Abstract
Background:
Previous studies have shown interference with HbA1c measurement from the 4 most common heterozygous Hb variants (HbAS, HbAE, HbAC, and HbAD) with some assay methods. Here we examine analytical interference from 49 different less common variants with 7 different HbA1c methods using various method principles.
Methods:
Hb variants were screened using the Bio-Rad Variant or Variant II beta thal short program, confirmed by alkaline and acid electrophoresis, and identified by sequence analysis. The Trinity ultra2 boronate affinity high-performance liquid chromatography (HPLC) method and Roche Tinaquant immunoassay were used as primary and secondary comparative methods, respectively, since these methods are least likely to show interference from Hb variants. Other methods included were the Tosoh G7 and G8, Bio-Rad D-10 and Variant II Turbo, Diazyme Enzymatic, and Sebia Capillarys 2 Flex Piercing. To eliminate any inherent calibration bias, results for each method were adjusted using regression verses the ultra2 with nonvariant samples. Each method’s calibration-adjusted results were compared and judged to be acceptable if within the 99% prediction interval of the regression line for nonvariant samples.
Results:
Almost all variant samples were recognized as such by the ion-exchange HPLC methods by the presence of abnormal peaks or results outside the reportable range. For most variants, interference was seen with 1 or more of the ion-exchange methods. Following manufacturer instructions for interpretation of chromatograms usually, but not always, prevented reporting of inaccurate results.
Results:
Laboratories must be cautious about reporting results when the presence of a variant is suspected.
Keywords: accuracy, HbA1c, hemoglobin variants, interference
Glycated hemoglobin (GHB), reported as HbA1c, is a biochemical marker that is routinely used in the management of diabetes mellitus to monitor long term glycemic control and assess the risk of developing complications.1,2 GHB is hemoglobin that has been irreversibly modified by addition of glucose through a nonenzymatic process and provides a weighted average of blood glucose concentration over the erythrocyte lifespan. HbA1c is a specific GHB that is modified at the N-terminal valine of the Hb beta chains. Although some methods measure HbA1c specifically and others measure all glycated Hb species, all results are now standardized to report HbA1c. Treatment goals for HbA1c have been established, and more recently the test has been recommended for use in diagnosing diabetes.3-5 Therefore, accurate and precise measurement of HbA1c is extremely important. The presence of hemoglobin variants can adversely affect the accuracy of some HbA1c methods depending on the variant.6-8
The most common hemoglobin (Hb) variants worldwide in descending order of prevalence are HbS, HbE, HbC, and HbD. In the United States HbS is the most prevalent variant followed by HbC and HbE. There are many reports in the literature showing that the presence of these common Hb variants affects the accuracy of some HbA1c assays.9-11 Subjects who are heterozygous for any of these Hb variants are usually asymptomatic and have normal red cell survival.12-13 Thus, a physician may be unaware that his or her patient with diabetes has 1 of these variants in the heterozygous form. Results from some methods can alert the clinician that the Hb variant is present but may or may not give accurate HbA1c results. Other methods do not show the presence of the variant and may or may not provide accurate results. The worst-case scenario, of course, is the case where the variant is not indicated and the HbA1c result is inaccurate. The effect of each variant must be examined with each specific HbA1c method.
In addition to the 4 common Hb variants, a large number of less common or rare variants, including over 300 beta chain variants, have been described.14 It is difficult to study all possible variant Hbs with all methods but it is useful to know how the different methods perform with most variants and whether or not the presence of the variant can be detected. A Hb variant interferes with the HbA1c determination in several ways. If the amino acid substitution causes a change in the net charge of the Hb (as with Hbs S, C, D, and E), then it may cause interference with methods such as ion-exchange high-performance liquid chromatography (HPLC) or electrophoresis. If there is a substitution at a glycation site, this could alter the rate of glycation and affect certain methods. If the variant causes a reduced erythrocyte lifespan, the HbA1c (or total GHB) would be falsely lowered, regardless of the method used. Each variant Hb needs to be evaluated to determine the extent of the interference with each method.
In this study 49 rare variants were identified by sequencing of over 90 samples; for some variants there was only 1 sample available. Samples were analyzed by several HbA1c methods of different types (ion-exchange HPLC, boronate affinity HPLC, immunoassay, and enzymatic) and results were compared to 2 comparative methods for HbA1c, which are unlikely to have interference. Accuracy of HbA1c was assessed as well as the likelihood of an inaccurate result being reported if manufacturer instructions are followed.
Methods
Samples
This study was approved by the Institutional Review Board of the University of Utah Health Sciences Center, where the samples originated. Whole blood samples from individuals homozygous for HbA (n = 100) and heterozygous for rare variants (n ~ 90) were collected in ethylenediaminetetraacetic acid (EDTA)-containing tubes. Hb variants were screened using the Variant or Variant II beta thalessemia short program (Bio-Rad Laboratories, Hercules, CA), confirmed by alkaline and acid electrophoresis, and identified by sequence analysis. Samples with HbF>10% (based on Tosoh G8 HbF peak) and samples with ultra2 (Trinity Biotech) results <4% or >12% HbA1c were excluded. All study samples were divided into small aliquots (100 µL each) at ARUP Laboratories and stored at −70°C until they were shipped to participating sites (University of Missouri, Bio-Rad Laboratories) for HbA1c analysis.
Comparative Methods
In many previous studies that have evaluated 1 or more of the 4 common variants, a boronate affinity HPLC method was used as the Reference or Comparative method because it is unlikely to have interference (based on the fact that glycated and non-GHB are separated regardless of hemoglobin species) and there are data showing lack of interference based on comparisons to the IFCC Reference Method which measures the ratio of glycated to nonglycated beta-chain terminal hexapeptides.15,16 Since there are no data showing lack of interference with rare variants, a secondary comparative method was also chosen for the current study. The Tinaquant immunoassay reagents have been shown to produce accurate results in the presence of common heterozygous variants Hb S, C, E, and D.6 In the current study, a NGSP Secondary Reference Laboratory in Columbia, Missouri, performed analyses using the ultra2 (Trinity Biotech, Kansas City, MO). In-house whole blood calibrators were used for calibration to NGSP; in all other ways the ultra2 method was used following manufacturer’s instructions. The Tinaquant Gen.2 reagents were used on the Cobas c501 (Roche Diagnostics, Indianapolis, IN) at ARUP Laboratories in Salt Lake City, Utah, following the manufacturer’s instructions.
Test Methods
The methods evaluated included 4 ion-exchange HPLC methods (G7 and G8 HPLC, Tosoh Biosciences, San Francisco, CA; D-10 and Variant II Turbo 2.0 HPLC, Bio-Rad Laboratories), 1 enzymatic method (Direct Enzymatic HbA1c on the Roche Cobas c501, Diazyme Laboratories, Poway, CA; Roche Diagnostics, Indianapolis, IN), and 1 capillary-electrophoresis method (Capillarys 2 Flex Piercing capillary electrophoresis, Sebia, Lisse, France). All methods were operated and results interpreted following manufacturer’s instructions with the exception of using whole blood (NGSP) calibrators for the G7 and G8. Not all samples were analyzed with the Capillarys 2 due to late availability of this method for the study. For HPLC and CE methods, manufacturer instructions include the manufacturer’s criteria for an acceptable chromatogram, or in the case of CE, an acceptable electropherogram. For example, for both Bio-Rad and Tosoh methods, there are total area limits that are considered acceptable. Depending on the particular HPLC method there may also be acceptable limits for HbF or labile HbA1c peaks as well as limits for other peak areas. For some methods specific Hb variants may be flagged or indicated as a variant peak and some of these peaks are considered acceptable.
Data Analysis
Our objective was to determine whether methods would report inaccurate results. To that end, we classified analyses with respect to whether or not they were “reportable” and/or “accurate.” To eliminate any inherent calibration bias, variant results for each method were adjusted using regression of each method’s results for HbAA samples against ultra2 results (adjusted %HbA1c). The accuracy limits for the variant samples were defined by the 99% prediction interval of the regression line for adjusted AA sample results.
However, if results from the secondary comparative method (Tinaquant) were outside the 99% prediction interval compared to the primary comparative method (ultra2), and this discordance could not be explained by the position of the variant substitution (eg, a variant with substitution within 4 amino acids from the N-terminus of the beta chain that would be predicted to interfere with the Tinaquant assay), the sample was excluded from further data analysis.
Variant results were classified as “reportable” if the result would have been reported following the manufacturers instructions. For all the test methods, except for the enzymatic method, chromatograms (for HPLC) or electropherograms (for CE) often allowed for determination of the presence of a Hb variant. In some cases, the variant might also be tentatively identified. Depending on the specificity of the manufacturer’s instructions, HbA1c results may or may not be considered “reportable.” For the Direct Enzymatic HbA1c and the Tinaquant HbA1c all results would be considered reportable if within assay range limits.
Results
For most of the rare variants included in the present study, the Tinaquant immunoassay results matched those from the ultra2 boronate affinity, that is, calibration-adjusted Tinaquant results were within the 99% prediction interval of the regression line for nonvariant samples. There were only 8 (8/49) variants where results were outside this acceptance range and in 3 of these cases (Hb Deer Lodge, Hb Okayama, and Hb Raleigh) the substitution was near the N-terminal of the beta chain and would therefore be predicted to interfere with the Tinaquant. Samples from these 3 variants were included in the evaluation of other methods and the ultra2 result was assumed to be accurate. Five other variants where the substitution was not close to the N-terminus of the beta chain and where there was discordance between Tinaquant and ultra2 results (Hb Buffalo, Hb G Coushatta, Hb Volga, Hb Zurich and a variant that has not yet been described) were excluded from further analysis since more data are needed to explain these discrepancies before the ultra2 result is accepted as being accurate.
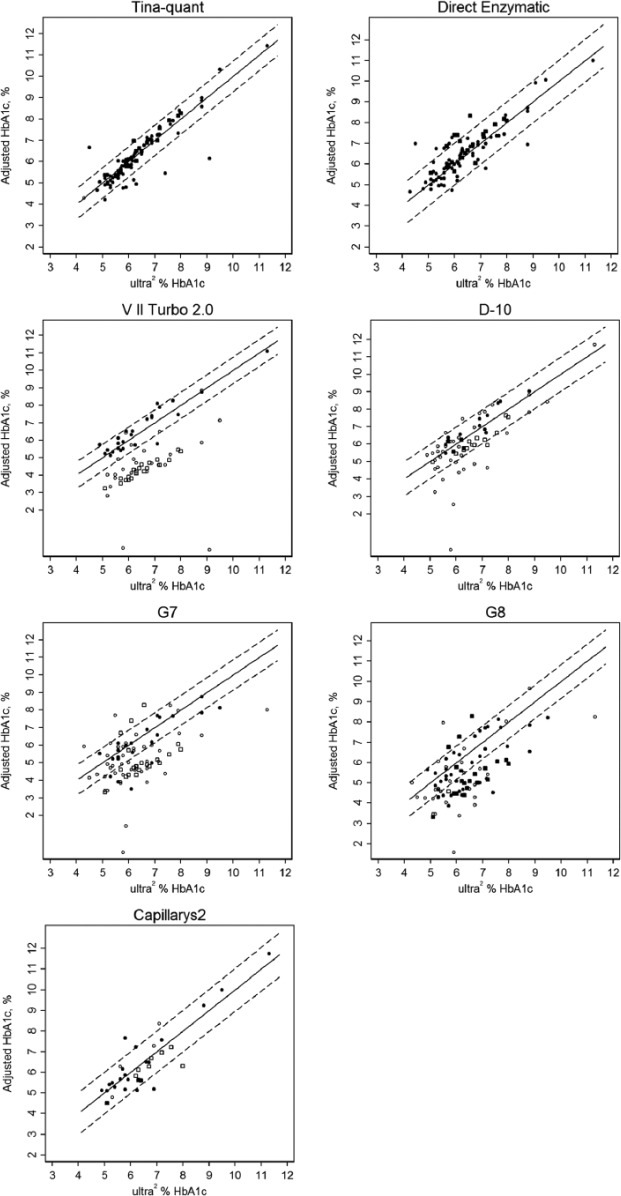
The calibration-adjusted variant results for each method compared to ultra2 are shown in Figure 1 (A-F). The regression line and the 99% prediction interval (based on AA samples) are shown; all the results within this interval are considered accurate. For the ion-exchange methods, open symbols are those results that would not be reported based on manufacturer recommendations. Some results (indicated in the figure legend) that do not appear on the graph would likely not be reported due to a very high result (eg, >30% HbA1c) or no reported result. The results of most concern are the closed symbols that are outside the 99% prediction interval since these represent inaccurate results that would likely be reported.
Figure 1.
Calibration-adjusted HbA1c results for rare Hb variant samples versus ultra2: Solid lines mark the equivalence (y = x); dashed lines indicate the 99% prediction intervals based on comparisons of HbA1c values of AA samples. Open circles are results that would not be reported; solid circles are results that would be reported; open and closed squares are for Hb J Baltimore specifically. HbA1c results for several variants were >12% and do not appear on the graphs. Hb Camperdown (n = 1), Hb Hope (n = 10), Hb Hopkins II (n = 1), Hb I (n = 3), Hb Raleigh (n = 1), and Hb Sherwood Forest (n = 1) were >15% on the VII Turbo 2.0. Results for Hb Camperdown (n = 1), Hb Hope (n = 10), and Hb Raleigh (n = 1) were >30% on the D-10. Hb Okayama (n = 1) and Hb Raleigh (n = 2) were >45% on the G7 and G8. Note: solid symbols outside the 99% prediction interval represent inaccurate results that would likely be reported.
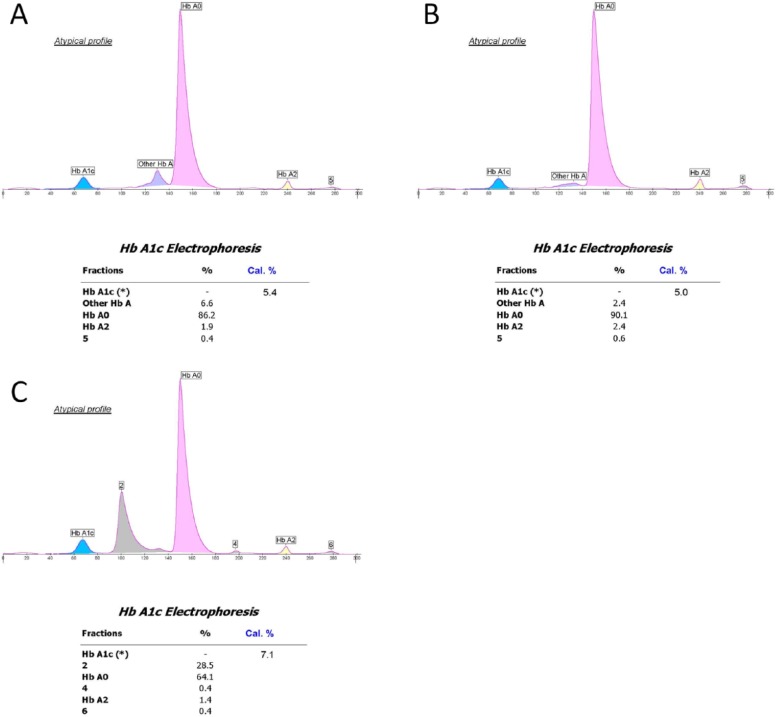
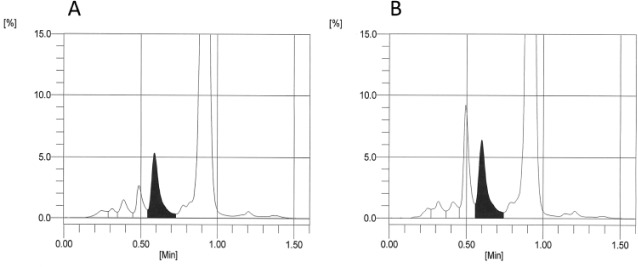
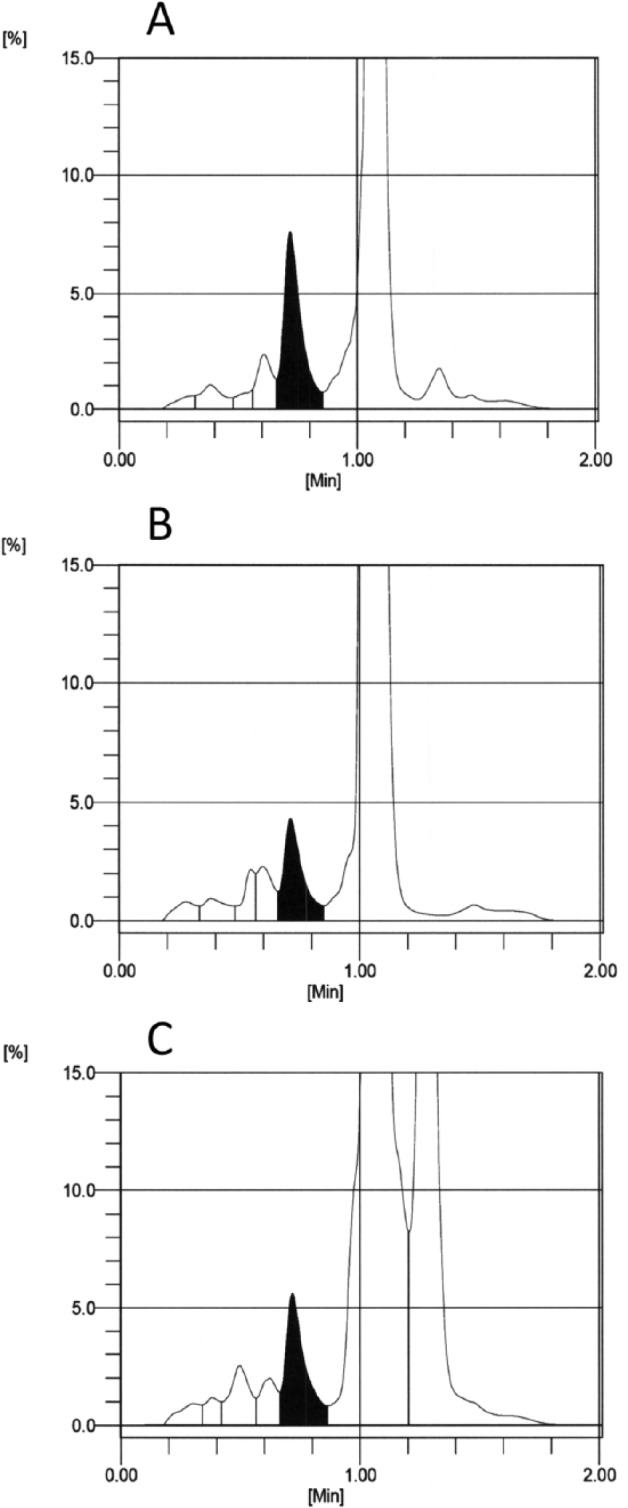
Table 1 lists the 44 Hb variants that were included for evaluation by the test methods. The 5 variants with discordant results between the ultra2 and Tinaquant that could not be explained by the position of the variant substitution are not included. For each method and variant, the number of inaccurate results (outside 99% prediction interval) that would have been reported and the total number of samples are indicated. For example, the notation for Hb J-Baltimore for the G8 is 17/19 indicating that there were 19 samples with Hb J Baltimore analyzed by the G8 and 17 of these gave results that were inaccurate (were outside the 99% prediction interval) but would have been reported. This occurred because there was no indication that a variant was present in these 17 sample chromatograms. An example is shown in Figure 2; the G8 chromatogram for the sample with Hb J Baltimore (Figure 2B) is not easily distinguished from a normal HbAA chromatogram (Figure 2A). Similarly, a sample with Hb J-Broussais (not shown) also appears to be normal with no indication of the variant. However, in the case of Hb J-Broussais, 2 samples were tested and the results for both were accurate (0/2), thus the notation 0/2 in Table 1. In all, 29 inaccurate results from 9 variants would likely have been reported for the G8. For the Variant II Turbo 2.0 and G7 very few inaccurate results (3 for G7, 2 for Turbo 2.0) would have been reported if the manufacturer’s instructions were followed. For the D-10, no inaccurate results would have been reported. Another example (Figure 3) shows G7 chromatograms where results from Hb I (A) and Hb Khartoum (B) would have been reported following manufacturer instructions even though, in both cases, the results were inaccurate. The Hb I chromatogram appears normal with no indication of a variant and Hb Khartoum appears to elute in the HbD window which is reportable according to manufacturer instructions. Of the 18 rare variants analyzed by the Capillarys 2 Flex, inaccurate results would have been reported for only 2. These electropherograms are shown in Figure 4; the electropherogram for Hb Silver Springs (Figure 4B) is fairly typical of that for HbAA in the present study (Figure 4A). The electropherogram from a sample containing Hb J-Broussais (Figure 4C) contains an abnormal Hb peak but would be considered reportable. It should be noted that most of the electropherograms from samples with normal Hb (AA) in this study included a small degradation peak and/or a slight slope in the baseline between the “other HbA” and Hb A0 peaks similar to those shown in Figure 4; these are not expected to affect the HbA1c result. For the enzymatic method, variant Hbs cannot be detected (only numerical results are provided), and for Hb J Baltimore 4 out of 19 results were inaccurate and would have been reported.
Table 1.
Proportion of Inaccurate Results That Would Be Reported for Each Rare Variant.
| Variant Name | Tinaquant | Direct Enzymatic | G7 | G8 | V II Turbo 2.0 | D-10 | Cap-2 Flex Piercing |
|---|---|---|---|---|---|---|---|
| Hb Andrew Minnesota | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Austin | 0/2 | 1/2 | 0/2 | 1/2 | 0/2 | 0/2 | — |
| Hb Camden | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Camperdown | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Charolles/Manitoba | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Cowtown | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Deer Lodgea | 1/1 | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | — |
| Hb D–Iran | 0/1 | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | 0/1 |
| Hb Ethiopia | 0/1 | 0/1 | 0/1 | 0/2 | 0/1 | 0/1 | — |
| Hb Fannin-Lubbock I | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb GCopenhagen | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb GNorfolk | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb GPhiladelphia | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Hb Hope | 1/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | — |
| Hb Hopkins–II | 0/3 | 1/3 | 0/3 | 0/2 | 0/3 | 0/2 | 0/2 |
| Hb Hoshida | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb I | 0/3 | 0/3 | 1/3 | 2/3 | 0/3 | 0/3 | 0/2 |
| Hb Inkster | 0/2 | 1/2 | 0/2 | 0/2 | 0/2 | 0/2 | — |
| Hb J-Baltimore | 1/19 | 4/19 | 0/19 | 17/19 | 0/19 | 0/16 | 0/10 |
| Hb J-Bangkok | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Jackson | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 |
| Hb J–Broussais | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 2/2 |
| Hb J–Toronto | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Khartoum | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb Lepore | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Malmo | 0/1 | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | — |
| Hb Manitoba–I | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb N Baltimore | 0/3 | 0/3 | 0/3 | 2/3 | 0/2 | 0/2 | — |
| Hb NewVariant–1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb NewVariant–3 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
| Hb NewVariant–4 | 0/1 | 0/1 | / | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb O Indonesia | 0/1 | 1/1 | 0/1 | 0/1 | 0/1 | — | — |
| Hb Okayamaa | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | — | — |
| Hb Osu Christianborg | 0/2 | 0/2 | 0/2 | 2/2 | 0/2 | 0/2 | 0/2 |
| Hb Park Ridge | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Hb Queens | 0/1 | 1/1 | 0/1 | 0/1 | — | — | — |
| Hb Raleigha | 3/3 | 0/3 | 0/3 | 0/3 | 0/1 | 0/1 | 0/1 |
| Hb Rambam | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | 0/1 | — |
| Hb Riyadh | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | — |
| Hb Russ | 0/2 | 0/2 | 0/2 | 0/2 | 1/2 | 0/2 | 0/1 |
| Hb Setif | 0/2 | 1/2 | 0/2 | 0/1 | 0/2 | — | — |
| Hb Sherwood Forest | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — | — |
| Hb Silver Springs | 0/2 | 0/2 | 0/2 | 2/2 | 0/2 | 0/2 | 2/2 |
| Hb St Luke’s | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | — |
These variants have substitutions near the N-terminal of the β chain and would interfere with Tinaquant.
Figure 2.
G8 chromatograms from samples with normal (AA) hemoglobin (A) and with Hb J-Baltimore (B). The presence of the variant is not detected.
Figure 3.
G7 chromatograms from samples with normal (AA) hemoglobin (A) and with Hb I (B) and Hb Khartoum (C). In (B) the variant is not detected. In (C) the variant appears in the same area window as HbD and would be reported.
Figure 4.
Capillarys 2 electropherograms from samples with normal (AA) hemoglobin (A) and with Hb Silver Spring (B) and Hb J-Broussais (C). Note that many of the samples that were frozen prior to analysis, including samples with normal hemoglobin show the “atypical profile” flag due in part to identification of a degradation peak after HbA2 and/or slightly raised baseline between the “other HbA” and “HbA0.”
Conclusions
Hb variants can interfere with HbA1c methods for a variety of reasons.7 Depending on the patient population for a particular laboratory, this can be a concern. For most (41/49) of the rare variants in the present study, the Tinaquant immunoassay results matched ultra2 boronate affinity results (within 99% prediction interval of the regression line for calibration-adjusted nonvariant results). Three of the discordant variants had substitutions close to the β N-terminus, and these would be expected to give inaccurate results from the Tinaquant method. For 1 of these, Hb Raleigh, although the ultra2 may be analytically accurate, the result is artificially lowered and would not be useful for estimating glycemic control due to the substitution of alanine for valine at the N-terminus of the beta chain (the N-terminal valine is the predominant Hb glycation site). For at least 1 of the samples from 8 of the 44 variants included, Diazyme results were inaccurate; this method does not detect the presence of variants and all results would be reported. Following manufacturer instructions and training for interpretation of ion-exchange HPLC chromatograms and CE electropherograms, careful examination usually, but not always, prevented reporting of inaccurate results. Inaccurate G8 results for several variants (10/44) would have been reported either because the variant could not be detected or because the variant peak appeared like a common variant that would be considered reportable.
To ensure that accurate HbA1c results are obtained for all patients, it is important to know if a patient has a hemoglobin variant and how that variant can affect his or her HbA1c results. Laboratories must be cautious about reporting results when the presence of a variant is suspected. Manufacturers may need to reexamine their criteria for accepting results, update their software to flag potential variants, and/or improve the analytical characteristics of their methods in an effort to reduce the likelihood of incorrect HbA1c results being reported. As with any laboratory test, any result that seems discordant with clinical impression should be investigated further.
Acknowledgments
We would like to thank Julie Myers, Steve Tanaka, and Bio-Rad Laboratories for performing the analyses on the VII Turbo and D-10. We would also like to thank William Roberts, who initiated this project.
Footnotes
Abbreviations: EDTA, ethylenediaminetetraacetic acid; GHB, glycated hemoglobin; Hb, hemoglobin; HPLC, high-performance liquid chromatography; NGSP, National Glycohemoglobin Standardization Program.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was funded by a grant from Roche Diagnostics.
References
- 1. Diabetes Control and Complications Trial Research Group. The effect of intensive treatment of diabetes on the development and progression of long-term term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993:329:977-986. [DOI] [PubMed] [Google Scholar]
- 2. UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998;352:837-853. [PubMed] [Google Scholar]
- 3. American Diabetes Association. Standards of medical care in diabetes. Diabetes Care. 2014;37(suppl 1):S5-S15. [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization. Use of Glycated Haemoglobin (HbA1c) in the Diagnosis of Diabetes Mellitus: Abbreviated Report of a WHO Consultation. 2011. [PubMed] [Google Scholar]
- 5. Diabetes Federation, Clinical Guidelines Task Force. Global Guideline for Type 2 Diabetes. 2012. Available at: http://www.idf.org/sites/default/files/IDF-Guideline-for-Type-2-Diabetes.pdf. Accessed August 16, 2014.
- 6. Little RR, Roberts WL. A review of variant hemoglobins interfering with hemoglobin A1c measurement. J Diabetes Sci Technol. 2009;3:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bry L, Chen PC, Sacks DB. Effects of hemoglobin variants and chemically modified derivatives on assays for glycohemoglobin. Clin Chem. 2001;47:153-163. [PubMed] [Google Scholar]
- 8. Weykamp CW, Penders TJ, Muskiet FA, van der Slik W. Influence of hemoglobin variants and derivatives on glycohemoglobin determination, as investigated by 102 laboratories using 16 methods. Clin Chem. 1993;39:1717-1723. [PubMed] [Google Scholar]
- 9. Little RR, Rohlfing CR, Hanson S, et al. Effects of hemoglobin E and D traits on glycated hemoglobin (HbA1c) measurements by twenty-three methods. Clin Chem. 2008;54:1277-1282. [DOI] [PubMed] [Google Scholar]
- 10. Mongia SK, Little RR, Rohlfing CL, et al. Effects of hemoglobin C and S traits on the results of 14 commercial glycated hemoglobin assays. Am J Clin Pathol. 2008;130:136-140. [DOI] [PubMed] [Google Scholar]
- 11. Lin CN, Emery T, Little RR, et al. Effects of hemoglobin C, D, E, and S traits on measurements of HbA1c by six methods. Clin Chim Acta. 2012;413:819-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bunn FH, Forget BG. Hemoglobin: Molecular, Genetic and Clinical Aspects. Philadelphia, PA: Saunders; 1986 [Google Scholar]
- 13. Bain BJ. Haemoglobinopathy Diagnosis. Malden, MA: Blackwell; 2006. [Google Scholar]
- 14. Huisman THJ, Carver MFH, Efremov GD. A Syllabus of Human Hemoglobin Variants. Augusta, GA: Sickle Cell Anemia Foundation; 1996. Available at: http://globin.bx.psu.edu/html/huisman/variants/. [Google Scholar]
- 15. Little RR, Vesper H, Rohlfing CL, Ospina M, Sekineh SP, Roberts WL. Validation by a mass spectrometric reference method of use of boronate affinity chromatography to measure glycohemoglobin in the presence of hemoglobin S and C traits. Clin Chem. 2005;51:264-265. [DOI] [PubMed] [Google Scholar]
- 16. Connolly S, Hanson S, Higgins T, Rohlfing C, Little R. Assessment of the validity of Trinity Biotech ultra2 hemoglobin A1c results in the presence of HbE or HbD Punjab trait. Clin Chem. 2013;59(suppl):A161. [DOI] [PubMed] [Google Scholar]