Abstract
CHO1 is a kinesin-like motor protein essential for cytokinesis in mammalian cells. To analyze how CHO1 functions, we established RNAi and genetic rescue assays. CHO1-depleted cells reached a late stage of cytokinesis but fused back to form binucleate cells because of the absence of the midbody matrix in the middle of the intercellular bridge. Expression of exogenous CHO1 restored the formation of the midbody matrix and rescued cytokinesis in siRNA-treated cells. By analyzing phenotypes rescued with different constructs, it was shown that both motor and stalk domains function in midbody formation, whereas the tail is essential for completion of cytokinesis after the midbody matrix has formed. During the terminal stage of cytokinesis, different subregions of the tail play distinctive roles in stabilizing the midbody matrix and maintaining an association between the midbody and cell cortex. These results demonstrate that CHO1 consists of functionally differentiated subregions that act in concert to ensure complete cell separation.
INTRODUCTION
Cytokinesis is the last step of cell division, during which segregated chromosomes are permanently separated into daughter cells. For faithful transmission of the genetic material, initiation of cytokinesis must be tightly coupled with the progression of chromosome movement. Experimental data suggest that the mitotic spindle, which drives the separation of chromosomes, is also responsible for determining where and when cytokinesis will occur (Rappaport and Rappaport, 1974; Rappaport, 1986). Two structures of the mitotic spindle, spindle asters (Rappaport, 1961) and the central spindle, have been implicated in induction of cleavage furrows. Although the central spindle appears to be essential in triggering cytokinesis in Drosophila (Adams et al., 1998; Giansanti et al., 1998; Somma et al., 2002; Somers and Saint, 2003) and mammalian cells (Cao and Wang, 1996), the same structure is largely dispensable for the furrow initiation in other systems, such as marine invertebrate eggs (Rappaport, 1961; Rappaport and Rappaport, 1983) and Caenorhabditis elegans (Raich et al., 1998; Jantsch-Plunger et al., 2000; Severson et al., 2000). Nonetheless, the presence of a central spindle has been shown to be essential for completion of cytokinesis in most systems (Wheatley and Wang, 1996; Raich et al., 1998; Jantsch-Plunger et al., 2000; Severson et al., 2000; Chen et al., 2002, Alsop and Zhang, 2003).
Central spindles consist of highly bundled microtubule arrays formed between separating chromosomes. During anaphase, an electron-dense material appears and surrounds microtubule bundles at the midzonal region of the spindle where antiparallel microtubules overlap (Buck and Tisdale, 1962; McIntosh and Landis, 1971). With the progression of cytokinesis, areas of the dense material coalesce into a single midbody matrix, which subsequently positions at a midpoint of the intercellular bridge. The midbody matrix has been shown to be a stable structure (Mullins and McIntosh, 1982) important in gluing microtubules together and stabilizing them at the midbody (Salmon et al., 1976). As disorganization of the midbody matrix closely correlates with regression of cleavage furrows (King and Akai, 1971; Mullins and Biesele, 1977; Matuliene and Kuriyama, 2002), the midbody matrix has been implicated in completion of cytokinesis. However, the molecular structure and function of the midbody matrix have not yet been elucidated.
Using monoclonal antimitotic spindle antibodies, we identified a matrix component of the midbody in Chinese hamster ovary cells (Sellitto and Kuriyama, 1988). The protein, named CHO1, turned out to be a plus-end-directed motor protein present in a wide range of organisms from humans (MKLP1: Nislow et al., 1992) and Drosophila (PAVKLP: Adams et al., 1998) to C. elegans (ZEN-4: Raich et al., 1998). All those molecules are composed of well-conserved three subdomains: the mechanochemical motor domain at the N terminus, the central α-helical coiled-coil stalk, and the C-terminal globular tail. However, CHO1 represents a unique isotype, in which an actin-binding sequence encoded by exon 18 is included in the middle of the tail domain (Kuriyama et al., 2002). Members of the MKLP1 subfamily (Miki et al., 2001) have been shown to function in cytokinesis in different species (Adams et al., 1998; Powers et al., 1998; Raich et al., 1998; Chen et al., 2002; Matuliene and Kuriyama, 2002). In humans and C. elegans, the motor proteins interact with a GAP for Rho family GTPases, which facilitates microtubule bundling and formation of the spindle midzone and midbody (Mishima et al., 2002). The homologous complex in Drosophila further interacts with Pebble RhoGEF, suggesting that the motor protein exists as a larger multi-protein complex (Somers and Saint, 2003).
In our previous studies, we analyzed the CHO1 function in mammalian cells by expressing ATP-binding mutant proteins (Matuliene and Kuriyama, 2002). Mutated CHO1 failed to accumulate at the midzonal region, resulting in disorganization of the midbody matrix and inhibition of cytokinesis. To extend our analysis of CHO1, we developed RNAi and genetic rescue assays, which allowed us to dissect the function of CHO1 subdomains. Here we report that CHO1 consists of functionally distinctive multiple subregions that act in concert to ensure complete separation of two daughter cells at the end of cytokinesis.
MATERIALS AND METHODS
Preparation of cDNA Constructs
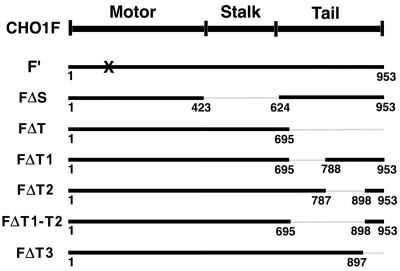
The full coding sequence of CHO1 in pBluescript was obtained by immunoscreening of a CHO expression library (Kuriyama et al., 1994). Figure 1 summarizes the constructs used in this study: 1) Full-coding sequence CHO1 (CHO1F), 2) Full-coding sequence CHO1 with mutation in the ATP-binding site (CHO1F′). 3-8) Full-coding sequences of CHO1 with deleted: 3) stalk domain (CHO1FΔS), 4) most of the tail domain (CHO1FΔT), 5) the tail subregion (T1), which interacts with actin filaments and is encoded by exon 18 (CHO1FΔT1), 6) the tail subregion (T2) encoded mainly by exons 19 and 20 (CHO1FΔT2), 7) T1 and T2 subregions (CHO1FΔT1-T2), and 8) nuclear localization signal-containing subregion, T3 (CHO1FΔT3). In all of those constructs, four silent nucleotide changes were introduced in the region complementary to siRNA. The region AGTAATACA, corresponding to nucleotide positions 151-159 of CHO1, was mutated to TCTAACACG, leaving the amino acid sequence unchanged. Mutations were introduced using Quick Change Site-Directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Three amino acids in the ATP-binding motif of CHO1 were mutated from GKT to AAA (amino acid positions 117-119), as previously described (Matuliene and Kuriyama, 2002). CHO1 constructs were subcloned into a multicloning site of the eukaryotic expression vector pCMV-HA (Matuliene et al., 1999) and/or pEGFP-C1 (Clontech, Palo Alto, CA), fusing the HA epitope tag and/or GFP to the N terminus of CHO1 constructs.
Figure 1.
A map of CHO1 constructs used for RNAi-rescue experiments. The number of amino acids, encoded by each construct, is identified beneath the bold lines. All constructs have four nucleotide changes in the region complementary to siRNA used for RNAi assays and are tagged with the HA epitope at the N terminus. X indicates the mutation in the ATP-binding site in the motor domain.
Mutations in the nuclear localization signals (NLS) of CHO1, summarized in Figure 9A, were introduced as above, and the constructs were subcloned into pCMV-HA. To fuse a foreign NLS to a CHO1F(NLS1,2)′ construct, HA-tagged CHO1F(NLS1,2)′ was subcloned into the multicloning site of pCMV/myc/nuc vector (Invitrogen, Carlsbad, CA) fusing three SV40 virus-derived NLS sequences to the C terminus of the construct. The NLS-containing vector was obtained from Dr. N. Kikyo (University of Minnesota, Minneapolis, MN).
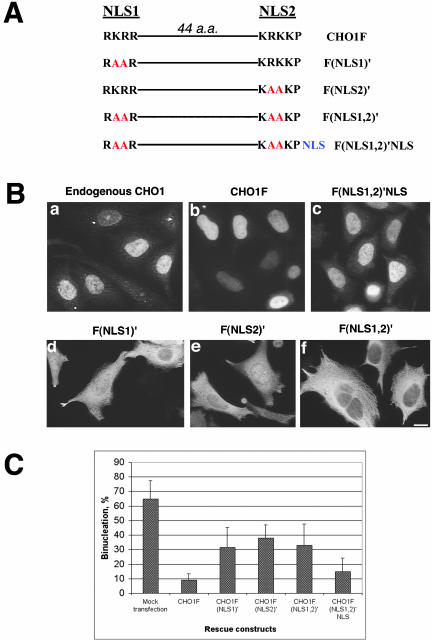
Figure 9.
Nuclear localization is important for the function of CHO1 in cytokinesis. (A) Schematic representation of NLS-containing region in wild-type CHO1 and its NLS mutants. Amino acid changes within the sequence of the first NLS (NLS1), the second NLS (NLS2), and both NLS (NLS1,2) are shown in red. NLS shown in blue represents three SV40-derived nuclear localization signals fused to the C terminus of CHO1F(NLS1,2)′ construct to make CHO1F(NLS1,2)′NLS construct. (B) Endogenous CHO1 and exogenous CHO1F localize to the nucleus in interphase cells (a and b). Mutation in NLS1 and NLS2 significantly reduce the localization of CHO1 to the nucleus (d and e). Although nuclear localization is still detected in some cells expressing CHO1F(NLS1)′ and CHO1F(NLS2)′ constructs, cytoplasmic localization is more prominent in the majority of cells. Mutations introduced in both NLS completely inhibit nuclear targeting of CHO1 (f). Foreign NLS fused to the C terminus of CHO1F(NLS1,2)′ construct restores the CHO1 localization to the nucleus (c). (C) Rescue histogram shows a threefold increase in the level of binucleation after alteration of any one or both NLS. Frequency of binucleation is reduced twofold by fusing the viral NLS to the C terminus of CHO1F(NLS1,2)′ mutant, indicating the importance of nuclear localization for the function of CHO1 in cytokinesis. Cells were stained with polyclonal anti-CHO1 (B-a) and anti-HA antibody (B-b to f). Bar, 10 μm.
Cell Culture and Transfection
CHO cells were cultured as monolayers in Ham's F-10 medium containing 10% fetal bovine serum (FBS) and 15 mM HEPES at pH 7.4 (Matuliene et al., 1999). To enrich mitotic populations, CHO cells were exposed to 0.05 μg/ml nocodazole for 5-7 h to arrest them at prometaphase. After the drug was carefully washed out from the culture, cells were either subjected to time-lapse video microscopy or cultured at 37°C in a CO2 incubator for 20-80 min before fixation with methanol at -20°C. Fixation at different time points within a 20-80-min period allowed us to observe cells at the different mitotic stages.
For RNAi assays, small inhibitory RNA (siRNA) corresponding to the nucleotide positions 138-156 of CHO1 (relative to the start codon) were synthesized previously described (Matuliene and Kuriyama, 2002) and introduced into CHO cells by transfection. 0.84 μg of siRNA were mixed with 6 μl of LipofectAMINE reagent (Life Technologies, Gaithersburg, MD) and applied to the CHO cells grown on coverslips in 3.5-cm culture plates. After 2 h, the transfection medium was replaced with fresh FBS-containing Ham's F-10 medium, and the cells were further cultured at 37°C for 28-30 h before fixation. Mock transfections were performed in an identical manner to siRNA transfections, except that siRNA was omitted.
For rescue assays, CHO cells were first transfected with pCMV-HA vector or CHO1 constructs by incubating with a mixture of 1-2 μg DNA and 6 μl LipofectAMINE as above. After 2 h, the transfection medium was replaced with fresh FBS-containing Ham's F-10 medium, and the cells were further cultured for 2-3 h. Cells were next transfected with siRNA, and 2 h later, the transfection medium was replaced with fresh F-10 medium. Cells were cultured for 28-30 h before fixation.
Immunofluorescence Staining and Immunoblot Analysis
Immunofluorescence staining was performed as previously described (Kuriyama et al., 1994). The following primary antibodies were used: rat monoclonal anti-HA (dilution 1:100; Roche Diagnostics, Indianapolis, IN), rabbit polyclonal anti-HA (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-chicken α-tubulin (1:500; Amersham, Arlington Heights, IL), rabbit polyclonal anti-CHO1 (1:100; Kuriyama et al., 1994), affinity-purified rabbit polyclonal anti-CHO1, raised against the 870-896 amino acid region of CHO1 (final concentration 10 μg/ml; Kuriyama, unpublished results). After washing out excess primary antibodies, cells were stained with secondary antibodies at dilution of 1:200 (fluorescein-conjugated anti-rat IgG, anti-mouse IgG + IgM, anti-rabbit IgG, Texas red-conjugated anti-rabbit IgG, and anti-mouse IgG). To visualize DNA, cells were treated with DAPI at 1 μg/ml for 2-5 min. Microscopic observations were made on a Nikon Eclipse TE300 inverted microscope (Garden City, NY) equipped with epifluorescence optics. Images were captured with an Olympus 3 CCD cooled digital camera OLY-750 (Olympus America, Inc., Melville, NY).
For immunoblot analysis, mock- and siRNA-transfected cells were collected 30 h after transfection, washed with cold PBS, resuspended in an SDS-sample buffer and run on denaturing 8.5% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane and incubated with monoclonal anti-CHO1 (Sellitto and Kuriyama, 1988) and monoclonal anti-α-tubulin antibodies, followed by alkaline phosphatase-conjugated secondary anti-mouse IgM and IgG antibody (Jackson ImmunoResearch, West Grove, PA).
Live cell observation
CHO cells were cultured on photoetched coverslips (Bellco Glass Co., Vine-land, NJ) or in glass-bottom, 3.5-cm culture dishes (MatTek Corp., Ashland, MA). After RNAi and RNAi-rescue with GFP-CHO1F or GFP-CHO1FΔT3 constructs, cells were cultured in a CO2 incubator for 18-20 h. Before starting time-lapse recording, the culture medium was replaced by FBS-containing Ham's F-10 medium without NaHCO3. Dishes were wrapped with parafilm and placed on a microscopic stage, which was maintained at 37°C by a stage temperature controller (Fryer Company Inc., Edina, MN) and an air curtain incubator (Nicholson Precision Instruments Inc., Bethesda, MD). Time-lapse images were taken using the ImagePro Plus software package (Media Cybernetics LP, Silver Spring, MD). Phase-contrast and fluorescence images were recorded at 1- and 3-min intervals with the exposure times of 0.05 and 0.1-0.5 s, respectively.
Statistical Analysis and Histograms
To determine the average level of cell binucleation shown in the histograms (see Figures 4C, 5B, 6, and 9C), each RNAi or RNAi-rescue experiment was repeated 3-6 times, and each time the percentage of binucleate cells was determined in 200 of randomly selected cells (RNAi, mock rescue, and vector rescue) or HA-positive cells (rescue with different CHO1 constructs). Data included in the same histogram were obtained from the rescue assays performed at the same time in an identical manner. The average level of binucleation was calculated from the data of three independent experiments, and the standard deviations were calculated for the p value of 0.05.
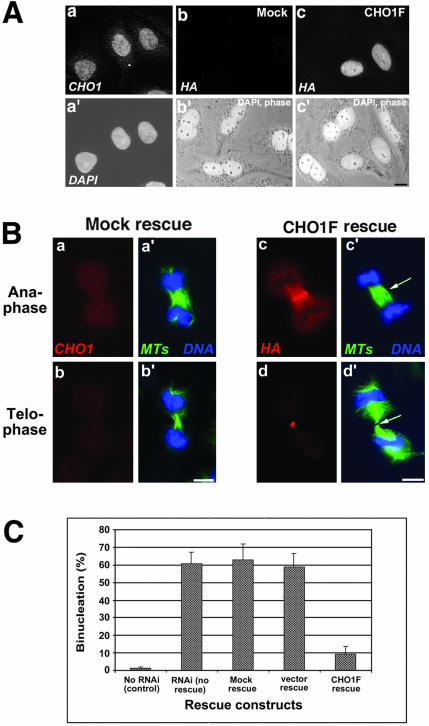
Figure 4.
Exogenous CHO1F rescues cytokinesis in siRNA-transfected cells. (A) Exogenous CHO1F localizes to the nucleus similarly to endogenous CHO1 and reduces binucleation in siRNA-treated cells. a and a′: Localization of endogenous CHO1 seen by polyclonal anti-CHO1 antibody staining (a) and DAPI (a′). b and b′: Mock-rescued cells are not recognized by HA antibody (b) and are binucleate. c and c′: CHO1F-rescued cells are stained by monoclonal anti-HA antibody (c) and contain a single nucleus, in contrast to nonexpressing binucleate cells seen in the same field. (B) Exogenous CHO1F localizes to the midzone/midbody region (c) in an identical manner to endogenous CHO1 (unpublished data) and restores the organization of midzone and midbody matrix in siRNA-treated cells (arrows in c′ and d′). This is in contrast with mock-rescued cells, which still have midzones composed of disorganized microtubule arrays (a′ and b′). Cells are stained with polyclonal anti-CHO1 (a and b), polyclonal anti-HA (c and d), anti-α-tubulin (a′-d′; green) antibodies and DAPI (a′-d′; blue). Bars, 10 μm. (C) Histograms show the percentage of binucleate cells in control (no RNAi) cells, not rescued RNAi-affected cells, and cells rescued with mock, pCMV-HA vector alone, and CHO1F construct 30 h after transfection with siRNA. Binucleation in CHO1F-expressing cells is reduced sixfold in comparison with other types of rescue or no rescue at all.
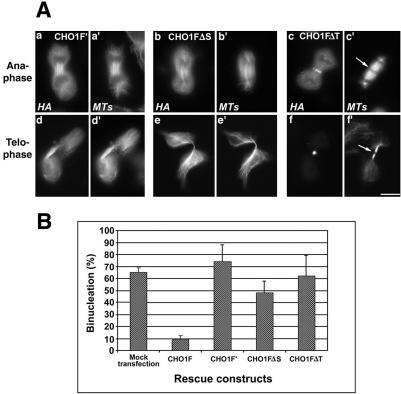
Figure 5.
ATP-binding mutant of CHO1 (CHO1F′) and CHO1 constructs lacking stalk (CHO1FΔS) or tail (CHO1FΔT) domains failed to rescue cytokinesis in endogenous CHO1-depleted cells. (A) CHO1F′ (a and d) and CHO1FΔS (b and e) constructs decorate the spindle microtubules throughout mitosis, but lack the ability to concentrate at the midzone/midbody region. In contrast to CHO1FΔT (c and f), CHO1F′ and CHO1FΔS constructs do not facilitate formation of the midbody matrix, based on continuous α-tubulin staining in the middle of the intercellular bridge (d′ and e′). CHO1FΔT localizes at the midzone (c) and midbody (f) and facilitates formation of the dense matrix (arrows in c′ and f′). Cells were stained with polyclonal anti-HA (a-f) and anti-α-tubulin (a′-f′) antibodies. Bar, 10 μm. (B) Histograms show no substantial decrease in the level of binucleation after rescue with CHO1F′, CHO1FΔS, and CHO1FΔT constructs.
Figure 6.
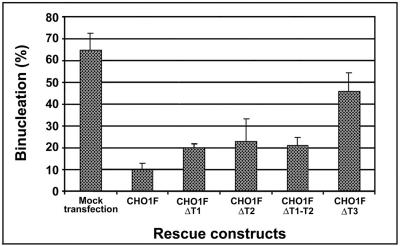
CHO1 constructs lacking different regions of the tail domain show different abilities to rescue cytokinesis in endogenous CHO1-depleted cells. Histograms show the percentage of binucleation in rescued cells. CHO1 lacking T1 (CHO1FΔT1) or T2 (CHO1FΔT2), or both (CHO1FΔT1-T2) are half as efficient in the rescue of cytokinesis, whereas the construct lacking NLS region (CHO1FΔT3) is one quarter as efficient in comparison to the wild-type CHO1F.
Quantitation of Protein Expression
To evaluate the specificity of RNAi rescue effects, it is important to compare the level of exogenous protein expression in siRNA-treated cells. Because CHO1 constructs used for the rescue assays showed different subcellular localization during interphase (nucleus and/or cytoplasm), we analyzed protein amounts in synchronized prometa/metaphase cells. Control cells, mock-rescued cells, and cells rescued with different CHO1 constructs summarized in Figures 1 and 9A were synchronized, fixed at metaphase stage, and then immunostained with anti-HA antibody to detect exogenous protein and with polyclonal anti-CHO1 antibody to detect CHO1 in cells. Fluorescence intensity was measured using the ImagePro Plus software package (Media Cybernetics, Silver Spring, MD). The amount of exogenous CHO1 expressed in rescued cells was compared with that of endogenous CHO1 in control cells by quantifying the fluorescence intensity corresponding to CHO1 antibody in 30 randomly selected control and mock-rescued cells and in 30 HA-positive cells rescued with each CHO1 construct. The average fluorescence intensity derived from control cells was used as 100% of endogenous CHO1 expression. The lowest fluorescence intensity detected in mock-rescued cells was used as 0% of CHO1 expression.
Figures S1 and S2 show that the amounts of CHO1 detected in rescued cells by both anti-CHO1 (Figure S1) and anti-HA antibodies (Figure S2) were comparable among 12 different rescue constructs analyzed in this study. On average, ∼42% of rescued cells had the expression level lower or equal to control CHO1 (100% = 1), ∼46% of cells fell into the range between 100 and 300% (1-3) of endogenous CHO1 expression, and in ∼12% of cells the expression level was higher than 300% (≥3) of endogenous level of CHO1 (Figure S1). Most of the rescued cells at all levels of protein expression showed normal bipolar spindle formation and mitotic progression with the rare exception (<5%) of cells expressing extremely high levels of exogenous protein. Those cells tended to have nonspecific mitotic defects and were not used for the study of CHO1 function. Intensity of HA fluorescence quantified as arbitrary units was divided into three categories: low (9-40 U), medium (41-70 U), and high (71-100 U) expression. Figure S2 indicates that the mount of exogenous proteins is comparable among twelve rescue constructs.
RESULTS
RNAi Directed against CHO1 Inhibits the Terminal Stage of Cytokinesis
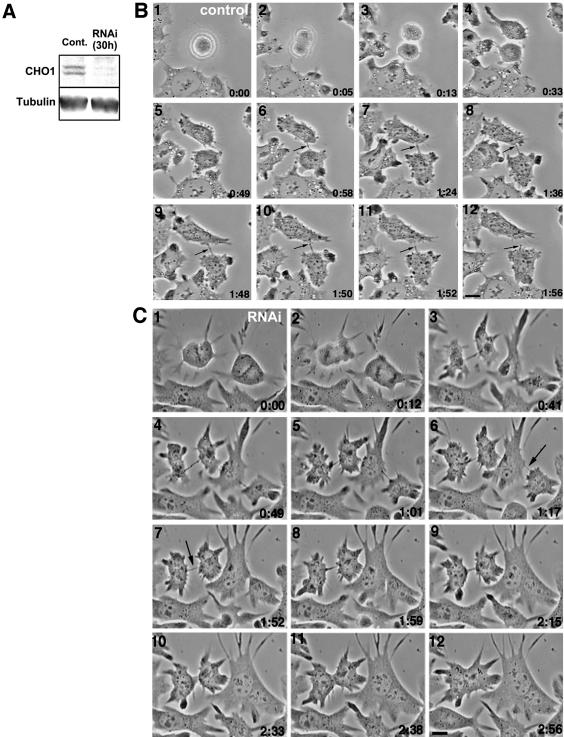
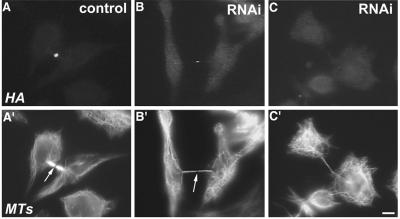
To study the function of CHO1 in cytokinesis, we performed RNAi assays by transfecting cells with siRNA specific for CHO1. Both immunoblotting analysis (Figure 2A) and a quantitation of immunofluorescence intensity (Matuliene and Kuriyama, 2002) showed that siRNA-treated cells expressed significantly reduced amounts of CHO1 (<10% of the control level). To observe how cytokinesis is affected, we monitored siRNA-treated cells by time-lapse phase-contrast microscopy. Both mock- (Figure 2B) and siRNA-transfected cells (Figure 2C) underwent normal mitosis and initiation of cytokinesis, although RNAi-affected cells seemed to be more adhesive to the substratum and less round than control cells (compare Figure 2B, frames 1-3, and 2C, frames 1 and 2). Within 40 min after initiation of cytokinesis, cells formed intercellular bridges between two separating cells. In control cells, a dense dot corresponding to the midbody matrix was clearly seen in the middle of the bridge (arrows in Figure 2B, frames 6-12), which was pinched off at one side of the midbody (frame 12) to separate two daughter cells. In contrast, the bridges formed in CHO1-depleted cells were thinner than those in controls and devoid of a distinguishable dark midbody dot at the center (arrows in Figure 2C, frames 6 and 7). Immunofluorescence staining showed that these bridges contained fewer microtubules and a faint (Figure 3, B and B′; arrow) or almost invisible (Figure 3, C and C′) midbody matrix, compared with the control (Figure 3, A and A′; arrow). Strikingly, instead of breaking off at one point, the intercellular bridges in RNAi-affected cells gradually widened up (frames 6-8 and 9-11 in Figure 2C) and eventually disappeared as two cells fused back together within 1-2 h. The similar phenotype was observed in >70% of cells monitored by time-lapse microscopy (Table 1). These results indicate that CHO1 is required for organization of the midbody matrix, which is essential for complete separation of two daughter cells at the end of cell division. It should be, however, pointed out that in 14.5% (9 of 62) of cells, the cleavage furrows regressed immediately after the initiation of cytokinesis, and 3.2% (2 of 62) of cells indicated no sign of initiation of cytokinesis (Table 1). These results suggest that, although CHO1 is essential for the terminal stage of cytokinesis, the motor protein could also play a role in initiation and/or ingression of cleavage furrows.
Figure 2.
RNAi against CHO1 causes depletion of CHO1 protein and inhibition of cytokinesis in CHO cells. (A) Cells were transfected with siRNA (RNAi) or transfection reagent alone (control) and analyzed by immunoblotting for the presence of CHO1 protein 30 h after transfection. Immunoblotting was performed using monoclonal anti-CHO1 and anti-α-tubulin antibodies for the loading control. CHO1 antibodies reveal two isoforms, MKLP1 and CHO1, coexpressed in a single cell (Kuriyama et al., 2002). (B) Successful division of mock-transfected cells monitored by time-lapse phase-contrast microscopy. A dark midbody dot indicated by arrows is seen in the middle of the intercellular bridge until complete separation of the daughter cells. (C) Time-lapse phase-contrast microscopy of RNAi-affected cells. Cells reach the late stage of cytokinesis, but fuse back within 1-2 h after formation of the intercellular bridge in which the dark midbody structures are generally missing (arrows). Bars, 10 μm.
Figure 3.
RNAi against CHO1 affects the thickness and the length of intercellular bridges and the formation of midbody matrix in dividing cells. Control cell (A) and siRNA-transfected cells (B and C) were fixed at the late stage of cell division and stained with polyclonal anti-CHO1 (A-C) and anti-α-tubulin (A′-C′) antibodies. The control daughter cells are connected by a thick intercellular bridge with a well-defined midbody matrix, which cannot be penetrated by anti-α-tubulin antibody and, therefore, appears as a dark region (arrow in A′). When CHO1 is depleted from the midbody region (B and C), the intercellular bridges become longer and thinner (B′ and C′), and the midbody matrix is faint (arrow in B′) or hardly detectable in the cells with the lowest amounts of CHO1 expression (C′). Bar, 10 μm.
Table 1.
RNAi against CHO1 inhibits cytokinesis in CHO cells
| Cells | Compete cytokinesis, % | No initiation of cytokinesis, % | Early furrow regression, % | Late furrow regression, % |
|---|---|---|---|---|
| Control | 100 (20) | |||
| siRNA-transfected | 9.5 (6) | 3.2 (2) | 14.5 (9) | 72.6 (45) |
To inhibit expression of CHO1, cells were transfected with siRNA specific for the CHO1 sequence. Mock-transfected or nontransfected cells were used as a control. Non-synchronized or partially synchronized cells were recorded by phase-contrast time-lapse microscopy. The number of observed cells is indicated in parentheses. In the Early furrow regression column, resumption of cleavage furrows occurs before a thin intercellular bridge is formed. The Late furrow regression usually takes place in cells in which the nuclear envelope has already been reformed, but two daughter cells are still connected with a cytoplasmic bridge.
Exogenous CHO1F Rescues the Defect of Cytokinesis in CHO1-depleted Cells
To confirm the specificity of RNAi effects, we determined whether progression of cytokinesis could be rescued by exogenous CHO1 expressed in siRNA-treated cells. The assay was done by first transfecting CHO cells with one of the following constructs: no DNA (mock transfection), pCMV-HA vector alone (control), and HA-tagged, full-length CHO1F, in which a short stretch of sequence, complementary to siRNA oligonucleotide, was mutated to prevent destruction of exogenous mRNA. After a 2-h recovery from the first transfection, cells were transfected again with siRNA to inhibit expression of endogenous CHO1. Immunofluorescence staining revealed no detectable levels of CHO1 in mock- or vector-rescued cells fixed 28-30 h later (Figure 4, A-b and B-a and -b). Those cells displayed disorganized midzone/midbodies, which is typical of RNAi-affected cells (Figure 4B-a′, b′). In contrast, ∼60% of the cells transfected with the CHO1F construct expressed HA-tagged exogenous protein, which was targeted to the same location as endogenous CHO1; interphase nuclei (Figure 4, A-a and -c) and the midzone/midbody region in mitotic cells (Figure 4B-c and -d). The expression of exogenous CHO1F in mitotic cells restored both organization of midzone region (Figure 4B-c′) and formation of the stem body/midbody matrix, which was easily detected by the presence of dark regions in tubulin staining (arrows in Figure 4B-c′ and -d′). Importantly, binucleation in interphase cells expressing CHO1F was reduced ∼sixfold in comparison with control cells (Figure 4A-c and -c′ compared with 4-b and -b′, and 4C). It is worth mentioning that >85% of rescued cells expressed the CHO1 amount comparable to that endogenous CHO1 detected in control cells (Figure S1). These results demonstrate that CHO1F expression in siRNA-treated cells compensates for the depletion of endogenous protein, thus proving not only the specificity of RNAi inhibition but also grounds for further study of CHO1 constructs in endogenous CHO1-depleted background.
Rescue Assays Reveal the Functional Differentiation Between Three Domains of CHO1
Using the genetic rescue approach, we evaluated the functional properties of CHO1 domains by expressing each of the following rescue constructs: 1) CHO1F′, an ATP-binding site mutant of CHO1; 2) CHO1FΔS, a construct with deleted stalk domain; and 3) CHO1FΔT, a construct lacking the tail domain (Figure 1). Figure 5A-a and -d shows siRNA-treated cells expressing an HA-tagged CHO1F′ construct. Consistent with our previous observations (Matuliene and Kuriyama, 2002), CHO1F′ was able to bind and bundle microtubules, but did not concentrate at the midline of the central spindles. The mutant protein was seen along the entire spindle fibers throughout mitosis, and it did not facilitate the formation of the midbody matrix (Figure 5A-a′ and -d′). Quantitative analysis showed that the level of binucleation was not reduced, compared with mock-rescued cells (Figure 5B). Rather, the number of binucleate cells increased in CHO1F′-expressing cells, which could be due to a dominant negative effect of the mutant construct on cytokinesis (Matuliene and Kuriyama, 2002). Similarly to CHO1F′, a CHO1 construct lacking the stalk domain (CHO1FΔS) was also unable to properly localize to the midzone/midbody (Figure 5A-b and -e). Although CHO1FΔS can interact with spindle microtubules through its motor domain (Matuliene and Kuriyama, 2002), it did not translocate to the midzonal region during anaphase and failed to facilitate the formation of the midbody matrix (Figure 5A-b′ and -e′). Figure 5B shows that no substantial rescue of cytokinesis was detected in CHO1FΔS expressing cells. These results indicate that both the motor and stalk domains are required for organization of the central spindle and midbody matrix. In contrast to CHO1F′ and CHO1FΔS, the CHO1 construct lacking the tail domain (CHO1FΔT) was targeted to the midzone/midbody (Figure 5A-c and -f), where it restored the organization of microtubule bundles and midbody matrix (arrows in c′ and f′). Nevertheless, the rescue construct was unable to reduce the percentage of binucleate cells, indicating that the tail is required for the terminal stage of cytokinesis, which occurs after midbody formation.
The Tail Domain of CHO1 Is Essential for Completion of Cytokinesis
To determine the role of the CHO1 tail in cytokinesis, we further analyzed different subregions of the tail sequence (Figure 1). The CHO1 tail is encoded by exons 17-23 (Kuriyama et al., 2002), and we divided it into three subregions: T1, T2, and T3. T1 (amino acids 696-787) is capable of interacting with actin filaments in vivo and in vitro (Kuriyama et al., 2002). It is encoded by exon 18 (amino acids 691-793) and can be removed by alternative splicing. Two isoforms, with (CHO1) and without (MKLP1) the actin-binding sequence, have been shown to be coexpressed in a single cell (Kuriyama et al., 2002). T2 (amino acids 788-897) corresponds to the highly conserved tail sequence encoded by exons 19 (amino acids 794-870) and 20 (amino acids 871-899). This region corresponds to the most conserved sequence of the CHO1 tail and covers the major part of the G-protein Arf-binding sequence identified by yeast two-hybrid screen (Boman et al., 1999). T3 (amino acids 898-953) is a short stretch of 56 amino acids containing two nuclear localization signals (NLS1 and NLS2) and is mainly encoded by the last three exons (21-23: amino acids 900-953) at the C terminus. The first NLS is encoded at the beginning of exon 21 (RKRR, amino acids 900-903) and the second NLS is encoded by an entire exon 23 (KRKKP, amino acids 949-953; see Figure 9A).
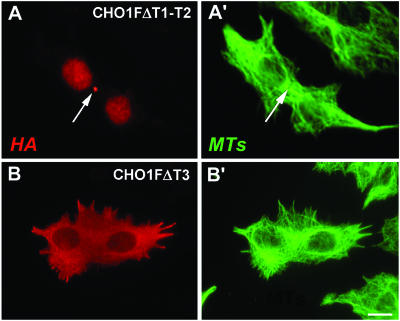
We expressed CHO1 constructs lacking T1 (CHO1FΔT1), T2(CHO1FΔT2), both T1 and T2 (CHO1FΔT1-T2), or T3 (CHO1FΔT3) for RNAi rescue assays. As summarized in Figure 6, constructs lacking either T1 or T2, or both, were at least two times less efficient in rescuing cytokinesis than the full-length CHO1F. Because CHO1FΔT1 corresponds to a shorter version of CHO1 (MKLP1), these results suggest that MKLP1 alone is insufficient for completion of cytokinesis. This is consistent with our previous observation that the actin-binding sequence is essential for cytokinesis as the antibody specific to this particular region of CHO1 caused regression of cleavage furrows in microinjected cells (Kuriyama et al., 2002). To our surprise, the least efficient rescue of cytokinesis was detected in cells expressing CHO1FΔT3 construct (Figure 6). In those cells, binucleation was reduced from ∼65% to only ∼46%, suggesting that the T3 subregion is largely responsible for the function of the tail domain in cytokinesis. Despite its inability to rescue the defect of cytokinesis, CHO1FΔT3 was properly localized at the spindle midzone and efficiently restored the midbody matrix structure (unpublished data).
The Tail Subregions Reveal Distinctive Roles in Cytokinesis
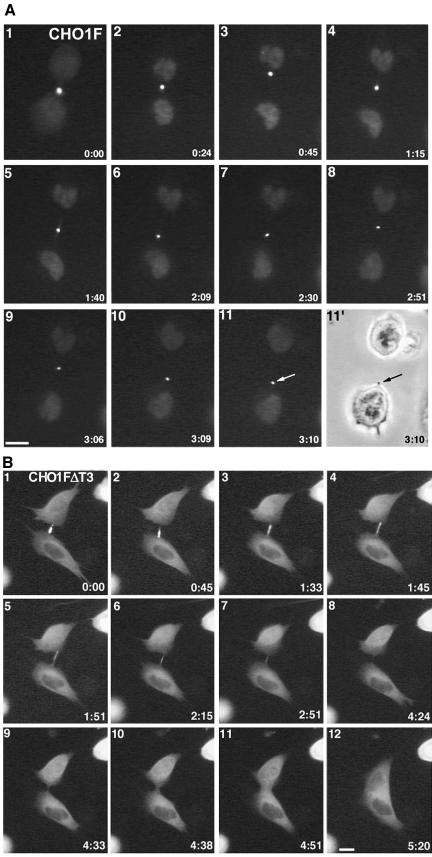
To identify what causes the failure of cytokinesis in CHO1FΔT3 cells, we expressed GFP-tagged rescue constructs and monitored the cell division pattern using time-lapse fluorescence microscopy. Figure 7 shows that siRNA-treated cells expressed GFP-CHO1F (A) and GFP-CHO1FΔT3 (B) in the middle of the intercellular bridges (see also supplemental video GFP-CHO1FΔT3). Although the midbodies in CHO1F-rescued cells (n = 6) remained intact until the end of cell division (arrows in frames 11 and 11′ of Figure 7A), the majority (9 of 12) of CHO1FΔT3-expressing cells failed to maintain the integrity of the dense midbody structure. In those cells, the midbody slowly deteriorated as fluorescent dots gradually disappeared, which ultimately led to fusion of two daughter cells within 3-5 h after midbody formation (Figure 7B, supplemental video-GFP-CHO1FΔT3). It should be noted that no midbodies and midbody matrices were detected inside the fused cells (Figures 7B-12 and 8, B and B′). These results strongly suggest that the T3 subregion is required for stabilization of the midbody during cytokinesis.
Figure 7.
The NLS-containing region of the CHO1 tail is required for stabilization of the midbody matrix. (A) The midbody is stable in a GFP-CHO1F-rescued cell monitored by the time-lapse fluorescence microscopy. At the end of cytokinesis, the intercellular bridge is pinched off at one side of the midbody, and the midbody is incorporated into one of the daughter cells (arrows). (B) Time-lapse fluorescence video and assembled micrographs shows that the midbody in a GFP-CHO1FΔT3-rescued cell gradually disintegrates, causing almost completely separated two daughter cells to merge again. Note that full-length CHO1F localizes to the nucleus after the nuclear envelope is formed (frames 2-11 in 7A), whereas the construct-lacking NLS region remains in the cytoplasm throughout interphase (B). Bars, 10 μm.
Figure 8.
Structural stability of the midbody/midbody matrix formed in cells expressing CHO1FΔT1-T2 (A and A′) and CHO1FΔT3 (B and B′). Cells were double-stained with anti-HA (A and B) and anti-α-tubulin antibodies (B and B′). Although the midbody remains inside the fused cells rescued with CHO1FΔT1-T2 construct, the CHO1FΔT3-containing midbody disintegrates. Bars, 10 μm.
When rescued with CHO1FΔT1-T2, the CHO1-depleted cells formed the midbody of normal appearance. However, the midbody became detached from the cell cortex during the last stage of cytokinesis, resulting in the fusion of two daughter cells. Unlike CHO1FΔT3, CHO1FΔT1-T2 was able to maintain the midbody matrix structure during the process of furrow regression, allowing us to detect the distinct midbody matrix between two nuclei in the fused cell (arrows in Figure 8, A and A′). The intact midbody structure was also detected in PtK1 cells after abortive cytokinesis caused by microinjection of antibodies specific to T1 (Kuriyama et al., 2002) or T2 (unpublished data) subregions of the CHO1 tail. Taken together, these results demonstrate functional differentiation between the regions of the tail domain: although the T1-T2 region appears to facilitate the interaction of the midbody with the cortex, T3 is required to maintain the integrity of the midbody matrix until the complete separation of the daughter cells.
Nuclear Localization Signals Are Important for the Function of CHO1 in Cytokinesis
T3 is responsible for nuclear targeting of CHO1. To examine whether the malfunctioning of the CHO1FΔT3 construct during cytokinesis is directly related to its inability to localize to the nucleus during interphase, we expressed mutant proteins in which two amino acids within first NLS (NLS1), a second NLS (NLS2), and both NLS (NLS1,2) in the full-length CHO1 sequence were changed to alanines, creating CHO1F(NLS1)′, CHO1F(NLS2)′, and CHO1F(NLS1,2)′ constructs, respectively (Figure 9A). These constructs, used in the rescue assay, showed that mutations in either of two NLS strongly affects localization of CHO1 to the nucleus. Although in some cells a fraction of the mutated protein still could be seen within the nucleus, the majority of the mutated protein showed cytoplasmic localization in most cells (Figure 9, B-d and -e), suggesting that two functional NLS are required for efficient nuclear localization of CHO1. Consistent with these results, mutation in both NLS caused a complete inhibition of nuclear localization (Figure 9B-f). Intriguingly, none of NLS mutants showed an efficient rescue of cytokinesis (Figure 9C). More than 30% of cells, expressing mutant proteins were found to be binucleate, suggesting that nuclear localization itself is important for the function of CHO1 in cytokinesis. However, the extent of binucleation did not reach the level seen in CHO1FΔT3-rescued cells (>45%, Figure 6), implying that the entire NLS-containing region has other functions besides facilitating the nuclear localization of CHO1. To confirm the importance of nuclear localization in cytokinesis, foreign NLS, derived from SV40 virus, was fused to the C terminus of CHO1F(NLS1,2)′, and a new construct CHO1F(NLS1,2)′NLS (Figure 9A) was used in the rescue assay. During interphase, CHO1F(NLS1,2)′NLS was found to localize to the nucleus (Figure 9B-c) similar to endogenous CHO1 (Figure 9B-a) and exogenous CHO1F (Figure 9B-b), indicating that the viral NLS is sufficient to drive the NLS mutant to the nucleus. At the same time, the number of binucleate cells was clearly reduced by the expression of the CHO1F(NLS1,2)′NLS. Figure 9C shows that the efficiency of cytokinesis rescue by the CHO1F(NLS1,2)′NLS construct was at least doubled after the addition of the foreign NLS, suggesting that NLS is important for the CHO1 function in cytokinesis.
DISCUSSION
In this study we used the RNAi and genetic rescue assay to analyze the function of the kinesin-like protein, CHO1. The study showed that CHO1 is a multifunctional protein that acts at different stages of cytokinesis. Although motor and stalk domains are required for organization of the midzone and the midbody matrix, the tail domain is a prerequisite for maintaining both the structural integrity of the midbody matrix and midbody interaction with the cortex until the final step of cytokinesis: abscission of the intercellular bridge.
Role of CHO1 in Cytokinesis
The homologues of MKLP1/CHO1 studied thus far function in cytokinesis in diverse organisms (Adams et al., 1998; Powers et al., 1998; Raich et al., 1998; Chen et al., 2002; Matuliene and Kuriyama, 2002). However, there is some controversy regarding the stage of cytokinesis in which these proteins are involved. In C. elegans, the role of ZEN-4 appears to be restricted to completion of cytokinesis and maintenance of cell separation (Powers et al., 1998; Raich et al., 1998; Jantsch-Plunger et al., 2000; Severson et al., 2000). In contrast, Drosophila PAV-KLP has been suggested to be essential for assembly of the contractile ring and initiation of cytokinesis (Adams et al., 1998; Somma et al., 2002; Somers and Saint, 2003). Somers and Saint (2003) have recently proposed that the complex of PAV-KLP and RacGAP50C plays a role in positioning of Pebble RhoGEF at the equator of the cell cortex through direct interaction between RhoGAP and RhoGEF. This in turn results in formation of a ring of activated Rho, ultimately leading to organization of the actomyosin contractile ring. In the case of C. elegans, Dechant and Glotzer (2003) have provided evidence that cleavage furrows are formed in the vicinity of local areas where microtubule density becomes minimal as a result of aster separation during anaphase and by bundling of microtubules into the central spindle. Thus cytokinesis can be initiated in the absence of organized central spindles provided that the length of mitotic spindles is normal and the asters are separated to a normal extent. However, the experimental evidence has suggested that depletion of PAV-KLP profoundly affects the organization of central spindles and also shortens the spindle in Drosophila S2 cells (Somma et al., 2002). This might be one reason why the motor protein affects furrow initiation in Drosophila.
In mammalian cells, we observed that CHO1 depletion by siRNA treatment mostly affects the late stage of cytokinesis. As shown in Figures 3C and 4B-b and -b′, even cells with no detectable endogenous CHO1 could achieve advanced cytokinetic furrows. This may favor the idea that CHO1 is not required for initiation of cytokinesis in mammalian cells. However, we cannot rule out the possibility that the trace amount of CHO1 remained in siRNA-treated cells might be sufficient to induce furrow formation. We also noticed that more extensive depletion of CHO1 tended to cause faster regression of cleavage furrows, leaving the possibility that 100% depletion of CHO1 might result in inhibition of furrow initiation. Indeed, 2 of 64 cells treated with siRNA indicated no sign of furrow initiation (Table 1). Further analysis is required to determine whether CHO1 is essential for initiation of cytokinesis in mammalian cells.
Not only the presence or absence of CHO1, but also the degree of structural disorganization of the midzone might be critical for the initiation of cytokinesis. Although RNAi against CHO1 caused the disorganization of central spindles, some microtubule bundles still remained in most of the CHO1-depleted cells. This could be attributed to the presence of additional motor proteins with similar function to CHO1 at the spindle midzone. It has recently been reported that Rab6-KIFL (MKLP2: Hill et al., 2000; Neef et al., 2003) and PMM1 (Abaza et al., 2003) at the central spindle contain the microtubule bundling activity and are required for cytokinesis. Simultaneous elimination of all three motors may result in complete disorganization of central spindles and inhibition of cleavage furrow formation.
Dissecting the Function of CHO1 Subdomains
Roles of Motor and Stalk Domains Analysis of the ATP-binding mutant of CHO1 (CHO1F′) revealed that the ATP-based motility of the protein is essential for organization of central spindles and midbodies by translocating CHO1 to the spindle midzone. The mechanochemical motor domain covering amino acids 1-423 might also be important for formation of the midbody matrix by bundling midzonal microtubules. This could be facilitated by the ATP-independent microtubule-binding site located at the C-terminal region of the CHO1 motor domain at the N terminus (amino acids 277-421; Matuliene and Kuriyama, 2002). Because the stalk domain facilitates dimerization of CHO1, the motor-stalk construct can have multiple microtubule-binding sites, allowing the protein to bundle microtubules. Mishima et al. (2002) have shown that the stalk is also responsible for interaction with RhoGAP, which is essential for both proper localization of the protein complex at the midzone and the microtubule-bundling activity. Consistent with these results, we found that the CHO1 construct lacking the stalk domain revealed improper distribution in the central spindle and midbodies and showed no ability to bundle microtubules in siRNA-treated cells (Figure 5A-b, b′ and e, e′). In contrast, the motor-stalk construct was able to show correct localization, microtubule-binding activity, and midbody matrix formation (Figure 5A-c, c′ and f, f′). From these results, we conclude that the motor and stalk domains are sufficient for organization of functional central spindles and midbodies. To complete cytokinesis, however, cells must express the tail sequence.
Role of T1 and T2 Subregions of the Tail The tail domain of CHO1 can be subdivided into several distinctive subregions: T1, T2, and T3. Exon 18 encoding T1 is capable of interaction with actin filaments in vivo and in vitro (Kuriyama et al., 2002). T2 contains the highly conserved sequence encoded by exons 19 and 20, and it partially overlaps with the Arf-binding domain (Boman et al., 1999). Exons 21-23 express ∼50 amino acids containing two nuclear localization signals. The protein lacking T1 and/or T2 is half as efficient as wild-type CHO1 in rescue of cytokinesis (Figure 6). Because G-protein Arf functions in membrane trafficking (Donaldson et al., 1995; Boman and Kahn, 1995), the T2-containing region of CHO1 could be involved in membrane deposition during cytokinesis (Skop et al., 2001). Another possibility is that either T1 and T2, or both facilitate interaction between the midbody microtubules and the cell cortex. Indeed, immunofluorescence microscopy showed the presence of dense midbody structures inside CHO1FΔT1-T2-rescued cells after the failure of cytokinesis. This suggests that the midbody remained intact, but it detached from the cell cortex to cause the regression of cleavage furrows (Figure 8, A and A′). As daughter cells move apart, the connecting intercellular bridge becomes a subject of intense mechanical tension. During this tug-of-war, the midbody matrix must maintain tight association with the cortex and/or membrane to prevent daughter cells from merging again. Because the actin cytoskeleton is in the cortex and Arfs are involved in both membrane trafficking and actin dynamics (Norman et al., 1998; Donaldson, 2003), association between the midbody and the cell cortex could be mediated through the actin/Arf-binding domain of CHO1. In support of this hypothesis, PAV-KLP has recently been demonstrated to locate at the cortical equatorial ring during cytokinesis in Drosophila cells (Somers and Saint, 2003; Minestrini et al., 2003). We also detected the interaction of CHO1 with the cortex in cytochalasin D-treated mitotic CHO cells (unpublished results). This close proximity between the motor proteins and cell cortex strongly supports the role of CHO1 in connecting the central spindle/midbody to the cortex/membrane during the late stage of cell division. It is noteworthy that deletion of either T1 or T2, or both, resulted in almost identical phenotypes, suggesting functional collaboration between these two regions.
Role of the NLS-containing Region Besides interaction with the cell cortex, we determined a novel function of the CHO1 tail: stabilization of the midbody structure during the late stage of cytokinesis. In cells rescued by CHO1FΔT3, a thick and bright midbody gradually disintegrates in the intercellular bridge (Figure 7B), whereas the same structure remains stable in the cell expressing GFP-CHO1F (Figure 7A). To ensure completion of cytokinesis, midbody microtubules must be tightly glued together by the matrix until the inter-cellular bridge breaks. If the connection between matrix components becomes loose, the midbody structure may not be strong enough to resist the ripping forces generated between two daughter cells and will soon start to disintegrate. Time-lapse studies showed that small fluorescent particles in the middle of the intercellular bridge detached from the midbody, moved away along the intercellular bridge, and subsequently disappeared as the midbody moved back and forth daughter cells (Figure 7B). These observations suggest that matrix components in CHO1FΔT3-rescued cells are connected too weakly to maintain intact the dense midbody structure. Isolated midbodies contain at least 158 different molecules (Skop et al., 2002); some of those might be responsible for maintaining the structural integrity of the midbody by connecting molecules in the midbody. Possible candidates might be PRC1, Rab6-KIFL, or MPP1, which were shown to bundle midzone microtubules (Jiang et al., 1998; Mollinari et al., 2002; Abaza et al., 2003; Neef et al., 2003). One possible scenario is that the T3 sequence of CHO1 is directly involved in association of CHO1 with those midbody components.
Alternatively, the function of T3 subregion could be mediated through the nuclear localization signal. Loss of NLS may cause the change in association of CHO1 with the importin complex during mitosis, which in turn alters the CHO1 activity by affecting the distribution of CHO1 complex controlled by Ran GTPase (Quimby and Dasso, 2003). Another possibility is that the CHO1 function in cytokinesis may be under direct control of actual localization of CHO1 to the nucleus during interphase. As shown in Figure 9, CHO1 with a mutation in NLS was incapable of both nuclear localization and efficient rescue of cytokinesis. Introduction of SV40 virus-derived NLS restored the localization of CHO1 to the nucleus and reduced the level of binucleation in a rescue assay from ∼32% to ∼15%. However, because the extent of binucleation after CHO1F(NLS1,2)′ rescue did not reach the level seen in CHO1ΔT3-rescued cells (>45%, Figure 6), and the presence of exogenous NLS did not bring the level of binucleation down to 10% as in CHO1F-rescued cells, it seems likely that the NLS-containing region also has other function(s) in addition to facilitating the nuclear localization of CHO1. Although the role of NLS in cytokinesis is still under investigation, we speculate that CHO1 may have to enter the nucleus to undergo a certain modification. Interestingly, both NLS sequences are strictly conserved among all vertebrate homologues of CHO1, identified in human, mouse, chicken and zebrafish (Kuriyama et al., 2002). Moreover, all three proteins: CHO1/MKLP1, CYK-4/MgcRacGAP, and Rho GEF ECT2, which appear to exist in a complex (Somers and Saint, 2003), have an NLS and are localized to the nucleus in mammalian cells (Tatsumoto et al., 1999; Hirose et al., 2001; Mishima et al., 2002), suggesting the importance of nuclear localization. Therefore, CHO1 may have to establish interactions with those molecules to become fully active during cytokinesis.
It is important to point out that nuclear localization of the motor proteins in invertebrate cells does not seem as critical as in mammalian cells. Although PAV-KLP in Drosophila has a functional NLS, corresponding to NLS1 in vertebrates, nuclear localization of the motor protein appears to be essential only for the female germ line development (Minestrini et al., 2002), but not for embryonic cell division (Minestrini et al., 2003). In C. elegans, ZEN-4 lacks not only NLS but also the entire C-terminus corresponding to the NLS-containing region. Thus, it is possible that the motor proteins in vertebrates have acquired novel functions that may not be necessary for development of invertebrate organisms, or the T3 function might be performed by other proteins in the same pathway. A clear precedent for such differences between the organisms is the presence of the actin-interacting region in the tail domain of CHO1. Even though this region is absent in invertebrate homologues, both antibody injection (Kuriyama et al., 2002) and the rescue assay (this study) suggest the importance of this region for cytokinesis in mammalian cells.
Model of CHO1 Functions in Cytokinesis
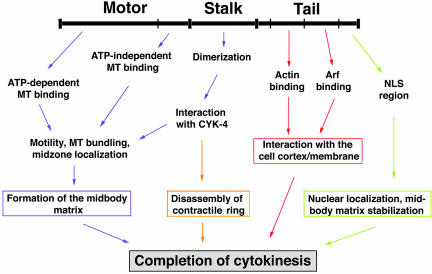
From the results obtained from this study, we summarized the role of CHO1 in cytokinesis (Figure 10). At the onset of anaphase, CHO1 translocates to the equatorial region of mitotic spindles, where it bundles antiparallel polar microtubules and participates in formation of the central spindle and the midbody matrix. This function is facilitated by multiple microtubule binding sites in the motor domain (Matuliene and Kuriyama, 2002), stalk-mediated protein dimerization, and interaction with Cyk-4/MgcRacGAP (Mishima et al., 2002). Because the complex of PAV-KLP and RacGAP50C has been shown to directly interact with Pebble RhoGEF (Somers and Saint, 2003) in Drosophila, it is possible that a similar protein complex in mammalian cells could activate Rho1 at the cortex and participate in signaling events required for ingression of cleavage furrows. Once the furrowing and the midbody matrix formation are completed and the Rho GAP activity of CYK-4/MgcRacGap is activated by Aurora B kinase (Minoshima et al., 2003), the protein complex may participate in the inactivation of Rho1 at the cortex and disassembly of the contractile ring. At the same time, the tail T1 and T2 subregions facilitate the interaction between the midbody and the cell cortex/membrane, while the NLS-containing T3 region ensures the structural integrity of the midbody until the successful abscission of the intercellular bridge. Further studies will be required to better understand the complex pathway of cytokinesis and determine molecular components and signaling events.
Figure 10.
A model of CHO1 functions in cytokinesis.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM55735) and the University of Minnesota Graduate School Grant-in-Aid (19354) to R.K. J.M. was supported by the Frieda Marta Kunze Predoctoral Fellowship from the University of Minnesota Graduate School.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0888. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0888.
We dedicate the article to the memory of our collaborator and colleague, Dr. Annette L. Boman.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Abaza, A., Soleilhac, J.M., Westendorf, J., Piel, M. Crevel, I., Roux, A., and Pirollet, F. (2003). M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J. Biol. Chem. 278, 27844-27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, R.R., Tavares, A.A.M., Salzberg, A., Bellen, H.J., and Glover, D.M. (1998). pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12, 1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop, G.B., and Zhang, D. (2003). Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J. Cell Biol. 162, 383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., and Kahn, R.A. (1995). Arf proteins: the membrane traffic police? Trends Biochem. Sci. 20, 147-150. [DOI] [PubMed] [Google Scholar]
- Boman, A.L., J. Kuai, J., Zhu, X., Chen, J., Kuriyama, R., and Kahn, R.A. (1999). Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil. Cytoskel. 44, 119-132. [DOI] [PubMed] [Google Scholar]
- Buck, R.C., and Tisdale, J.M. (1962). The fine structure of the mid-body of the rat erythroblast. J. Cell Biol. 13, 109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L.G., and Wang, Y.L. (1996). Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell 7, 225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.C., Zhou, Y., and Detrich, H.W., III. (2002). Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol. Genomics 8, 51-66. [DOI] [PubMed] [Google Scholar]
- Dechant, R., and Glotzer, M. (2003). Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell 4, 333-344. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., Radhakrishna, H., and Peters, P.J. (1995). The ARF GTPases: defining roles in membrane traffic and organelle structure. Cold Spring Harb. Symp. Quant. Biol. 60, 229-234. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G. (2003). Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573-41576. [DOI] [PubMed] [Google Scholar]
- Giansanti, M.G., Bonaccorsi, S., Williams, B., Williams, E.V., Santolamazza, C., Goldberg, M.K., and Gatti, M. (1998). Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 12, 396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, E., Clarke, M., and Barr, F.A. (2000). The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19, 5711-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, K., Kawashima, T., Iwamoto, I., Nosaka, T., and Kitamura, T. (2001). MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276, 5821-5828. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger, V., Gonczy, P., Romano, A., Schnabel, H., Hamill, D., Schnabel, R., Hyman, A.A., and Glotzer, M. (2000). CYK-4, a Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149, 1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Jimenez, G., Wells, N.J., Hope, T.J., Wahl, G.M., Hunter, T., and Fukunaga, R. (1998). PRC1, a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2, 877-885. [DOI] [PubMed] [Google Scholar]
- King, R.C., and Akai, H. (1971). Spermatogenesis in Bombyx mori. I. The canal system joining sister spermatocytes. J. Morphol. 134, 47-56. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., Dragas-Granoic, S., Maekawa, T., Vassilev, A., Khodjakov, A., and Kobayashi, H. (1994). Heterogeneity and microtubule interaction of the CHO1 antigen, a mitosis-specific kinesin-like protein. Analysis of subdomains expressed in insect Sf9 cells. J. Cell Sci. 107, 3485-3499. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., Gustus, C., Uetake, Y., Terada, Y., and Matuliene, J. (2002). CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J. Cell Biol. 156, 783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene, J., R. Essner, R., Ryu, J.-H., Hamaguchi, Y., Baas, P.W., Haraguchi, T., Hiraoka, Y., and Kuriyama, R. (1999). Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J. Cell Sci. 112, 4041-4050. [DOI] [PubMed] [Google Scholar]
- Matuliene, J., and Kuriyama, R. (2002). Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell 13, 1832-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, J.R., and Landis, S.C. (1971). The distribution of spindle microtubules during mitosis in cultured mammalian cells. J. Cell Biol. 49, 468-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Setou, M., Kaneshiro, K., and Hirokawa, N. (2001). All kinesin superfamily protein, KLF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98, 7004-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini, G., Harley, A.S., and Glover, D.M. (2003). Localisation of Pavarotti-KLP in living Drosophila embryos suggests roles in re-organising the cortical cytoskeleton during the mitotic cycle. Mol. Biol. Cell 14, 4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini, G., Mathe, E., and Glover, D.M. (2002). Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J. Cell Sci. 115, 725-736. [DOI] [PubMed] [Google Scholar]
- Minoshima Y. et al. (2003). Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 4, 549-560. [DOI] [PubMed] [Google Scholar]
- Mishima, M., Kaitna, S., and Glotzer, M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41-54. [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.P., Jiang, W., Schoehn, G., Hunter, T., and Margolis, R.L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, J.M., and Biesele, J.J. (1977). Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73, 672-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, J.M., and McIntosh, J.R. (1982). Isolation and initial characterization of the mammalian midbody. J. Cell Biol. 94, 654-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef, R., Preisinger, C., Sutcliffe, J., Kopajtich, R., Nigg, E.A., Mayer, T.U., and Barr, F.A. (2003). Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162, 863-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., Lombillo, V.A., Kuriyama, R., and McIntosh, J.R. (1992). A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 359, 543-547. [DOI] [PubMed] [Google Scholar]
- Norman, J.C., Jones, D., Barry, S.T., Holt, M.R., Cockcroft, S., and Critchley, D.R. (1998). ARF1 mediates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fiber formation in intact and permeabilized Swiss 3T3 fibroblasts. J. Cell Biol. 143, 1981-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, J., Bossinger, O., Rose, D., Strome, S., and Saxton, W. (1998). A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8, 1133-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby, B.B., and Dasso, M. (2003). The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15, 338-344. [DOI] [PubMed] [Google Scholar]
- Raich, W.B., Moran, A.N., Rothman, J.H., and Hardin, J. (1998). Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9, 2037-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport, R. (1961). Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 148, 81-89. [DOI] [PubMed] [Google Scholar]
- Rappaport, R., and Rappaport, B.N. (1974). Establishment of cleavage furrows by the mitotic spindle. J. Exp. Zool. 189, 189-196. [DOI] [PubMed] [Google Scholar]
- Rappaport, R., and Rappaport, B.N. (1983). Cytokinesis: effects of blocks between the mitotic apparatus and the surface on furrow establishment in flattened echinoderm eggs. J. Exp. Zool. 227, 213-227. [Google Scholar]
- Rappaport, R. (1986). Establishment of the mechanism of cytokinesis in animal cells. Int. Rev. Cytol. 105, 245-281. [DOI] [PubMed] [Google Scholar]
- Salmon, E.D., Goode, D., Maugel, T.K., and Bonar, B.D. (1976). Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J. Cell Biol. 69, 443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto, C., and Kuriyama, R. (1988). Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J. Cell Biol. 106, 431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., Hamill, D.R., Carter, J.C., Schumacher, J., and Bowerman, B. (2000). The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10, 1162-1171. [DOI] [PubMed] [Google Scholar]
- Somers, W.G., and Saint, R. (2003). A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29-39. [DOI] [PubMed] [Google Scholar]
- Somma, M.P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. (2002). Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13, 2448-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., Bergmann, D., Mohler, W.A., and White, J.G. (2001). Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11, 735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., Liu, H., Yates, J., Meyer, B.J., and Heald, R. (2002). Functional proteomic analysis of the mammalian midbody reveals conserved cell division components. Mol. Biol. Cell 13, 3a. [Google Scholar]
- Tatsumoto, T., Xie, X., Blumenthal, R., Okamoto, I., and Miki, T. (1999). Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phase, and involved in cytokinesis. J. Cell Biol. 147, 921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, S.P., and Wang, Y. (1996). Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135, 981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.