Abstract
Release of cytochrome c from the mitochondrial intermembrane space is critical to apoptosis induced by a variety of death stimuli. Bid is a BH3-only prodeath Bcl-2 family protein that can potently activate this efflux. In the current study, we investigated the mitochondrial localization of Bid and its interactions with mitochondrial phospholipids, focusing on their relationships with Bid-induced cytochrome c release. We found that Bid binding to the mitochondria required only three of its eight helical structures (α4-α6), but not the BH3 domain, and the binding could not be inhibited by the antideath molecule Bcl-xL. Membrane fractionations indicated that tBid bound to mitochondrial outer membranes at both contact and noncontact sites. Bid could interact with specific cardiolipin species on intact mitochondria as identified by mass spectrometry. Like the binding to the mitochondria, this interaction could not be blocked by the mutation in the BH3 domain or by Bcl-xL. However, a cardiolipin-specific dye, 10-N-nonyl acridine orange, could preferentially suppress Bid binding to the mitochondrial contact site and inhibit Bid-induced mitochondrial cristae reorganization and cytochrome c release. These findings thus suggest that interactions of Bid with mitochondrial cardiolipin at the contact site can contribute significantly to its functions.
INTRODUCTION
The Bcl-2 family proteins regulate apoptosis at the level of mitochondria and consist of both antideath and prodeath members (Gross et al., 1999). Bid belongs to the BH3-only subgroup of the prodeath molecules (Wang et al., 1996), which serve as sentinels to diverse apoptotic signals (Kelekar and Thompson, 1998; Huang and Strasser, 2000). Bid can be activated by multiple proteases, including caspases (Li et al., 1998; Luo et al., 1998), granzyme B (Barry et al., 2000; Wang et al., 2001), lysosomal enzymes (Stoka et al., 2001; Reiners et al., 2002), and calpains (Chen et al., 2001; Mandic et al., 2002), in response to various death stimuli. The C-terminal part of the truncated Bid is able to translocate to the mitochondria and to induce release of apoptogenic factors, including cytochrome c and Smac/DIABLO (reviewed in van Gurp et al., 2003), which are critical to the initiation and promotion of cell death (Green and Reed, 1998; Budihardjo et al., 1999). The Bid-mediated mitochondrial events are also important to the successful completion of the death program in the so-called type II cells, such as hepatocytes, after death receptor engagement (Yin et al., 1999; Zhao et al., 2001; Li et al., 2002).
Release of the apoptogenic factors is one of the key events of mitochondria activation during apoptosis, for which the mechanisms are still largely elusive. Because of its potent effects in activating mitochondria, Bid has been studied as a model molecule to understand how mitochondria could be activated by the BH3-only prodeath Bcl-2 family proteins. Bid can induce oligomerization of Bak or Bax, leading to mitochondrial outer membrane permeabilization (Eskes et al., 2000; Wei et al., 2000). The BH3 domain of Bid is required for its interaction with Bak or Bax (Eskes et al., 2000; Wei et al., 2000) and is also the target of the antideath Bcl-2 or Bcl-xL (Wang et al., 1996; Cheng et al., 2001). However, other mechanisms can be involved. Bid can cause mitochondrial cristae reorganization, which mobilizes a major portion of cytochrome c for a maximal release (Scorrano et al., 2002). In addition, acidic phospholipids, particularly cardiolipin, may also participate in the function of Bid based on in vitro liposome permeabilization study (Schendel et al., 1999; Kudla et al., 2000; Lutter et al., 2000; Zha et al., 2000; Esposti et al., 2001; Zhai et al., 2001; Epand et al., 2002). Only cardiolipin has been implicated in the in vivo function of Bid in one study, in which binding of Bid to the mitochondria derived from cells with diminished cardiolipin synthesis was significantly reduced, accompanied with a reduced cytochrome c release (Lutter et al., 2000). These studies, however, have not demonstrated that Bid could actually interact directly with cardiolipin on intact mitochondria and have not shown how cardiolipin may contribute to Bid-induced mitochondria permeabilization.
Localization of Bid in the mitochondria has been shown to be restricted in the mitochondria contact site by immunoelectron tomography (Lutter et al., 2001). However, it is not known how Bid could induce mitochondrial outer membrane permeabilization at this site. For example, if Bid induced Bak or Bax oligomerization at the contact site, the process might lead to permeabilization of both outer and inner membranes, because the two membranes are closely juxtaposed at this unique site. However, permeabilization of inner membranes has not been observed (Kluck et al., 1999; Van Loo et al., 2002).
To address these issues, we carried out an in-depth analysis of the binding of Bid to the mitochondria, the distribution of Bid in the mitochondrial subcompartments, the interactions of Bid with cardiolipin on intact mitochondria and the effects of such interactions on Bid localization, Bid-induced Bak oligomerization and cristae reorganization, and Bid-induced cytochrome c release. Our study indicates that Bid-cardiolipin interaction at mitochondrial contact site could contribute significantly to Bid-induced mitochondrial permeabilization.
MATERIALS AND METHODS
Expression and Purification of Recombinant Proteins
Recombinant Bid proteins were prepared as described previously (Kim et al., 2000). Briefly, murine full-length Bid (α1-8, amino acid 1-195), wild-type tBid (α3-8, amino acid 60-195), mutant tBid (amino acid 60-195, G94E), Bid α3-6 (amino acid 60-166), and Bid α3-5 (amino acid 60-145) were amplified by polymerase chain reaction and cloned into pET23dw, an expression vector modified from pET23d(+) with the polyhistidine tag. Human Bcl-xL ΔTM (amino acid 1-209) was also cloned in pET23dw as described previously (Kim et al., 2000). All the constructs were expressed in Escherichia coli BL21(DE3) and purified using His-Bind nickel-agarose affinity column chromatography. However, truncated Bid (α4-6, amino acid 105-166) was fused to enhanced green fluorescent protein (EGFP) in the vector pEGFP-c1 (BD Biosciences Clontech, Palo Alto, CA). The fusion protein was analyzed in vivo with confocal microscopy.
Preparation of Mitochondria
Murine liver mitochondria were isolated as described previously (Kim et al., 2000) with buffer A (250 mM mannitol, 70 mM sucrose, 0.5 mM EGTA, 5 mM HEPES-NaOH, pH 7.2) and resuspended in buffer B (250 mM sucrose, 10 mM HEPES-NaOH, pH 7.5, 2 mM KH2PO4, 5 mM sodium succinate, 25 μM EGTA, 0.1 mM phenylmethylsulfonyl fluoride). This preparation was used in all studies except for the mass spectrometric analysis. For the latter, isolated mitochondria were resuspended in buffer A and further purified by a sucrose step gradient consisting of 2 ml each of 1.2 and 1.6 M sucrose through centrifugation at 40,000 × g for 1 h at 4°C. The mitochondria, which show as a brownish band at the interface of 1.2 and 1.6 M sucrose, were recovered, washed once with buffer A, and resuspended in buffer B.
Mitochondrial Membrane Fractionation with Digitonin
This was performed as reported previously (Greenawalt, 1974) and modified (Ohlendieck et al., 1986; Hovius et al., 1990). Briefly, 10 mg of mitochondria (0.5 mg/ml) were treated as indicated in the figure legend. The mitochondria were then pelleted by centrifugation at 10,000 × g for 15 min and resuspended in buffer A. Freshly prepared 2% digitonin in buffer A was added to the mitochondrial suspension to a final ratio of 0.2, 0.3, or 0.4% (wt/wt, digitonin/mitochondrial protein). The mixtures were then gently rotated (∼35 rpm) at 4°C for 15 min to solubilized the outer membranes. After the centrifugation at 10,000 × g for 15 min, the outer membrane fraction was collected from the supernatant, and the inner membrane fraction was collected from the pellets. These fractions were analyzed by SDS-PAGE followed by immunoblot with antibodies against VDAC (mAb4; Calbiochem, San Diego, CA), COX IV (clone 20E8-C12; Molecular Probes, Eugene, OR), Bid (Wang et al., 1996), Bak (Up-state Biotechnology, Lake Placid, NY), or Bcl-xL (Upstate Biotechnology).
Mitochondrial Membrane Fractionation by Linear Sucrose Gradient Centrifugation
This was mainly based on the swell-shrink-sonicate procedure described previously (Ohlendieck et al., 1986; Adams et al., 1989; Pon et al., 1989; Ardail et al., 1990) with modifications. Briefly, mitochondria suspended in buffer B (0.5 mg/ml) were treated as indicated in the figure legends before being washed and resuspended in 0.5 ml of hypotonic buffer C (10 mM KH2PO4, pH 7.4, 1 mM EDTA) at a concentration of 10 mg/ml. The mitochondria were allowed to swell at 4°C with gentle rotation for 15 min. The suspension was then admixed with 121 μl of 2.3 M sucrose per 0.5 ml of volume so that the final concentration of the sucrose was 0.45 M. After 15 min of incubation at 4°C, the shrunken mitochondria were sonicated (Sonic Vibracell, 3 × 10 s at 40% amplitude or 5-W output). The mitochondria suspension was centrifuged at 10,000 × g for 10 min to remove debris and intact mitochondria. The supernatants were then loaded onto a sucrose linear gradient (1.8-1.4 M, 4.0 ml) prepared in buffer C, and centrifuged at 100,000 × g in a SW60Ti rotor (Beckman Coulter, Fullerton, CA) for 20 h at 4°C. Fractions were then collected from the bottom of the gradient. Thus, the heavier inner membranes were eluted first, followed by the lighter outer membranes. Fractions were sequentially labeled and each contained ∼100 μl of sample. The sucrose concentration of each fraction was derived from linear regression analysis with the first fraction set at 1.8 M and the last fraction (40) set at 1.4 M. Protein concentrations were determined. A 20-μl sample containing similar amounts of proteins for each fraction was analyzed by SDS-PAGE followed by immunoblot with antibodies against VDAC (mAb4; Calbiochem), COX IV (clone 20E8-C12; Molecular Probes), Bid (Wang et al., 1996), or Bak (Upstate Biotechnology).
Analysis of Mitochondrial Targeting, Cytochrome c Release, and Bak Oligomerization
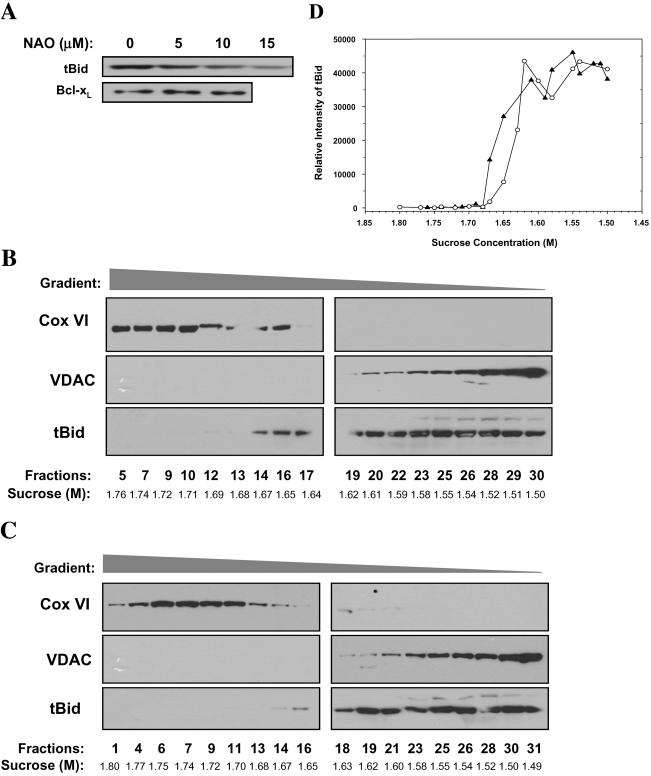
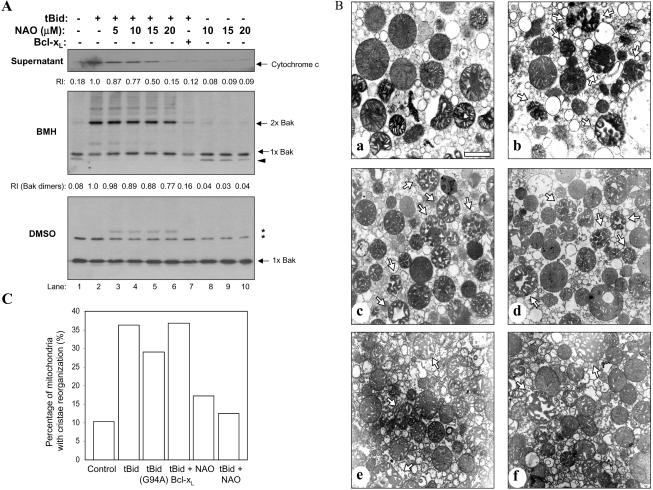
Isolated mitochondria (0.5 mg/ml) in buffer B were incubated with various recombinant proteins as indicated in the figure legends for 1 h at 30°C. The supernatants were separated by centrifugation at 10,000 × g for 15 min at 4°C and analyzed for cytochrome c release by immunoblot with an anti-cytochrome c antibody (BD Biosciences PharMingen, San Diego, CA). For analysis of protein insertion into the membrane, the mitochondrial pellets were resuspended in buffer B containing 0.1 M Na2CO3, pH 11.5, and incubated on ice for 30 min. The mitochondria were repelleted by centrifugation at 100,000 × g for 30 min and analyzed by immunoblot for Bid (Wang et al., 1996) or Bcl-xL (Upstate Biotechnology). To analyze cytochrome c release induced by tBid, mitochondria were suspended in buffer B with 4 mM MgCl2 and treated with tBid for 60 min at 30°C. The supernatant were then separated and analyzed by immunoblot for cytochrome c. To determine Bak oligomerization, the mitochondria were treated with tBid as described above. After centrifugation, the mitochondria were resuspended in buffer B and incubated with the homobifunctional cross-linker bismaleimidohexane (BMH) (10 mM) or the solvent dimethyl sulfoxide (DMSO) for 60 min at room temperature followed by SDS-PAGE and immunoblot for Bak (Upstate Biotechnology) (Wei et al., 2000). To determine the effect of 10-N-nonyl-acridine orange (NAO) on cytochrome c release and Bak oligomerization, mitochondria were pretreated with NAO (5-15 μM) for 10 min at 30°C before being treated with various recombinant proteins as described above.
Analysis of tBid-Lipid Interactions by Mass Spectrometry
Purified mitochondria were treated with wild-type tBid or mutant tBid (G94E) (0.1 μg/ml) in the absence or presence of Bcl-xLΔTM (13 μg/ml) for 1 h at 30°C. Lipids were extracted from untreated or treated mitochondria and analyzed by mass spectrometry as described previously (Matsko et al., 2001). Briefly, mitochondria were extracted in chloroform:methanol (2:1, vol/vol) overnight at 4°C under a N2 atmosphere in the presence of butylated hydroxytoluene. Sodium chloride (0.15M) was added and the sample was vortexed and centrifuged. The chloroform layer was removed and the lipids were dried under a stream of N2 and stored under a N2 atmosphere at -20°C until analysis. Lipids were analyzed by direct infusion into a Quattro II triple quadrupole mass spectrometer (Micromass, Manchester, United Kingdom) equipped with an electrospray ionization source. The electrospray probe was operated at a voltage differential of -3.5 keV in the negative ion mode. Mass spectra were obtained by scanning in two mass/charge ranges, 400-950 Da/e and 1200-1800 Da/e, every 1.6 s and summing individual spectra. Source temperature was maintained at 70°C. Scanning in the range of 400-950 Da/e in the negative ion mode affords identification of the major phospholipid species, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidic acid (PA), phosphatidylinositol (PI), and phosphatidylglycerol (PG), running as singly-charged (M-H)- ions. Some species may also run as chloride adducts. In addition, the 400-950 Da/e range also detects cardiolipin (CL) that runs as the doubly charged (M-2H)2-species due to its additional phosphate. Due to is higher mass, cardiolipin may also be detected in the singly-charged (M-H)- state in the range of 1200-1800 Da/e. Daughter ion spectra were obtained by selecting the ion of interest and performing daughter ion scanning in Q3 at 400 Da/s. Collision gas (argon) and collision energy were adjusted accordingly. The mass spectrometer was operated at unit resolution in the ms mode and slightly below unit resolution in the tandem ms mode.
Electron Microscopy
This was performed as described previously (Scorrano et al., 2002) with modification. Briefly, About 150 μl of mitochondria (0.5 mg/ml) in buffer B with 4 mM MgCl2 were treated with NAO (10 μM) or buffer for 5-10 min, washed, and then incubated with wild-type or G94E mutant tBid (0.3 μg/ml) for 10 min at 30°C. Alternatively, mitochondria were preincubated with Bcl-xLΔTM (20 μg/ml) for 10 min before tBid was added for another 10 min. Mitochondria were then separated by centrifugation at 10,000 × g for 10 min and washed three times with isotonic potassium phosphate buffer to remove sucrose, which otherwise would interfere with the microscopy. The pellets were then fixed with 1.25% glutaraldehyde at room temperature for 1 h, rinsed in cacodylate buffer, postfixed with 1% osmium tetroxide, dehydrated, embedded, and thin-sectioned for electron microscopic analysis at the Electron Microscopy Facility (Department of Pathology, University of Pittsburgh School of Medicine).
RESULTS
Binding of tBid to the Mitochondria Does Not Require the BH3 Domain, but Requires the α4, α5, and α6 Helices
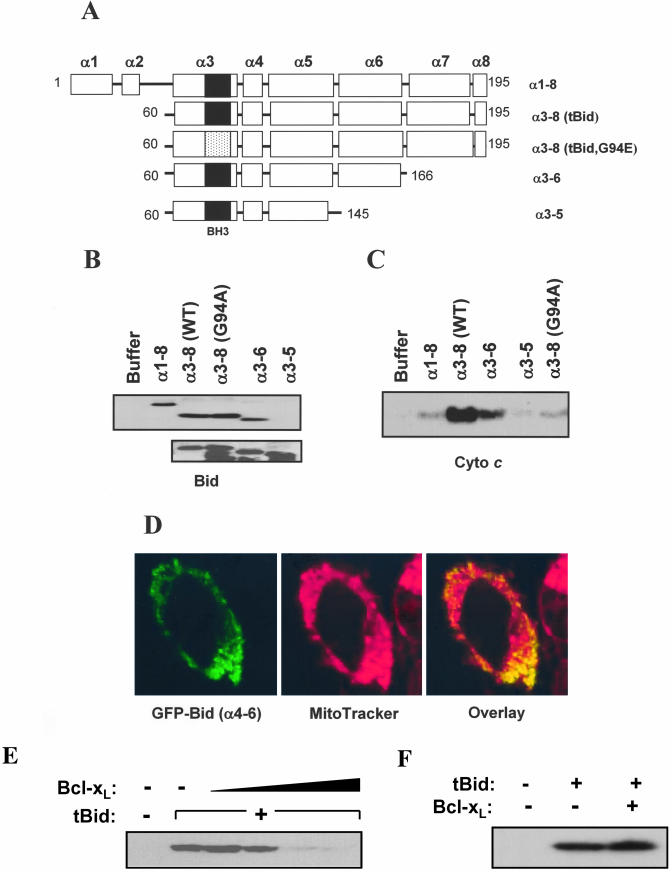
To study the interaction of Bid with the mitochondria and the subsequent events, we first defined the region of Bid responsible for the binding to the mitochondria. Bid is composed of eight alpha helices, namely, α1 to α8 (Chou et al., 1999; McDonnell et al., 1999). We constructed several deletional Bid mutants, encompassing different α helices (Figure 1A). Purified recombinant Bid proteins were then incubated with isolated mouse liver mitochondria. The membrane-inserted Bid on alkali-treated mitochondria and the cytochrome c released into the supernatant were determined by immunoblot analysis (Figure 1, B and C).
Figure 1.
Different structures of tBid are required for membrane binding and cytochrome c release. (A) Schematic diagram of various recombinant Bid molecules. Designation of α helices in relation with the deletional mutants is shown. The solid box in the α3 helix indicates the wild-type BH3 domain, whereas the dotted box in the same helix indicated the BH3 domain with a point mutation (G94E). (B) Binding of Bid to the mitochondria depends on specific α helices. Various Bid proteins (1 μg/ml) were added to isolated mitochondria (0.5 mg/ml) and incubated for 1 h at 30°C. Mitochondria were separated, treated with alkali, and analyzed by immunoblot with an anti-Bid antibody (Wang et al., 1996) for Bid targeting (top). This antibody was able to recognize the various truncated recombinant Bid mutants in purified form (bottom). (C) Supernatants isolated from Bid-treated mitochondria as in B were analyzed by immunoblot for released cytochrome c. (D) Bid binding to the mitochondria can be BH3-domain independent. Bid (α4-6)-EGFP was transfected into HeLa cells for 8 h in the presence of 2 mM of z-VAD-fmk (left, green). Mitochondria were identified by MitoTracker (100 nM; middle, red). Localization of Bid to the mitochondria is indicated by orange (right) by confocal microscopy. (E) Bcl-xL inhibits tBid-induced cytochrome c release. Mitochondria (0.5 mg/ml) were incubated with tBid (0.1 μg/ml) and Bcl-xL (3.3, 6.7, 13, or 26 μg/ml) for 1 h at 30°C. The supernatants were separated and analyzed by immunoblot for cytochrome c release. (F) Bcl-xL does not interfere with tBid binding to the mitochondria. Mitochondria were pretreated with Bcl-xL (27 μg/ml) and then tBid (0.1 μg/ml). The treated mitochondria were separated, alkali-treated, and analyzed by immunoblot with the anti-Bid antibody.
Full-length Bid (α1-α8) was able to bind to and insert into the mitochondria but its ability to induce cytochrome c release was minimal. Bid (α3-α8), the truncated form of Bid after caspase-8 cleavage (tBid) (Li et al., 1998; Luo et al., 1998), bound to the mitochondria equally well, and caused a much stronger cytochrome c release, indicating that the N-terminal α1 and α2 did not participate in membrane binding and was in fact inhibitory to the activity of Bid (Tan et al., 1999). The central hydrophobic helix 6 was required for Bid to interact with the mitochondria, thus α7 and α8, but not α6, could be dispensed. On the other hand, BH3 domain-containing α3 helix seemed to be required for the step subsequent to membrane insertion, i.e., cytochrome c release, but not for the binding itself, as indicated by the behavior of the BH3 domain mutant tBid (G94A) (Figure 1, B and C). None of these Bid recombinant proteins could be detected in the absence of mitochondria in the same fraction (our unpublished data). To further confirm that only helices 4-6 of Bid was required for its binding to the mitochondria, we constructed this mutant Bid (amino acid 105-166) for expression in E. coli. However, the protein was not stable and we were not able to conduct the in vitro experiment. Thus, we constructed Bid (α4-α6) in fusion with the green fluorescence protein (GFP) gene and transiently expressed the fusion molecule in HeLa cells. Confocal microscopy showed that GFP-Bid (α4-α6) was located on the mitochondria (Figure 1D), indicating that these three helices were structurally sufficient for Bid to localize to the mitochondria. However, because of the lack of the BH3 domain, GFP-Bid (α4-α6) would not be able to induce cytochrome c release (Lutter et al., 2000).
The above-mentioned studies suggested that the binding domain of Bid (α4-α6) and its BH3 domain (α3) could function separately. This notion was further supported by the observation that the antideath molecule Bcl-xL was able to suppress tBid-induced cytochrome c release (Figure 1E), but not the binding of tBid to the mitochondria (Figure 1F). Cytochrome c release by tBid is dependent on its BH3 domain, which is the binding target of Bcl-xL (Cheng et al., 2001).
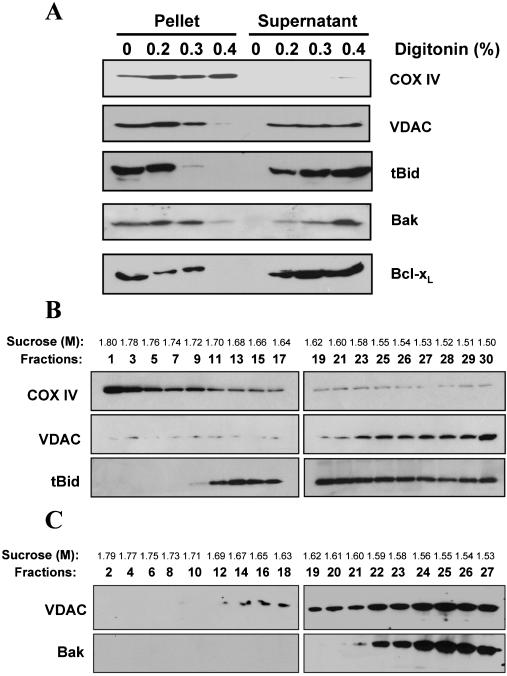
Localization of tBid on Both Contact and Noncontact Sites of the Mitochondria Outer Membranes Targeted tBid (α3-α8) remained mainly in the mitochondrial outer membranes, as indicated by limited digitonin solubilization. The digitonin-solubilized fraction was enriched with the outer membranes and the insoluble fraction was enriched with the inner membranes, as verified by the immunoblot analysis with antibodies against the outer membrane marker, voltage-dependent anion channel (VDAC), and the inner membrane marker, cytochrome c oxidase subunit IV (COX IV) (Figure 2A). tBid exhibited a digitonin solubilization pattern similar to that of VDAC, but not to that of COX IV, indicating its main presence in the outer membranes (Figure 2A). Similarly, exogenously targeted Bcl-xL and endogenous Bak were also found in the digitonin-soluble fraction containing the outer membranes (Figure 2A).
Figure 2.
Localization of tBid in the mitochondrial outer membranes. (A) Localization of tBid, Bak, and Bcl-xL in the mitochondrial outer membranes. Mitochondria were incubated with tBid (0.1 μg/ml) or Bcl-xL (13 μg/ml) for 1 h at 30°C before being treated with different amount of digitonin (wt/wt, 0-0.4%). Mitochondria were then separated by centrifugation. The supernatant, containing the outer membranes, and the pellet, containing the inner membranes, were analyzed by SDS-PAGE followed by immunoblot for the proteins indicated. (B) Distribution of tBid on different mitochondrial fractions. Mitochondria were incubated with tBid as in A, and then disrupted and fractionated on a sucrose linear gradient. Eluted fractions were analyzed by SDS-PAGE and immunoblot. (C) Mitochondria without tBid treatment were fractionated as in B and analyzed by immunoblot with antibodies against VDAC and Bak. For B and C, the sucrose concentration of each fraction is indicated.
A dichotomy in Bid location and Bid function was implicated in an immunoelectron tomography study finding Bid at the mitochondrial contact site (Lutter et al., 2001), as the formation of Bak or Bax oligomers at this site might lead to the leakage of matrix proteins, but not proteins in the inter-membrane space, due to the lack of intermembrane space. To address this issue, we separated membranes from tBid-treated mitochondria in a linear sucrose gradient, which segregated the inner membranes, the contact site and the outer membranes to fractions of heavier, intermediate, and lighter density, respectively. These fractions had been previously characterized extensively using classical outer membrane markers, such as monoamine oxidase and VDAC, and inner membrane markers, such as succinate dehydrogenase and COX IV (Ohlendieck et al., 1986; Adams et al., 1989; Pon et al., 1989; Ardail et al., 1990; Simbeni et al., 1991). Correspondingly, immunoblot analysis of the fractions showed distinguished distribution of COX IV and VDAC, corresponding to the inner and outer membranes, respectively (Figure 2B). tBid could be found in the lighter density fractions like VDAC, but it could be also found in the intermediate density fractions where the levels of both VDAC and Cox IV were low (Figure 2B). These fractions were previously characterized to be enriched with mitochondrial contact site (Ohlendieck et al., 1986; Adams et al., 1989; Pon et al., 1989; Simbeni et al., 1991). On the other hand, Bak was mainly found in the lighter density fractions of normal mitochondria (Figure 2C).
These results suggest that tBid may be present in the two subcompartments of the mitochondria, the contact site and the noncontact site. Because Bak was mainly found in the lighter density fractions corresponding to the noncontact sites, we also speculate that its interaction with Bid as documented in previous studies (Wei et al., 2000; Scorrano et al., 2002) may occur mainly at this subcompartment. On the other hand, those tBid localized at the intermediate fractions (contact sites) might be involved in a different type of function. To address this possibility, we performed the following experiments investigating the interaction of Bid with lipids on the mitochondria.
tBid Binds to Mitochondrial Cardiolipin at Mitochondrial Contact Sites
Previous studies had shown that Bid was able to bind to anionic phospholipids, particularly cardiolipin, in artificial systems, such as in liposomes or on solid surface (Schendel et al., 1999; Kudla et al., 2000; Lutter et al., 2000; Epand et al., 2002). Although Bid targeting to the mitochondria isolated from cardiolipin-deficient cells was reduced significantly (Lutter et al., 2000), direct evidence of Bid interaction with cardiolipin in intact mitochondria was still missing. Cardiolipin is enriched in the inner membranes and the outer membranes at the contact site (Ardail et al., 1990; Simbeni et al., 1991). We thus decided to investigate whether tBid could be associated with cardiolipin on intact mitochondria.
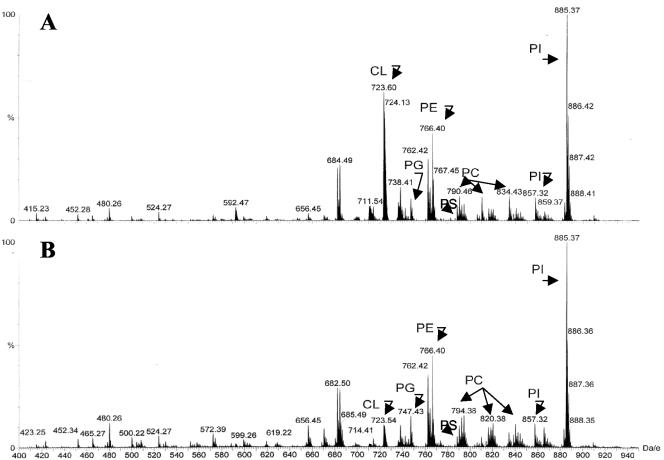
Mitochondria were first incubated with the recombinant proteins under the same conditions that led to their integration into the mitochondrial membranes (Figure 1B). Total mitochondrial lipids were then extracted and analyzed by electrospray ionization mass spectrometry. If a particular type of lipid was associated with the recombinant proteins, the protein would compromise the lipid's ability to ionize properly and the signals for the particular lipid species would be reduced. On the other hand, the Bid-binding lipids may partition into the hydrophilic phase with Bid and its signals would be also reduced in the hydrophobic phase, which was used for lipid extraction and analysis. Lipid scans were conducted in two mass/charge (Da/e) ranges, 400-950 and 1200-1800. The 400-950 Da/e range affords identification of the major phospholipid species, which run as singly charged entities. This range also detects any doubly charged species of cardiolipin, that run as (M-2H)- ions. Scanning in this range showed that the only mass ion that had significant changes upon Bid-treatment was the 723.6 Da/e species. Daughter ion analysis indicated that this was a doubly charged cardiolipin species. There were no significant signal changes in other major phospholipids including PG, PE, PC, PS, and PI (Figure 3, A and B).
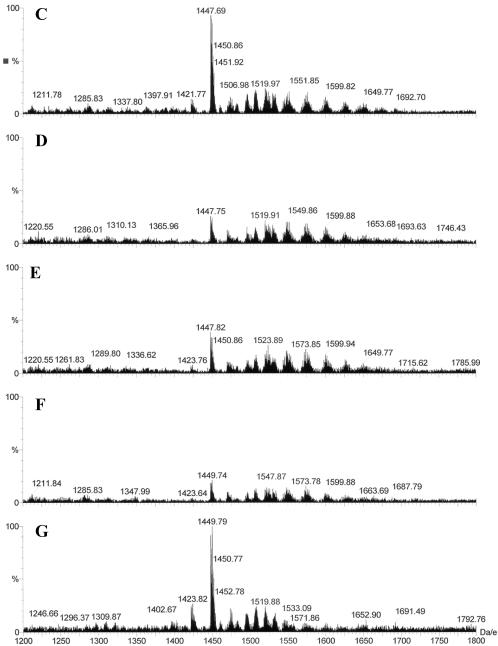
Figure 3.
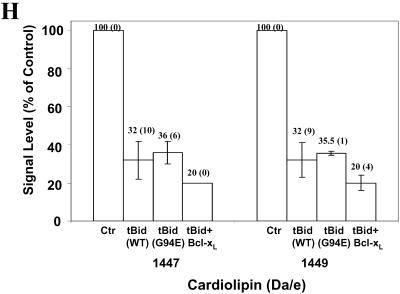
Binding of tBid to mitochondrial cardiolipin. (A-G) Mitochondria were incubated with buffer (A and C), tBid (0.1 μg/ml) (B and D), G94A mutant tBid (0.1 μg/ml) (E), tBid plus Bcl-xL (13 μg/ml) (F), or Bcl-xL (13 μg/ml) (G) for 30 min, and then purified for lipid extraction. Extracted lipids were analyzed by electrospray ionization mass spectrometry. Lipid scans in the 400-950 Da/e range (A and B) and in the 1200-1800 Da/e range (C-G) were conducted. Typical profiles are shown. Note the reduction in the signals of certain cardiolipin species in B (723.6 Da/e) and D-F (1447-1452 Da/e). CL, PC, PE, PS, PI, and PG are spelled out in text. (H) Relative signal levels of the two major affected cardiolipin species are shown based on the scan in the range of 1200-1800 Da/e. The cardiolipin signal level of the control sample (Ctr) was invariably set at 100% in each experiment, to which the signal levels of the treated groups were compared. The numbers shown on the top of each bar are mean (SD). Data are representative of two to five experiments performed.
Scanning in the 1200-1800 Da/e range would detect any cardiolipin species that carries only a single negative charge (M-H)- in the negative ion mode. Cardiolipin contains two phosphates, each capable of losing one hydrogen upon negative ionization. Electrospray ionization itself promotes multiple charging, hence both minus (-1) and (-2) charge states can exist. We found that signals of several species with mass/charge ratios around 1447-1452 were dramatically reduced in mitochondria treated with wild-type tBid (Figure 3, C and D). These species were identified as cardiolipin by daughter ion analysis. One of the species with a Da/e ratio of 1447 represented the singly charged version of the 723.6 mass ion detected in the 400-950 Da/e range. The decrease in signal intensity of these cardiolipin species were consistently observed, although the extent of the change varied among experiments. The average reduction of signals was >50% for the two species (Figure 3H).
These results seemed to be most compatible with the interpretation of Bid being tightly associated with these cardiolipin species, which compromised the signal detection by mass spectrometry. In support of this argument, the Bid-induced signal reduction of the selected cardiolipin species was not affected by the G94E BH3 mutation (Figure 3E), in agreement with the mitochondria binding results (Figure 1). Bcl-xL alone did not seem to have the same effect (Figure 3G), indicating some specificity of the Bid-cardiolipin inter-action. Intriguingly, treatment of mitochondria with both tBid and Bcl-xL could further reduce the cardiolipin signals (Figure 3F). This may suggest either that Bcl-xL could further attenuate the cardiolipin signal only in the presence of tBid, or that the tBid-Bcl-xL complex had a higher affinity to cardiolipin than tBid alone.
The interaction between tBid and cardiolipin on intact mitochondria could be further confirmed by the use of a cardiolipin-specific dye NAO (Petit et al., 1992). Pretreatment of mitochondria with NAO in low concentrations noticeably inhibited the membrane targeting of tBid, but not Bcl-xL, to the mitochondria (Figure 4A), perhaps by blocking the access of tBid to cardiolipin. These results were consistent with the mass spectrometry finding.
Figure 4.
Interaction of tBid with cardiolipin contributes to its targeting to the mitochondrial contact site. (A) NAO inhibits binding of tBid, but not Bcl-xL, to the mitochondria. Mitochondria (0.5 mg/ml) were pretreated with different amounts of NAO, washed, incubated with tBid (0.1 μg/ml) or Bcl-xL (27 μg/ml) for 1 h at 30°C, alkali-treated, and analyzed by immunoblot with an anti-Bid or anti-Bcl-xL antibody. (B and C) NAO preferentially blocks tBid targeting to the contact site. Mitochondria were pretreated with buffer (B) or with NAO (10 μM) (C) for 10 min, washed, and then incubated with tBid (0.1 μg/ml) for 30 min at 30°C before disrupted and fractionated. Eluted fractions were quantified for protein concentrations and aliquots containing similar amount of proteins were separated by 12% SDS-PAGE and immunoblotted with anti-COX VI, VDAC, or Bid antibodies. For B and C, the sucrose concentration of each fraction is indicated. (D) Densitometry analysis of the Bid blots from B (without NAO, filled triangle) and C (with NAO, open circle). The relative intensity of the Bid signal is plotted against the sucrose gradient. Data were normalized to the amount of proteins loaded. Densitometric values of those fractions with immunoblot results compromised by gel artifacts (fractions 17 and 19 from B and fractions 28 and 31 from C) were not included for a clearer presentation. Note the significant reduction of tBid binding at 1.65-1.67M of sucrose gradient (fractions 14-16 in C). Data are representative of four experiments performed.
Among the several cardiolipin species that displayed signal reduction, we determined the fatty acyl chain content of two major species (1447 and 1449 Da/e) by collision-induced dissociation tandem mass spectrometry of the selected mass ions. Daughter ion analysis determined that 1447 Da/e contained the following fatty acyl chain composition: C18:2, C18:2, C18:2, C18:2; and 1449 Da/e: C18:2, C18:2, C18:2, C18:1. These compositions are consistent with those reported for mouse liver cardiolipins, which express relatively high linoleoyl content (Hoch, 1992).
Cardiolipins containing 18:2 acyl compositions are more concentrated in the mitochondrial contact site (Ardail et al., 1990). To determine whether tBid association with the contact site (Figure 2B) was mediated by the interaction with cardiolipin, mitochondria sequentially treated with NAO and tBid were subfractionated. Immunoblot analysis of the various fractions indicated that NAO preferentially blocked tBid targeting to membranes of intermediate density, or the contact sites (Figure 4, B and C), as confirmed by densitometry analysis based on equal amount of protein loading (Figure 4D). That the G94A mutant of tBid could bind to cardiolipin (Figure 3C) was also consistent with the finding that this mutant could target to the contact site as shown by immunoelectron tomography (Lutter et al., 2001). Targeting of tBid to the noncontact site might be also mediated by Bid-cardiolipin interactions, because a small amount of cardiolipin can also be found in this part of the outer membrane (Ardail et al., 1990; Simbeni et al., 1991). In addition, although mass spectrometry analysis did not detect signal alterations in other lipid species in tBid-treated mitochondria, the participation of other noncardiolipin lipid species in Bid-mitochondria interaction, particularly at the noncontact site, remains possible (Esposti, 2002).
Interaction of Bid with Cardiolipin Contributes to Mitochondrial Cristae Reorganization
NAO pretreatment also led to a significant suppression of tBid-induced cytochrome c release (Figure 5A), indicating that there was a functional consequence of tBid-cardiolipin interaction. This seemed to be less likely caused by a reduced tBid binding to the mitochondria, because a large amount of tBid was still present in NAO-treated mitochondria, particularly at the noncontact site (Figure 4C). Thus, cardiolipin may not only serve as a docking site for Bid but also as a mediator of functional alterations.
Figure 5.
NAO inhibits Bid-induced cytochrome c release and mitochondrial cristae reorganization, but not Bak oligomerization. (A) NAO inhibits tBid-induced cytochrome c release without significantly affecting Bak oligomerization. Mitochondria were treated with tBid (0.1 μg/ml) alone or with NAO or Bcl-xL (30 μg/ml) as indicated. The supernatants were separated by centrifugation and analyzed by immunoblot for cytochrome c (top). The mitochondria pellets were then washed and incubated with 10 mM BMH (middle), or the solvent DMSO (bottom) for 60 min. The pellets were then solubilized in the SDS-PAGE loading buffer and separated by SDS-PAGE. Immunoblot was conducted with the anti-Bak antibody. The BMH cross-linked product that migrated faster than the Bak monomer (indicated as 1 × Bak) in lanes 1, 8, 9, and10 may represent the intramolecular cross-linked Bak monomer (indicated by the arrowhead) (Wei et al., 2000). BMH cross-linked Bak dimers are indicated as 2 × Bak. Treatment with the solvent control DMSO did not resulted in any cross-linked forms of Bak, although nonspecific bands were observed as indicated by the asterisks. The relative intensities (RI) of cytochrome c (top) and Bak dimers (middle) as determined by densitometry are indicated. (B) NAO inhibits mitochondrial cristae reorganization. Mitochondria were treated and processed as described in the method section. Representative electron micrographs are shown: control (a), tBid only (0.3 μg/ml) (b), G94A mutant tBid only (0.3 μg/ml) (c), tBid plus Bcl-xL (20 μg/ml) (d), NAO (10 μM) (e), and tBid plus NAO (f). Arrows indicated the mitochondria with typical cristae reorganization. Bar, 1 μm. (C). Mitochondria with reorganized cristae were quantified and expressed as the percentage of total mitochondria counted (400-500 total per group). (B and C) Data are representative of three experiments performed.
A possible outcome is an enhanced membrane permeabilization. In vitro analysis indicates that cardiolipin, as well as other anionic phospholipids, is important for tBid to permeabilize liposomes (Schendel et al., 1999; Kudla et al., 2000; Epand et al., 2002). However, liposomes reconstituted with Xenopus mitochondria lipids cannot be permeabilized by caspase-8 activated Bid alone, even if the right amount of cardiolipin was present (Kuwana et al., 2002). Thus, permeabilization of membranes may not be the primary consequence of Bid-cardiolipin interactions, particularly on intact mitochondria.
Alternatively, tBid can cause mitochondria cristae reorganization, which is accompanied with the mobilization of a major portion of cytochrome c (Scorrano et al., 2002). In line with this notion, we found that although cristae remodeling induced by tBid could not be blocked by the G94A BH3 mutation or by Bcl-xL, it could be significantly suppressed by NAO at a low concentration (10 μm), indicating the participation of cardiolipin in this process (Figure 5, B and C). In contrast, tBid-induced Bak oligomerization was only slightly affected by NAO at high concentrations, although it was completely suppressed by Bcl-xL (Figure 5A). Cytochrome c release and cristae reorganization were thus more susceptible to NAO treatment than Bak oligomerization, indicating the role of cristae reorganization in cytochrome c release, as indicated previously (Scorrano et al., 2002). Together, our results suggest that Bid at different mitochondrial subcompartments could exhibit distinct activities, culminating in a coordinated maximal release of cytochrome c.
DISCUSSION
Bid contains eight alpha helices, including two central hydrophobic helices, α6 and α7, but no transmembrane domain (Chou et al., 1999; McDonnell et al., 1999). Previous study showed that only a region from α4 to α6 is required for Bid binding, excluding the requirement of one central hydrophobic helix, α7 (Lutter et al., 2000). We confirmed this observation in the current study. In addition, we found that the BH3 domain-containing α3 helix was not required for membrane binding but was needed for the cytochrome c releasing activity. Thus, the minimal functional unit of Bid would include the α3-α6 helices. The present study also indicates that the binding domain of Bid (α4-α6) and its BH3 domain (α3) can function separately. The binding of Bid to the mitochondria through the binding domains can lead to distinctive functional consequences independently of the BH3 domain (see below).
The truncated Bid is generally considered to be localized in the outer membranes of the mitochondria because it is able to permeabilize the outer membranes without affecting the permeability of the inner membranes (Kluck et al., 1999; Van Loo et al., 2002). Both limited digitonin solubilization and sucrose linear gradient fractionation studies now provide the actual evidence to support this notion. Furthermore, the fractionation study indicates that Bid can be present in both mitochondrial contact site and noncontact site. In addition, it is possible that some tBid proteins may diffuse into the inner membranes because they could be found in fractions with an even higher density, which are more enriched with COX IV (Figures 2 and 4). However, the significance of this observation has yet to be determined. The finding of tBid in the contact site of the outer membranes by the gradient fractionation method is consistent with the study by using immunoelectron tomography (Lutter et al., 2001). However, in the latter study, localization of tBid in the noncontact site was apparently not readily observable, perhaps reflecting the lower sensitivity of this method.
Notably, Bak was found mainly in the noncontact site (Figures 2 and 4). Because Bak can be activated by tBid (Wei et al., 2000), we speculate that tBid-Bak interaction may occur mainly at the noncontact site. On the other hand, our data indicate that tBid at the contact site is involved in the interaction with cardiolipin. Bid-cardiolipin interactions had been reported previously only in artificial conditions (Lutter et al., 2000; Zha et al., 2000; Epand et al., 2002). Through the use of electrospray ionization mass spectrometry and a cardiolipin-specific dye, NAO (Petit et al., 1992), we now found that Bid could indeed interact with cardiolipin on functional mitochondria and furthermore we were able to define the location where such interactions occurred by mitochondria fractionation analysis.
The mass spectrometry had been successfully used to analyze lipid-protein interactions (Elviri et al., 2001; de Brouwer et al., 2002; Demmers et al., 2003), and it allowed us to identify at least two cardiolipin species, whose signals were affected significantly by tBid. We consider that this signal reduction is due to Bid-cardiolipin interactions, which cause an inadequate ionization of the affected cardiolipin species. This interpretation is most compatible with other findings reported here and in the literature (Lutter et al., 2000; Kuwana et al., 2002). Although we cannot completely exclude other possibilities, the signal reduction does not seem to be due to a general degradation, because only selected cardiolipin species were affected (Figure 3).
Cardiolipin is predominantly present in the inner membranes but can also be found in the outer membranes in smaller amounts (Daum, 1985; Ardail et al., 1990; Hovius et al., 1990; Simbeni et al., 1991; Hoch, 1992; Schlame et al., 2000). It is much more concentrated in the outer membranes at the contact site (Ardail et al., 1990; Simbeni et al., 1991) and has a higher 18:2 acyl content (Ardail et al., 1990). Indeed, two of the cardiolipin species identified in this study (1447 and 1449 Da/e) were found to be enriched in linoleic acid (C18:2). Thus, the ability of tBid to bind to cardiolipin may allow tBid to localize to the contact site. In support of this notion, pretreatment of the mitochondria with the cardiolipin-specific dye NAO preferentially inhibited the binding of tBid to the mitochondria at the contact site (Figure 4). Furthermore, that the G94A mutant of tBid could still bind to cardiolipin (Figure 3C) is also consistent with the finding that it retained its ability to bind to mitochondria (Figure 1B) and with the finding that this mutant could be found at the contact site (Lutter et al., 2001). It has to be pointed out that although mass spectrometry analysis did not detect signal alterations in other lipid species in tBid-treated mitochondria, we cannot completely rule out the participation of other noncardiolipin lipids in tBid-mitochondria interaction (Esposti et al., 2001). However, interaction of tBid with cardiolipin is unique in that it contributes to tBid-induced cytochrome c release via a defined mechanism.
NAO has a high affinity to cardiolipin (Ka = 2 × 106 M-1), and each cardiolipin molecule can bind to two NAO molecules (Petit et al., 1992). NAO can also bind to two other monoacidic phospholipids, PI and PS, at a much lower affinity (Ka = 7 × 104 M-1) in an equal molar ratio (Petit et al., 1992). Comparison of the binding kinetics between cardiolipin-containing liposomes and mitochondria indicates that the main target of NAO in mitochondria was cardiolipin (Petit et al., 1992). NAO is thus very specific to cardiolipin. When mitochondria were pre-treated with NAO, it reduced tBid binding to the mitochondria at the contact site and also reduced cytochrome c release. This effect is likely resulted from the blockage of tBid-cardiolipin interaction by NAO, as suggested by the available data (Figures 2, 3, 4) (Petit et al., 1992). Furthermore, NAO pretreatment only affected Bid-induced mitochondrial cristae reorganization, but not Bak oligomerization (Figure 5), the two major mechanisms responsible for tBid-induced cytochrome c release (Wei et al., 2000; Scorrano et al., 2002). Previous studies have suggested a model in which Bid-induced Bak oligomerization is essential for outer membrane permeabilization and Bid-induced cristae reorganization is required to maximally mobilize cytochrome c for release (Scorrano et al., 2002). However, it is not known how Bid induces cristae reorganization, which is apparently independent of Bak and the BH3 domain of Bid (Scorrano et al., 2002; Figure 5). Our finding now suggests that cristae reorganization may be resulted from tBid-cardiolipin interaction at the mitochondrial contact site. At the present time, it is not clear how such an interaction could lead to the alteration of cristae structures. We speculate that this may be related to the nonlamellar tendency of cardiolipin due to its intrinsic negative curvature (Epand et al., 2002), which can render cardiolipin in the hexagonal phase. This conversion from lamellar to nonlamellar structure may be facilitated by the interaction with tBid and may lead to alterations of the cristae membrane structure.
In summary, we found that targeting of tBid to the mitochondria through its α4-α6 domain led to its localization to the mitochondrial outer membranes. Our data indicate that tBid could be distributed into different mitochondrial sub-compartments with distinctive functional consequences. In particular, tBid-cardiolipin interactions on functional mitochondria seemed to occur mainly in the contact site, which contributes to mitochondrial cristae reorganization and cytochrome c release. These data also imply that Bid-Bak interaction may occur at the noncontact site. Thus, our studies support a model in which Bid can initiate distinctive molecular events at specific mitochondrial subcompartments to induce a maximal cytochrome c release.
Acknowledgments
X.-M.Y. is in part supported by National Institutes of Health grants CA R01-83817 and NS R01-045252. A.A.A. is in part supported by National Institutes of Health grant R01-92389. T.-H.K. is supported in part by a grant from Chosun University. W.-X.D. is a recipient of American Liver Foundation Fellowship.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0864. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0864.
References
- Adams, V., Bosch, W, Schlegel, J., Wallimann, T., and Brdiczka, D. (1989). Further characterization of contact sites from mitochondria of different tissues: topology of peripheral kinases. Biochim. Biophys. Acta 981, 213-225. [DOI] [PubMed] [Google Scholar]
- Ardail, D., Privat, J.P., Egret-Charlier, M., Levrat, C., Lerme, F., and Louisot, P. (1990). Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265, 18797-18802. [PubMed] [Google Scholar]
- Barry, M., Heibein, J.A., Pinkoski, M.J., Lee, S.F., Moyer, R.W., Green, D.R., and Bleackley, R.C. (2000). Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 20, 3781-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budihardjo, I., Oliver, H., Lutter, M., Luo, X., and Wang, X. (1999). Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15, 269-290. [DOI] [PubMed] [Google Scholar]
- Chen, M., He, H., Zhan, S., Krajewski, S., Reed, J.C., and Gottlieb, R.A. (2001). Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. J. Biol. Chem. 276, 30724-30728. [DOI] [PubMed] [Google Scholar]
- Cheng, E.H., Wei, M.C., Weiler, S., Flavell, R.A., Mak, T.W., Lindsten, T., and Korsmeyer, S.J. (2001). BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8, 705-711. [DOI] [PubMed] [Google Scholar]
- Chou, J., Li, H., Salvesen, G., Yuan, J., and Wagner, G. (1999). Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96, 615-624. [DOI] [PubMed] [Google Scholar]
- Daum, G. (1985). Lipids of mitochondria. Biochim. Biophys. Acta 822, 1-42. [DOI] [PubMed] [Google Scholar]
- de Brouwer, A.P., Versluis, C., Westerman, J., Roelofsen, B., Heck, A.J., and Wirtz, K.W. (2002). Determination of the stability of the noncovalent phospholipid transfer protein-lipid complex by electrospray time-of-flight mass spectrometry. Biochemistry 41, 8013-8018. [DOI] [PubMed] [Google Scholar]
- Demmers, J.A., van Dalen, A., de Kruijff, B., Heck, A.J., and Killian, J.A. (2003). Interaction of the K+ channel KcsA with membrane phospholipids as studied by ESI mass spectrometry. FEBS Lett. 541, 28-32. [DOI] [PubMed] [Google Scholar]
- Elviri, L., Zagnoni, I., Careri, M., Cavazzini, D., and Rossi, G.L. (2001). Non-covalent binding of endogenous ligands to recombinant cellular retinol-binding proteins studied by mass spectrometric techniques. Rapid Commun. Mass Spectrom. 15, 2186-2192. [DOI] [PubMed] [Google Scholar]
- Epand, R.F., Martinou, J.C., Fornallaz-Mulhauser, M., Hughes, D.W., and Epand, R.M. (2002). The apoptotic protein tBid promotes leakage by altering membrane curvature. J. Biol. Chem. 277, 32632-32639. [DOI] [PubMed] [Google Scholar]
- Eskes, R., Desagher, S., Antonsson, B., and Martinou, J.C. (2000). Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20, 929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposti, M.D. (2002). The roles of Bid. Apoptosis 7, 433-440. [DOI] [PubMed] [Google Scholar]
- Esposti, M.D., Erler, J.T., Hickman, J.A., and Dive, C. (2001). Bid, a widely expressed proapoptotic protein of the Bcl-2 family, displays lipid transfer activity. Mol. Cell. Biol. 21, 7268-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309-1312. [DOI] [PubMed] [Google Scholar]
- Greenawalt, J.W. (1974). The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 31, 310-323. [DOI] [PubMed] [Google Scholar]
- Gross, A., McDonnell, J.M., and Korsmeyer, S.J. (1999). BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13, 1899-1911. [DOI] [PubMed] [Google Scholar]
- Hoch, F.L. (1992). Cardiolipins and biomembrane function. Biochim. Biophys. Acta 1113, 71-133. [DOI] [PubMed] [Google Scholar]
- Hovius, R., Lambrechts, H., Nicolay, K., and de Kruijff, B. (1990). Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim. Biophys. Acta 1021, 217-226. [DOI] [PubMed] [Google Scholar]
- Huang, D.C., and Strasser, A. (2000). BH3-Only proteins-essential initiators of apoptotic cell death. Cell 103, 839-842. [DOI] [PubMed] [Google Scholar]
- Kelekar, A., and Thompson, C.B. (1998). Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 8, 324-330. [DOI] [PubMed] [Google Scholar]
- Kim, T.H., Zhao, Y., Barber, M.J., Kuharsky, D.K., and Yin, X.M. (2000). Bid-induced cytochrome c release is mediated by a pathway independent of mitochondrial permeability transition pore and Bax. J. Biol. Chem. 275, 39474-39481. [DOI] [PubMed] [Google Scholar]
- Kluck, R.M., et al. (1999). The pro-apoptotic proteins, Bid and Bax, cause a limited permeabilization of the mitochondrial outer membrane that is enhanced by cytosol. J. Cell Biol. 147, 809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, G., Montessuit, S., Eskes, R., Berrier, C., Martinou, J.-C., Ghazi, A., and Antonsson, B. (2000). The destabilization of lipid membranes induced by the C-terminal fragment of caspase 8-cleaved Bid is inhibited by the N-terminal fragment. J. Biol. Chem. 275, 22713-22718. [DOI] [PubMed] [Google Scholar]
- Kuwana, T., Mackey, M.R., Perkins, G., Ellisman, M.H., Latterich, M., Schneiter, R., Green, D.R., and Newmeyer, D.D. (2002). Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331-342. [DOI] [PubMed] [Google Scholar]
- Li, H., Zhu, H., Xu, C.J., and Yuan, J. (1998). Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491-501. [DOI] [PubMed] [Google Scholar]
- Li, S., Zhao, Y., He, X., Kim, T.-H., Kuharsky, D.K., Rabinowich, H., Chen, J., Du, C., and Yin, X.-M. (2002). Relief of extrinsic pathway inhibition by the Bid-dependent mitochondrial release of Smac in Fas-mediated hepatocyte apoptosis. J. Biol. Chem. 277, 26912-26920. [DOI] [PubMed] [Google Scholar]
- Luo, X., Budihardjo, I., Zou, H., Slaughter, C., and Wang, X. (1998). Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481-490. [DOI] [PubMed] [Google Scholar]
- Lutter, M., Fang, M., Luo, X., Nishijima, M., Xie, X., and Wang, X. (2000). Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2, 754-761. [DOI] [PubMed] [Google Scholar]
- Lutter, M., Perkins, G.A., and Wang, X. (2001). The pro-apoptotic Bcl-2 family member tBid localizes to mitochondrial contact sites. BMC Cell Biol. 2, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandic, A., Viktorsson, K., Strandberg, L., Heiden, T., Hansson, J., Linder, S., and Shoshan, M.C. (2002). Calpain-mediated Bid cleavage and calpain-independent Bak modulation: two separate pathways in cisplatin-induced apoptosis. Mol. Cell. Biol. 22, 3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsko, C.M., Hunter, O.C., Rabinowich, H., Lotze, M.T., and Amoscato, A.A. (2001). Mitochondrial lipid alterations during Fas-and radiation-induced apoptosis. Biochem. Biophys. Res. Commun. 287, 1112-1120. [DOI] [PubMed] [Google Scholar]
- McDonnell, J., Fushman, D., Milliman, C., Korsmeyer, S., and Cowburn, D. (1999). Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonist and antagonists. Cell 96, 625-634. [DOI] [PubMed] [Google Scholar]
- Ohlendieck, K., Riesinger, I., Adams, V., Krause, J., and Brdiczka, D. (1986). Enrichment and biochemical characterization of boundary membrane contact sites from rat-liver mitochondria. Biochim. Biophys. Acta 860, 672-689. [DOI] [PubMed] [Google Scholar]
- Petit, J., Maftah, A., Ratinaud, M., and Julien, R. (1992). 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 209, 267-273. [DOI] [PubMed] [Google Scholar]
- Pon, L., Moll, T., Vestweber, D., Marshallsay, B., and Schatz, G. (1989). Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J. Cell Biol. 109, 2603-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners, J.J., Jr., Caruso, J.A., Mathieu, P., Chelladurai, B., Yin, X.M., and Kessel, D. (2002). Release of cytochrome c and activation of pro-caspase-9 following lysosomal photodamage involves Bid cleavage. Cell Death Differ. 9, 934-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel, S.L., Azimov, R., Pawlowski, K., Godzik, A., Kagan, B.L., and Reed, J.C. (1999). Ion channel activity of the BH3 only Bcl-2 family member, BID. J. Biol. Chem. 274, 21932-21936. [DOI] [PubMed] [Google Scholar]
- Schlame, M., Rua, D., and Greenberg, M.L. (2000). The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257-288. [DOI] [PubMed] [Google Scholar]
- Scorrano, L., Ashiya, M., Buttle, K., Weiler, S., Oakes, S.A., Mannella, C.A., and Korsmeyer, S.J. (2002). A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell 2, 55-67. [DOI] [PubMed] [Google Scholar]
- Simbeni, R., Pon, L., Zinser, E., Paltauf, F., and Daum, G. (1991). Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 266, 10047-10049. [PubMed] [Google Scholar]
- Stoka, V., et al. (2001). Lysosomal protease pathways to apoptosis. Cleavage of Bid, not pro-caspases, is the most likely route. J. Biol. Chem. 276, 3149-3157. [DOI] [PubMed] [Google Scholar]
- Tan, K.O., Tan, K.M., and Yu, V.C. (1999). A novel BH3-like domain in BID is required for intramolecular interaction and autoinhibition of pro-apoptotic activity. J. Biol. Chem. 274, 23687-23690. [DOI] [PubMed] [Google Scholar]
- van Gurp, M., Festjens, N., van Loo, G., Saelens, X., and Vandenabeele, P. (2003). Mitochondrial intermembrane proteins in cell death. Biochem. Biophys. Res. Commun. 304, 487-497. [DOI] [PubMed] [Google Scholar]
- Van Loo, G., et al. (2002). A matrix-assisted laser desorption ionization post-source decay (MALDI-PSD) analysis of proteins released from isolated liver mitochondria treated with recombinant truncated Bid. Cell Death Differ. 9, 301-308. [DOI] [PubMed] [Google Scholar]
- Wang, G.Q., Wieckowski, E., Goldstein, L.A., Gastman, B.R., Rabinovitz, A., Gambotto, A., Li, S., Fang, B., Yin, X.M., and Rabinowich, H. (2001). Resistance to granzyme B-mediated cytochrome c release in Bak-deficient cells. J. Exp. Med. 194, 1325-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Yin, X.M., Chao, D.T., Milliman, C.L., and Korsmeyer, S.J. (1996). BID: a novel BH3 domain-only death agonist. Genes Dev. 10, 2859-2869. [DOI] [PubMed] [Google Scholar]
- Wei, M.C., Lindsten, T., Mootha, V.K., Weiler, S., Gross, A., Ashiya, M., Thompson, C.B., and Korsmeyer, S.J. (2000). tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14, 2060-2071. [PMC free article] [PubMed] [Google Scholar]
- Yin, X.M., Wang, K., Gross, A., Zhao, Y., Zinkel, S., Klocke, B., Roth, K.A., and Korsmeyer, S.J. (1999). Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400, 886-891. [DOI] [PubMed] [Google Scholar]
- Zha, J., Weiler, S., Oh, K.J., Wei, M.C., and Korsmeyer, S.J. (2000). Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290, 1761-1765. [DOI] [PubMed] [Google Scholar]
- Zhai, D., Miao, Q., Xin, X., and Yang, F. (2001). Leakage and aggregation of phospholipid vesicles induced by the BH3-only Bcl-2 family member, BID. Eur. J. Biochem. 268, 48-55. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Li, S., Childs, E.E., Kuharsky, D.K., and Yin, X.-M. (2001). Activation of pro-death Bcl-2 family proteins and mitochondria apoptosis pathway in tumor necrosis factor-alpha-induced liver injury. J. Biol. Chem. 276, 27432-27440. [DOI] [PubMed] [Google Scholar]