et al., 1999), and meiosis (Iino et al., 1991) in a rRNA processing-independent pathway. However, the requirement of RNA catalysis for RNase III function in all of these functions has never been directly established. We speculate that other eukaryotic RNase IIIs may also contribute to cell functions independent of RNA catalysis.
How Does Rnt1p Affect the Cell Cycle and Nuclear Division?
By forming a stable RNP, Rnt1p could block or stimulate the function of nontranslated RNA, or regulate mRNA export to the cytoplasm. Although, Rnt1p has been shown to form a stable RNP complex with short RNA hairpins in vitro (Lamontagne et al., 2003), this phenomenon has yet to be demonstrated in vivo. Indeed, demonstrating the effects of Rnt1p-RNA interaction on the cell functions would be difficult because all mutations that affect RNA binding also impair RNA cleavage (Lamontagne et al., 2000; Tremblay et al., 2002) or perturb Rnt1p localization (our unpublished data). Alternatively, Rnt1p could influence the cell cycle by binding to proteins involved in this process. A two-hybrid screen by using Rnt1p as bait (Tremblay, 2002; Catala and Abou Elela, unpublished data) has identified an interaction with the microtubule binding protein Yeb1p/Bim1p (Miller et al., 2000; Schuyler and Pellman, 2001) and Tub4p, which is required for microtubule organization and nuclear division (Geissler et al., 1996; Spang et al., 1996). The interaction of Rnt1p with Yeb1p/Bim1p and Tub4p could explain how Rnt1p may influence microtubule function and nuclear positioning. Inactivation of Tub4p leads to the assembly of a short spindle, which does not elongate impairing the formation of a mitotic spindle (Spang et al., 1996). On the other hand, Yeb1p/Bim1p deletion impairs nuclear movement and positioning in the bud neck (Adames and Cooper, 2000; Miller et al., 2000; Schuyler and Pellman, 2001). The expression levels of Yeb1p/Bim1p and Tub4p are not significantly changed in Rnt1p cells, suggesting that Rnt1p does not affect the functions of these proteins by degrading their mRNA (Catala and Abou Elela, unpublished data). Mutations that specifically inhibit the interaction with these two proteins would reveal the impact of Rnt1p interaction with Yeb1p/Bim1p on cell division.
Nucleolar Proteins Link pre-rRNA Processing to Cell Cycle
The link between the cell cycle and rRNA processing is becoming clear as more rRNA synthesis and processing proteins are shown to be associated with the cell cycle. In addition to Net1p, which is involved in both RNA Pol I transcription and the regulation of cell exit from mitosis (Shou et al., 2001), it was recently shown that the rRNA processing RNase MRP is required for cell cycle progression at the end of mitosis (Cai et al., 2002). Deletion of this enzyme results in large-budded cells, dumbbell-shaped nuclei, and extended spindles. These defects are likely mediated by an accumulation of Clb2p. Rnt1p, another pre-rRNA processing protein, is also required in both the G1 and G2/M phases of the cell cycle (Figures 1 and 2). Intriguingly, a single mutation in Rnt1p N-terminal domain blocks Rnt1p exit to the nucleus and delays cells at the G2/M phase of the cell cycle without affecting RNA processing (Figure 8). We propose that by influencing both ribosome synthesis and the decision to exit mitosis, Rnt1p, Net1p, and MRP help synchronize ribosome production with cell division, thereby ensuring the presence of an adequate number of ribosomes to sustain both the mother and daughter cells.
Supplementary Material
Acknowledgments
We thank R.J. Wellinger for strains RWY12 and MLY30, M.E. Schmitt for the pTS417 vector, A. Amon for strains 1724 and 1730, M. Nomura for strains NOY260 and NOY265, and K. Nagahari for the pNS and pNS/NLS vectors. We also thank J.P. Gélugne and J.P. Aris for anti-Nhp2p and anti-Nop1p antibodies. We are indebted to Mark E. Schmitt for critical reading of this manuscript. We are grateful for R.J. Wellinger for stimulating discussions and ideas. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada and a grant from the Canadian Institute of Health Research. Support for the RNA group core was provided by a group grant from Canadian Institute of Health Research. S.A. is a Chercheur-Boursier Junior II of the Fonds de la Recherche en Santé du Québec. B.L. is supported by a grant from the Fonds de la Recherche en Santé du Québec-Fonds pour la Formation du Chercheurs et l'Aide à la Recherche.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0183. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0183.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Abou Elela, S., and Ares, M., Jr. (1998). Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17, 3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela, S., Igel, H., and Ares, M., Jr. (1996). RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85, 115-124. [DOI] [PubMed] [Google Scholar]
- Adames, N.R., and Cooper, J.A. (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149, 863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., and Tollervey, D. (1998). The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278, 67-78. [DOI] [PubMed] [Google Scholar]
- Altuvia, S., Locker-Giladi, H., Koby, S., Ben-Nun, O., and Oppenheim, A.B. (1987). RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 84, 6511-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris, J.P., and Blobel, G. (1988). Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107, 17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, D., and Slack, F. (2002). Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays 24, 119-129. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. (1995). Double-stranded RNA binding proteins and their substrates. Nucleic Acids Symp. Ser. 13-15. [PubMed]
- Blaszczyk, J., Tropea, J.E., Bubunenko, M., Routzahn, K.M., Waugh, D.S., Court, D.L., and Ji, X. (2001). Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9, 1225-1236. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Cai, T., Aulds, J., Gill, T., Cerio, M., and Schmitt, M.E. (2002). The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 161, 1029-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J.L., and Stearns, T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138, 629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J.L., and Stearns, T. (1999). Cytoskeletal dynamics in yeast. Methods Cell Biol. 58, 87-105. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Abou Elela, S., Ares, M., Jr., and Guthrie, C. (1997). Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11, 2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Legrain, P., and Jacquier, A. (1998b). Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284, 975-988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. (1998a). Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17, 3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., and Nasmyth, K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. (2003). RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11, 1279-1289. [DOI] [PubMed] [Google Scholar]
- Dionne, I., and Wellinger, R.J. (1996). Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93, 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durovic, P., and Dennis, P.P. (1994). Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acidocaldarius: similarities to eukaryotic rRNA processing. Mol. Microbiol. 13, 229-242. [DOI] [PubMed] [Google Scholar]
- Estojak, J., Brent, R., and Golemis, E.A. (1995). Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15, 5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov, V., Solovyev, V., Filippova, M., and Gill, S.S. (2000). A novel type of RNase III family proteins in eukaryotes. Gene 245, 213-221. [DOI] [PubMed] [Google Scholar]
- Geissler, S., Pereira, G., Spang, A., Knop, M., Soues, S., Kilmartin, J., and Schiebel, E. (1996). The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 15, 3899-3911. [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (1991). Guide to Yeast Genetics and Molecular Biology, San Diego, Academic Press.
- Hall, I.M., Noma, K., and Grewal, S.I. (2003). RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100, 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras, A., Henry, Y., Bousquet-Antonelli, C., Noaillac-Depeyre, J., Gelugne, J.P., and Caizergues-Ferrer, M. (1998). Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17, 7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and Zamore, P.D. (2002). RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12, 225-232. [DOI] [PubMed] [Google Scholar]
- Iino, Y., Sugimoto, A., and Yamamoto, M. (1991). S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J. 10, 221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231-5243. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans, D.A., Xiao, C.Y., and Lam, M.H. (2000). Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22, 532-544. [DOI] [PubMed] [Google Scholar]
- Kharrat, A., Macias, M.J., Gibson, T.J., Nilges, M., and Pastore, A. (1995). Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14, 3572-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, S.W., and Bass, B.L. (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Dichtl, B., and Tollervey, D. (1999). Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5, 909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne, B., and Abou Elela, S. (2001). Purification and Characterization of Saccharomyces cerevisiae Rnt1p Nuclease, San Deigo: Academic Press. [DOI] [PubMed]
- Lamontagne, B., Ghazal, G., Lebars, I., Yoshizawa, S., Fourmy, D., and Abou Elela, S. (2003). Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 327, 985-1000. [DOI] [PubMed] [Google Scholar]
- Lamontagne, B., Larose, S., Boulanger, J., and Abou Elela, S. (2001). The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 3, 71-78. [PubMed] [Google Scholar]
- Lamontagne, B., Tremblay, A., and Abou Elela, S. (2000). The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20, 1104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., Cheng, S.C., and Rose, M.D. (2000). Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11, 2949-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, A.W. (1999). Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23, 371-390. [DOI] [PubMed] [Google Scholar]
- Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. (2002a). Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost, P., Silverstein, R.A., Dishart, D., Walfridsson, J., Djupedal, I., Kniola, B., Wright, A., Samuelsson, B., Radmark, O., and Ekwall, K. (2002b). Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA 99, 16648-16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rotondo, G., and Frendewey, D. (1996). Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 24, 2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, L.R., and Barber, G.N. (2003). The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17, 961-983. [DOI] [PubMed] [Google Scholar]
- Schuyler, S.C., and Pellman, D. (2001). Search, capture and signal: games microtubules and centrosomes play. J. Cell Sci. 114, 247-255. [DOI] [PubMed] [Google Scholar]
- Seipelt, R.L., Zheng, B., Asuru, A., and Rymond, B.C. (1999). U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27, 587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., et al. (2001). Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol Cell 8, 45-55. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin, S.J., Carter, B.L.A., and Halvorson, H.O. (1974). Changes in the rate of ribosomal RNA synthesis during the cell cycle in Saccharomyces cerevisiae. Exp. Cell Res. 89, 127-138. [DOI] [PubMed] [Google Scholar]
- Spang, A., Courtney, I., Grein, K., Matzner, M., and Schiebel, E. (1995). The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128, 863-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Geissler, S., Grein, K., and Schiebel, E. (1996). gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 134, 429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasov, K., Perdomo, L.I., Evakine, E., and Nazar, R.N. (2002). RAC protein directs the complete removal of the 3′ external transcribed spacer by the Pac1 nuclease. Mol. Cell 9, 433-437. [DOI] [PubMed] [Google Scholar]
- Thomas, B.J., and Rothstein, R. (1989). Elevated recombination rates in transcriptionally active DNA. Cell 56, 619-630. [DOI] [PubMed] [Google Scholar]
- Tremblay, A. (2002). Etude de la fonction de la RNase III eucaryote et identification de ses partenaires cellulaires dans un criblage double-hybrides. M.S. thesis, Université de Sherbrooke, Sherbrooke.
- Tremblay, A., Lamontagne, B., Catala, M., Yam, Y., Larose, S., Good, L., and Abou Elela, S. (2002). A physical interaction between Gar1p and Rnt1pi is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 22, 4792-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, N., Oda, T., Kondo, M., Yano, K., Noguchi, T., and Muramatsu, M. (1998). Selection system for genes encoding nuclear-targeted proteins. Nat. Biotechnol. 16, 1338-1342. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. (2000). Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20, 441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., Hwang, E.S., and Amon, A. (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818-823. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833-1837. [DOI] [PubMed] [Google Scholar]
- Waddle, J.A., Karpova, T.S., Waterston, R.H., and Cooper, J.A. (1996). Movement of cortical actin patches in yeast. J. Cell Biol. 132, 861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind, M., Dodd, J., Vu, L., Kolb, J.M., Buhler, J.M., Sentenac, A., and Nomura, M. (1988). Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 3997-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Xu, H., Miraglia, L.J., and Crooke, S.T. (2000). Human RNase III is a 160-kDa protein involved in preribosomal RNA processing [In Process Citation]. J. Biol. Chem. 275, 36957-36965. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. (2002). Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21, 5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Frendewey, D., and Lobo Ruppert, S.M. (1999). Pac1p, an RNase III homolog, is required for formation of the 3′ end of U2 snRNA in Schizosaccharomyces pombe [In Process Citation]. RNA 5, 1083-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.