Abstract
Fungal APSES proteins regulate morphogenetic processes, including filamentation and differentiation. The human fungal pathogen Candida albicans contains two APSES proteins: the regulator Efg1p and its homologue Efh1p, described here. Overexpression of EFG1 or EFH1 led to similar phenotypes, including pseudohypha formation and opaque-white switching. An efh1 deletion generated no phenotype under most conditions but caused hyperfilamentation in an efg1 background under embedded or hypoxic conditions. This suggests cooperation of these APSES proteins in the suppression of an alternative morphogenetic signaling pathway. Genome-wide transcriptional profiling revealed that EFG1 and EFH1 regulate partially overlapping sets of genes associated with filament formation. Unexpectedly, Efg1p not only regulates genes involved in morphogenesis but also strongly influences the expression of metabolic genes, inducing glycolytic genes and repressing genes essential for oxidative metabolism. Using one- and two-hybrid assays, we further demonstrate that Efg1p is a repressor, whereas Efh1p is an activator of gene expression. Overall, the results suggest that Efh1p supports the regulatory functions of the primary regulator, Efg1p, and indicate a dual role for these APSES proteins in the regulation of fungal morphogenesis and metabolism.
INTRODUCTION
The APSES proteins represent a conserved class of transcriptional regulators that is unique to fungi, regulating cellular differentiation in the ascomycetes. In Saccharomyces cerevisiae, the Phd1 protein induces pseudohyphal growth, whereas the related Sok2 protein represses this growth form (Gimeno and Fink, 1994; Ward et al., 1996). The Asm1 protein promotes ascospore maturation in Neurospora crassa (Aramayo et al., 1996), and the StuA protein is required for conidiophore maturation in Aspergillus nidulans (Dutton et al., 1997). In the human fungal pathogen Candida albicans, the APSES protein Efg1p controls several morphogenetic processes. Efg1p regulates the yeast-to-hypha transition, it is required for the generation of chlamydospores, and it determines cell shape during white-opaque switching (Lo et al., 1997; Stoldt et al., 1997; Sonneborn et al., 1999a,b; Srikantha et al., 2000). Thus, all known APSES proteins regulate reversible interconversions between a spherical cell type (e.g., a budding yeast cell, conidiophore, ascospore, and chlamydospore) and an elongated filament (e.g., true hypha, pseudohypha, an opaque-form cell). In addition, as was shown for C. albicans Efg1p (Lo et al., 1997), APSES proteins may be involved in determining virulence of these organisms. However, the mechanisms by which APSES proteins stimulate cellular differentiation and regulate virulence are still unknown.
APSES proteins may act both as activators and repressors of gene expression because they stimulate reversible transitions between spherical and filamentous cells. The induction of true hyphae in C. albicans, by serum, for example, requires the presence of Efg1p. This led to the presumption that Efg1 acts as an activator of hypha-specific genes (Lo et al., 1997; Stoldt et al., 1997). Likewise, the generation of chlamydospores and of the white (yeast-like) cell form depends on Efg1p as a positive factor (Sonneborn et al., 1999a,b). However, EFG1 expression is rapidly repressed by negative autoregulation after the onset of hyphal development, and it has been shown that Efg1p, in conjunction with the Sin3p-dependent histone deacetylase complex, mediates this repression (Tebarth et al., 2003). In addition, EFG1 overexpression inhibits the formation of true hyphae, inducing pseudohyphae instead (Stoldt et al., 1997; Tebarth et al., 2003). An alternative pathway of hypha formation, which is operative in embedded or microaerophilic conditions, is repressed by Efg1p (Brown et al., 1999; Sonneborn et al., 1999a; Giusani et al., 2002). Overexpression of PHD1 in S. cerevisiae has the same effect as the deletion of its homologue SOK2 (Gimeno and Fink, 1994; Ward et al., 1996). Pseudohyphal growth is induced suggesting that Phd1p is an activator and Sok2p a repressor of pseudohypha formation and that the conserved APSES domain does not determine regulatory specificity. Genetic interactions with elements of a mitogen-activated protein kinase pathway also support an activator function of Phd1p (Lo et al., 1997), whereas Sok2p is known to repress meiosis via a defined regulatory sequence (Shenhar and Kassir, 2001). Interestingly, heterologous expression of Efg1p in S. cerevisiae suggests both repressor and activator functions for this regulator. EFG1 is able to complement a sok2 mutation (Shenhar and Kassir, 2001) and at high expression levels EFG1 induces pseudohyphal growth (Stoldt et al., 1997).
The multiple functions of Efg1p suggest that it might interact with coregulatory proteins that modulate its functional specificities. The conserved domain of ∼100 residues in APSES proteins contains a basic helix-loop-helix (bHLH) motif, which might dimerize with another bHLH protein as a prerequisite for their binding to E-boxes (Nasi et al., 2001). The bHLH domain of Efg1p has indeed been shown to bind E-box sequences in vitro (Leng et al., 2001), although in vivo gene regulation by an E-box motif has not been demonstrated for any APSES protein. Organisms often contain pairs of APSES proteins, for example, Phd1p and Sok2p in S. cerevisiae. Hence, we reasoned that a second APSES protein might exist in C. albicans and that this protein might modulate Efg1p function. Here, we report the identification of an Efg1p-homologue, Efh1p. We show that Efh1p does not have an obvious opposing role to Efg1p, as has been reported for Phd1p and Sok2p in S. cerevisiae (Gimeno and Fink, 1994; Ward et al., 1996). In contrast, Efh1p seems to modulate some of the functions of the main regulator Efg1p, which on a molecular level has the activity of a transcriptional repressor. We show that Efg1p and Efh1p not only regulate fungal morphogenesis but also favor a fermentative mode of metabolism of C. albicans. These results suggest that APSES proteins may coordinate morphogenetic and metabolic processes in fungi.
MATERIALS AND METHODS
Strains and Media
Strains are listed in Table 1. To isolate strains homozygous at the MTL locus, we isolated derivatives able to grow on sorbose as their sole carbon source (Magee and Magee, 2000). The identity of strains as MTLa was established by colony polymerase chain reaction (PCR) (primers MTLa-For, 5′-TTGAAGCGTGAGAGGCTAGGAG-3′; MTLa-Rev, 5′-ATCAATTCCCTTTCTCTTCGATTAGG-3′) or as MTLa (primers MTLα-For, 5′-TTCGAGTACATTCTGGTCGCG-3′; MTLα-Rev, 5′-TGTAAACATCCTCAATTGTACCCGA-3′). Phenotypic switching in homozygous MTL strains was monitored after pregrowth on modified Lee's medium (Lee et al., 1975), followed by plating on SC medium containing 5 μg/ml phloxine B and growth at 25°C for 7 d (Miller and Johnson, 2002). Strains were grown in YPD, SC, SCAA or on supplemented SD minimal medium (Sherman et al., 1986); YPS medium is identical to YPD but contains 2% sucrose as carbon source. S4D medium is SD medium containing 4% glucose. Serum plates contained 10% horse serum and 2% agar. Transformation was done by the spheroplast method (Sherman et al., 1986). To fully induce the PCK1 promoter cells were grown in SCAA medium for several generations to a final OD600 = 0.8-1.0 (Stoldt et al., 1997). For growth in microaerophilic conditions, plates were placed in an anaerobic jar by using CampyGen bags (Oxoid, Basingstoke, Hampshire, England) to generate an atmosphere of ∼6% oxygen. Cells were embedded in YPS-Agar essentially as described previously (Brown et al., 1999) except that a thin layer of YPS-agar (8 ml) was poured onto the cell-containing layer. This procedure prevented spreading of colonies on the agar surface and, by slowing filamentation, enhanced morphogenetic differences between efg1 single and efg1 efh1 double mutants.
Table 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Reference/source |
|---|---|---|
| C. albicans strains | ||
| SC5314 | prototrophic | Fonzi and Irwin (1993) |
| CAF2-1 | Fonzi and Irwin (1993) | |
| CAF2-1/01α | as CAF2-1, but MTLα/MTLα | This work |
| CAF2-1/01α-op | as CAF2-1/01α, but opaque growth form | This work |
| CAI4 | Δura3::imm434/Δura3::imm434 | Fonzi and Irwin (1993) |
| CA18 | ade2::hisG/ade2::hisG Δura3::imm434/Δura3::imm434 | Fonzi and Irwin (1993) |
| C4/c, C4/d | as CAI4, but EFH1/efh1Δ::FRT-SAP2p-FLP-URA3-FRT | This work |
| C4/c4, C4/d6 | as CAI4, but EFH1/efh1Δ::FRT | This work |
| C4/d6-3 | as CAI4, but efh1Δ::FRT/efh1Δ::FRT-SAP2p-FLP-URA3-FRT | This work |
| efh/02α | as C4/d6-3, but MTLα/MTLα | This work |
| efh/02α-op | as efh/02α, but opaque growth form | This work |
| C4/d63-1, C4/d63-3 | as CAI4, but efh1Δ::FRT/efh1Δ::FRT | This work |
| JKC19 | as CAI4, but cph1Δ::hisG/cph1Δ::hisG-URA3-hisG | Liu et al. (1994) |
| HLC52 | as CAI4, but efg1Δ::hisG/efg1Δ::hisG-URA3-hisG | Lo et al. (1997) |
| Bca09-4 | as CAI4, but efg1Δ::hisG/efg1Δ::hisG-URA3-hisG | Braun and Johnson (2000) |
| HLC67 | as CAI4, but efg1Δ::hisG/efg1Δ::hisG | Lo et al. (1997) |
| HLC67[pDB35]a | HLC67 carrying pDB35, made MTLa/MTLa | This work |
| HLC74 | as CAI4, but efg1Δ::hisG/efg1Δ::hisG (EFG1) | Lo et al. (1997) |
| HLC54 | as HLC52, but cph1Δ::hisG/cph1Δ::hisG | Lo et al. (1997) |
| H/1.22, H/1.26 | as HLC67, but efh1Δ::hisG/efh1Δ::hisG-URA3-hisG | This work |
| H/1.22-1, H/1.26-1 | as HLC67, but efh1Δ::hisG/efh1Δ::hisG | This work |
| CRC101-103 | as CAI8, but containing ADH1-lacZ::ADE2 | Russell and Brown, unpublished data |
| CRC104-106 | as CAI8, but containing lexA-ADH1-lacZ::ADE2 | Russell and Brown, unpublished data |
| Red3/6 | ade2/ade2, phenotypic switching strain | Srikantha et al. (2000) |
| Transformation vectors | ||
| pRC18 | URA3-marked CARS2-vector | Stoldt et al. (1997) |
| pBI-1 | PCK1-promoter in URA3-marked CARS2-vector | Stoldt et al. (1997) |
| pAPE(2)/ADE | PCK1-promoter-EFG1 fusion in CARS2 ADE2-vector | Sonneborn et al. (1999b) |
| pRC2312P-H | PCK1-promoter-EFG1 fusion in pBI-1 | Stoldt et al. (1997) |
| pDB35 | PCK1-promoter-EFH1 fusion in pBI-1 | This work |
| pBT-4 | CARS2 ADE2-vector | Sonneborn et al. (1999b) |
| pBT-44 | PCK1-promoter in CARS2 ADE2-vector | This work |
| pBT-145 | PCK1-promoter-EFH1 fusion in pBT-4 | This work |
| pTD13 | PCK1-promoter-HA-EFH1 fusion in pBI-1 | This work |
| pBT-34 | Major EFG1-promoter-LAC4 fusion in CARS2 URA3-vector | Tebarth et al. (2003) |
| p89a | Major EFG1-promoter-RLUC fusion in URA3-vector | This work |
| pLR6 | EFH1 promoter-RLUC fusion in URA3 vector | This work |
| CIp-LexA | URA3-marked integrating vector containing ACT1p-lexA | Russell and Brown, unpublished data |
| CIp-LexA-Efg1 | as CIp-LexA, but containing ACT1p-lexA-EFG1 | This work |
| CIp-LexA-Efh1 | as CIp-LexA, but containing ACT1p-lexA-EFH1 | This work |
| pGBD-C1 | TRP1 vector with GAL4-BD | James et al. (1996) |
| pGAD-C1 | LEU2 vector with GAL4-AD | James et al. (1996) |
| pDB17 | pGBD-C1 containing EFG1 | This work |
| pDB16 | pGAD-C1 containing EFG1 | This work |
| pDB34 | pGBD-C1 containing EFH1 | This work |
| pDB33 | pGAD-C1 containing EFH1 | This work |
| pSKM69 | pGBD-C1 containing efh1ΔN1 | This work |
| pSKM68 | pGAD-C1 containing efh1ΔN1 | This work |
| pCS5 | pGBD-C1 containing efh1ΔN2 | This work |
| pCS6 | pGAD-C1 containing efh1ΔN2 | This work |
| pCS7 | pGBD-C1 containing efh1ΔN3 | This work |
| pCS8 | pGAD-C1 containing efh1ΔN3 | This work |
EFH1 Deletion
The 5′region of EFH1 was PCR amplified using primers EFH-FLP1 (5′-TGG GTA CCG GCC TGA TTA GAA TAT GAT TTC CG-3′; KpnI, underlined) and EFH-FLP2 (5′-AAC TCG ACC GAG TGA CAA ACT AAT AGC AGAC-3′) and inserted into pUC18 (SureClone; Amersham Biosciences UK), resulting in plasmid pDB36. Similarly, the 3′ region was PCR amplified using primers EFH-FLP3 (5′-ATGCGGCCGCGGCCAATTTTCTTCAAATATTCTGG-3′; NotI, underlined) and EFH-FLP4 (5′-CAG AGCTCCTGGACCTTTCCTACCTGGATTCTC-3′; SacI, underlined) and subcloned to generate pDB37. A 1035 base pairs NotI/SacI fragment from pDB37 was cloned into NotI/SacI-digested pSFU1 (Morschhäuser et al., 1999) flanking the “Ura-flipper,” resulting in pDB40. A 1043-base pair KpnI fragment from pDB36 was cloned into the KpnI site of pDB40 to generate pDB43. The 4.89-kb SacI fragment from pDB43 was transformed into strain CAI4 to disrupt the first EFH1 allele. Removal of the FLP-URA cassette was carried out as described previously (Morschhäuser et al., 1999) and the procedure was repeated to disrupt the second allele.
The Ura-blaster method was used to make a double efh1 efg1 mutant (Fonzi and Irwin, 1993). First, the 1-kb KpnI fragment from pDB36 was cloned into p5921 to generate pSKM49. The 3′ region of EFH1 was PCR amplified using primers SKM-BH1-EFH3′-1 (5′-ATTGGATCCAATTTTCTTCAAATATTCTGG-3′) and SKM-BglII-EFH3′-2 (5′-ATTAGATCTACCTGGATTCTCTCAAC-3′) (BamHI, BglII, underlined) and cloned into pUC18, resulting in pSKM47. A 1-kb BamHI/BglII fragment from pSKM47 was cloned into pSKM49 to construct pSKM50. The SacI-SphI fragment from pSKM50 was used to disrupt EFH1 alleles in strain HLC67 (Lo et al., 1999). The genotype of each mutant was confirmed by southern blotting (see Supplemental Data, Figure 1).
EFH1 Plasmids
EFH1 was PCR amplified from genomic DNA by using primers AN1-BHIATG (5′-TTGGATCCATGAATGGTATTATGACG-3′) and AN3-Stp-BHI (5′-TTGGATCCGTTTATCATAATGTTTTGTG-3′ (BamHI site, underlined; coding region, italics). The PCR fragment was cloned into pUC18 to generate pDB30. EFH1 overexpression plasmids were constructed by insertion of the EFH1 BamHI-fragment downstream of the PCK1 promoter into the BglII site of plasmid pBI-1 (Stoldt et al., 1997) or of plasmid pBT-44 to generate pDB35 (URA3) and pBT-145 (ADE2), respectively. To express an HA-tagged version of Efh1p the BamHI fragment of pDB30 was subcloned first into the BamHI site of YCpIF17 (Foreman and Davis, 1994) and the HA-EFH1 fusion of this plasmid (pTD11) was subsequently amplified by primers HAEFH1(ATG)+HincII (5′-TTAGTTAACTTATATGAGTCGATACCCATAC-3′) and HAEFH(STOP)+ HincII(5′-TTAGTCAACTTCATAATGTTTTGTGAAC-3′) (HincII, underlined). The HincII-digested PCR-fragment was cloned into the filled-in BglII site of pBI-1 to generate pTD13.
To join the EFH1 promoter to the RLUC reporter, we amplified the 2-kb genomic region upstream of the EFH1 open reading frame (ORF) by PCR from genomic DNA of strain SC5314, by using primers EFH1p-5 (5′-TTAGGATCCAGTTTACCCGAAATCTGTG-3′) and EFH1p-3 (5′-TTAGGATCCGTGAATATACTTATAACGAG-3′) (BamHI, underlined). The BamHI-digested PCR fragment was cloned into pUK21; into the SpeI site of the resulting plasmid pLR3 we inserted the 1-kb NheI-XbaI fragment RLUC fragment of pRL-Null (Promega, Madison, WI) to construct pLR4. The 3.1-kb SpeI fragment of pLR4 was then inserted into the XbaI site of the URA3-plasmid p1367/1 (Losberger and Ernst, 1989), resulting in pLR6. pLR6 was integrated into the genomic EFH1 promoter by digestion with BstI 107I and transformation of strain CAI4. For comparison an integrating URA3 plasmid, pBT89a, containing a fusion of the major EFG1 promoter to RLUC (Tebarth et al., 2003) was cut with HpaI and integrated into the EFG1 promoter of strain CAI4.
Transcriptional Profiling
DNA microarrays containing 6039 C. albicans ORFs of strain SC5314 (∼98% of total) in duplicate were purchased from Eurogentec (Seraing, Belgium). Transcriptional profiling was performed on 50 ml of cells grown in YPD or SCAA medium at 30°C to an OD600 nm = 0.5; alternatively, cells were pregrown in YPD medium at 30°C, starved in water for 1 h at 30°C, and then diluted and incubated in 10% horse serum at 37°C for 30 min (germ tubes were beginning to form at this time point). The isolation of total RNA, preparation of Cy3- and Cy5-cDNA and pairwise hybridization to DNA microarrays were performed according to Galar Fungail standard operating procedures (www.pasteur.fr/recherche/unites/Galar_Fungail/) with minor modifications. cDNAs were purified by the Qiaquick PCR purification kit (QIAGEN, Valenica, CA) and eluted from columns by twice rinsing with 50 μl of water (42°C). The eluate was concentrated using Microcon-YM30 columns (Millipore, Billerica, MA) to a volume of 10 μl. Ten microliters each of Cy3- and Cy5-labeled cDNA and herring sperm DNA (10 mg/ml) were mixed, denatured at 95°C for 2 min, and the quickly chilled on ice. Eighty microliters of hybridization mixture was added, and the solution was allowed to float onto DNA microarrays slides, which were covered with a rimmed cover glass (Erie Scientific, Portsmouth, NH). Slides were placed in a hybridization chamber (Corning, Palo Lato, CA) and hybridized for 24 h at 42°C by immersion in a water bath. After washing and drying, microarrays were scanned by a FLA-8000 scanner (Fuji, Tokyo, Japan) at a resolution of 10 μm. Cy3- and Cy5-fluorescence signals were measured at 532 and 635 nm, respectively. For quantitation of spot and background fluorescence the program AIDA Array Metrix (Raytest, Pittsburgh, PA) was used.
Data from at least three independent experiments using independent cultures and including one dye-swap experiment were evaluated with the GeneSpring (Silicon Genetics, Redwood City, CA) software. The Cy3-/Cy5-ratios were first normalized based on the overall fluorescence intensity and then exported into Excel spreadsheets. Because each ORF was spotted twice on each array and at least three independent experiments were carried out, at least six signal intensity ratios reflecting the activity of a test sample to a reference sample were compared. The data were statistically analyzed using the SAM program (www-stat.stanford.edu/~tibs/SAM), which calculated q-values reflecting variation among the six ratios (a low q-value indicates a low level of variation). The predicted minimal false discovery rate (FDR) value in SAM evaluations in most cases had a value close to 5% (see Supplemental Data, Tables 1-8). For comparisons of data sets, a delta value was chosen corresponding to an FDR value of 5%; in one case (strain HLC52 grown in YPD medium), >1000 regulated genes were predicted by this parameter, and a delta value corresponding to a FDR of 1% was chosen. For Venn diagrams and for gene listings, we applied more stringent criteria, defining genes as significantly regulated if they were regulated by at least a factor of 1.5-fold. Cluster analyses and Venn diagrams were carried out with the GeneSpring program, by using standard value settings. Gene designations were according to the annotation of the Candida DB Web server (http://genolist.pasteur.fr/CandidaDB/).
One- and Two-Hybrid Analyses
Details of the C. albicans one-hybrid system are described elsewhere (Russell and Brown, unpublished data). Briefly, expression vectors encoding Staphylococcus LexA protein fusions to Efg1p or Efh1p were constructed by PCR amplification of the EFG1 or EFH1 ORFs and inserting them into CIp-LexA, resulting in plasmids CIp-LexA-Efg1 and CIp-LexA-Efh1, respectively. The 5′ PCR primers used introduced a (Gly)3-Pro-(Gly)2 linker between the amino-terminal LexA domain and the carboxy terminal domains of Efg1p or Efh1p. These plasmids were introduced into C. albicans reporter strains that carried pCR-lacZ (no lexA operator upstream of basic ADH1 promoter) or pCR-OplacZ (containing lexA operator) integrated into the ade2::hisG locus (Russell and Brown, unpublished data).
For two-hybrid experiments, plasmids encoding protein fusions to the Gal4p-DNA-binding domain (BD) or the Gal4p-transcriptional activation domain (AD) were constructed by inserting appropriate DNA fragments into plasmids pGBD-C1 and pGAD-C1, respectively (James et al., 1996). The 1.5-kb EcoRI/BamHI fragment of pUC19-EFG1 was used to construct EFG1 expression vectors pDB17 (BD) and pDB16 (AD). Similarly, the 2.1-kb BamHI fragment of pDB30 was used to construct EFH1 vectors pDB34 (BD) and pDB33 (AD). Subfragments of the pDB30 insertion fragment also were cloned into pGBD-C1 or pGAD-C1 to test the effect of C-terminal shortening of Efh1p: (a) deletion efh1ΔN1: the BamHI-BglII fragment generated pSKM69 (BD) and pSKM68 (AD); (b) deletion efh1ΔN2: the BamHI-PstI fragment generated pCS5 (BD) and pCS6 (AD); and (c) deletion efh1ΔN3: the BamHI-ScaI fragment generated pCS7 (BD) and pCS8 (AD). Plasmids were transformed pairwise into strain PJ69-4A (MATa trp1-901 leu2,3-112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) and assayed for prototrophy for histidine and adenine, as well as for β-galactosidase (LacZ) activity (James et al., 1996).
Reporter Gene Assays
RLUC activity was determined using the Dual-Luciferase Reporter Assay (Promega). To 50-100 μl of LARII reagent 10 μl of crude extract was added and after 10 s at room temperature, 50-100 μl of Stop&Glo reagent was used to stop the reaction. Immediately afterward, light emission was assayed in a luminometer (Fluorskan Ascent FL; Labsystem, Helsinki, Finland). β-Galactosidase activity in cells, expressed in Miller units, was determined by standard procedures on plates containing X-Gal or in liquid by using O-nitrophenyl β-d-galactopyranoside (ONPG) as indicators (Miller, 1972).
RESULTS
EFH1 Is Related to EFG1
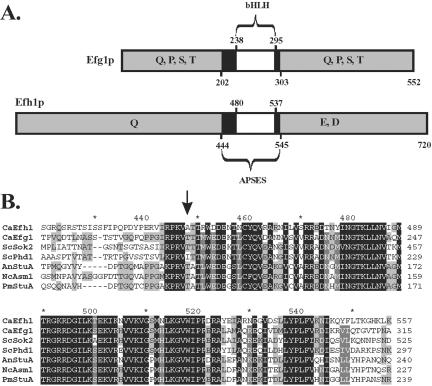
Analysis of C. albicans genomic sequences (http://www.sequence.stanford.edu/group/candida) by using the BLAST algorithm revealed a single EFG1 homologue (orf6.4659), which we named EFH1 (EFG1 homologue). The EFH1 ORF is 45% identical to the EFG1 ORF, and it encodes a hypothetical protein of 720 residues (81,373 Da), which is 23% identical to the 552 aa-Efg1 protein. Efh1p displays high similarity to Efg1p and other APSES proteins within a domain encompassing residues 443-546 (65% amino acid sequence identity) (Figure 1). This APSES domain contains a bHLH subdomain from residues 480-537. Efh1p lacks the putative protein kinase A phosphorylation target in the APSES domain corresponding to residue 206 in Efg1 (Bockmühl and Ernst, 2001). Outside the APSES domain, Efh1p and Efg1p share little sequence similarity except for the occurrence of glutamine-rich stretches. In Efh1p, these occur at residues 85-102 and 200-223. A unique feature of Efh1p is a stretch of acidic residues between residues 615 and 643.
Figure 1.
APSES proteins in C. albicans. (A) Schematic structures of Efg1 and Efh1 proteins. The location of bHLH and APSES domains and of regions enriched in the indicated amino acids is shown. (B) Comparison of APSES proteins. The arrow indicates the position of a potential phosphorylation site in Efg1p and Sok2p, which is absent in Efh1p and Phd1p. Proteins from the following species are compared: C. albicans (Efh1p/Efg1p), S. cerevisiae (Phd1p/Sok2p), A. nidulans (StuA), N. crassa (Asm1), and P. marneffei (StuA). Black boxes, identical residues; gray boxes, similar residues in a subset of APSES proteins.
Northern analyses revealed two low abundance EFH1 transcripts of ∼2.5 and 2.7 kb (our unpublished data), indicating that EFH1 is expressed at low levels. To compare the transcriptional activities of EFH1 and EFG1 loci, we integrated the RLUC reporter in place of the respective ORFs, by chromosomal integration of plasmids pLR6 and p89a to generate EFH1p-RLUC and EFG1p-RLUC fusions, respectively. Luciferase activities were ∼10-fold lower for the EFH1p-RLUC fusion compared with the EFG1p-RLUC fusion during growth in minimal medium (0.003 ± 0.001 vs. 0.025 ± 0.002 relative light units/μg protein, respectively). This suggests that the EFH1 promoter has an ∼10-fold lower activity compared with the EFG1 promoter under these conditions.
Phenotypes of efh1 Mutants
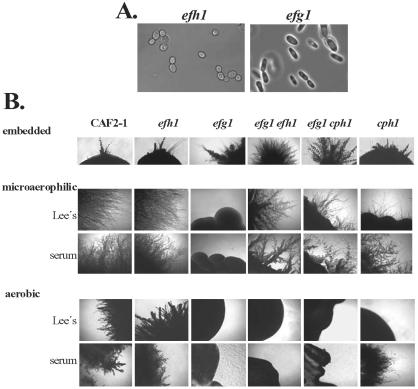
A homozygous efh1/efh1 mutant was constructed to examine the function of EFH1 (strain C4/d6-3). In addition, heterozygous EFH1/efh1 strains were obtained in this process (strains C4/c, d). The growth characteristics of these mutants were identical to the isogenic wild-type strain under numerous conditions, and no obvious antifungal resistance or stress phenotypes were detected. The cellular morphologies of efh1 mutants were similar to the wild-type strain, in contrast to the rod-like shape of efg1 mutants (Figure 2A). Hyphal development was normal on solid and in liquid media (Lee's, Spider, low-nitrogen and serum-containing media) and under embedded conditions (see below). In addition, chlamydospore formation was normal in cells lacking Efh1p. To test whether the inactivation of Efh1 effected phenotypic switching, we measured white-opaque switching in the homozygous MTLα/MTLα efh1/efh1 strain efh/02α. Isogenic wild-type and efh1 cells switched from white to opaque at a frequency of ∼5% as expected (Miller and Johnson, 2002). Likewise, opaque cells that arose in this experiment frequently switched back to the white form (our unpublished data). Thus, switching occurred normally in efh1 cells, in contrast to efg1 cells that seemed locked in an opaque-like state (Sonneborn et al., 1999b). Collectively, our data indicate that an efh1 mutation does not influence the various morphogenetic processes that are strongly affected by an efg1 mutation (morphogenesis and phenotypic switching).
Figure 2.
Morphological phenotypes of efh1 and efg1 mutants. (A) Cellular morphology of an efh1 mutant (strain C4/d63-1) and an efg1 mutant (strain HLC52) visualized by phase contrast microscopy. (B) Comparison of colony phenotypes. Strains CAF2-1 (wt), C4/d63-1 (Δefh1), HLC52 (Δefg1), H/1.22 (Δefh1 Δefg1), HLC54 (Δefg1 Δcph1), and JKC19 (Δcph1) were grown for 5 d at 24°C embedded in agar or at 30°C in a jar generating microaerophilic conditions or aerobically.
Sok2p and Phd1p have opposing regulatory functions in S. cerevisiae. Hence, we examined the possibility of genetic interactions between EFH1 and EFG1 by comparing the phenotypes of efh1 and efg1 single mutants with efh1 efg1 double mutants (strains C4/d63-1, HLC67, and H/1.22). During growth in standard hypha-inducing conditions the efh1 efg1 double mutant showed similar defects in hypha and chlamydospore formation to an efg1 single mutant, generating the rod-like cells typical of an efg1 mutant (our unpublished data).
We further tested morphogenesis under hypoxic or embedded conditions, in which Efg1p seems to repress a poorly defined alternative signaling pathway regulating hypha formation (Brown et al., 1999; Sonneborn et al., 1999a). When C. albicans cells were grown embedded in agar at 24°C, efg1 cells displayed increased hypha formation compared with isogenic wild-type or efh1 cells (Figure 2B). In contrast, efg1 cells did not form hypha in a jar generating microaerophilic conditions (∼6% oxygen), whereas efh1 cells formed hyphae like the wild-type parent under these conditions (Figure 2B).
We observed a synthetic phenotype between efh1 and efg1 mutations with regard to filament formation during growth under some embedding conditions (see MATERIALS AND METHODS). Compared with the efh1 and efg1 single mutants, the efh1 efg1 double mutants formed colonies with lateral hyphae faster and filamentation was more extensive. Furthermore, under microaerophilic conditions the efg1 efh1 double mutant filamented vigorously, like wild-type and efh1 cells, whereas the efg1 single mutant did not filament; also, in these conditions, an efg1 mutation did not block filamentation occurring in an cph1 genetic background. (Figure 2B). We interpret this to mean that the simultaneous inactivation of Efg1p and Efh1p (or Cph1p) derepresses an alternative pathway of filamentation in an oxygen-dependent manner. Under aerobic conditions Efg1p is required to form hyphae. However, this requirement for Efg1p is released by the additional inactivation of Efh1p, but only under low-oxygen or embedding conditions. In more hypoxic conditions (i.e., in a stream of nitrogen), however, the alternative pathway was almost fully induced in an efg1 mutant, and its excessive filamentation was not greatly increased by an efh1 mutation (our unpublished data). Thus, it seems that an efh1 phenotype is observed especially in intermediate conditions, in which neither the standard nor the alternative pathways of filamentation are fully induced.
EFH1 Overexpression Phenotypes
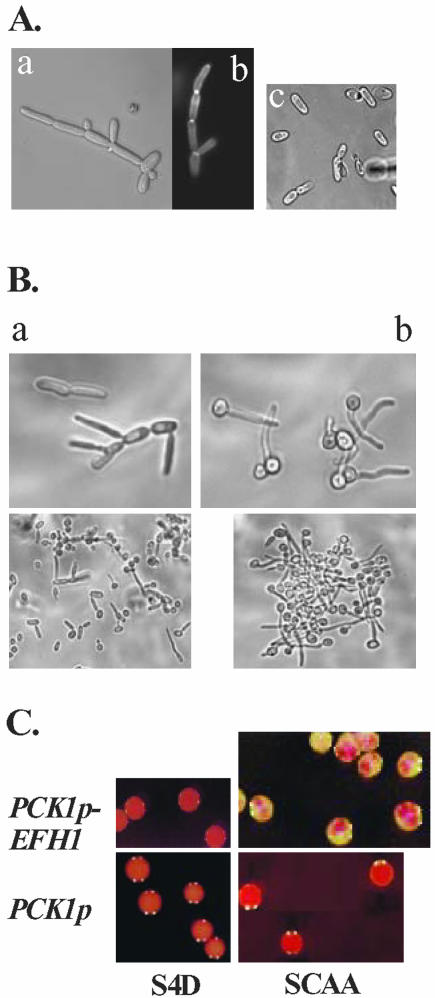
The deletion of the EFH1 locus alone had no obvious effect upon morphogenesis in C. albicans. Hence, we investigated whether EFH1 overexpression would have an effect on this process. Strains expressing EFH1 under the regulatable PCK1 promotor grew as regular or slightly elongated yeast cells under repressing conditions, but they formed pseudohyphae after induction of the PCK1 promoter. These cells budded apically and had calcofluor white-stainable constrictions at cellular junctions (Figure 3A). Interestingly, EFH1 overexpression did not lead to pseudohyphal formation in an efg1 genetic background (HLC67) (Figure 3A, c). However, EFG1 overexpression produced pseudohyphae in both an efg1 and an efh1 background (our unpublished data). Thus, although overexpression of either EFG1 or EFH1 induces pseudohyphal development, Efg1p would seem to be the main regulator of this process, because the EFH1-overexpression phenotype requires the presence of Efg1p.
Figure 3.
Phenotypes of EFH1 overexpression. (A) Generation of pseudohyphae. A pDB35-transformant of strain CAI4 (a and b) or of efg1 mutant HLC67 (c) was grown in SCAA medium before microscopy. Intercellular junctions are visualized by calcofluor white fluorescence (b). (B) Inhibition of true hypha formation. A pDB35-transformant of strain CAI4 was pregrown in SCAA medium (a) or SD medium (b) and shifted to the same medium containing 10% serum and grown for 60 min at 37°C. At low microscopic magnification (bottom photographs), the aggregate-formation by hyphae of SD-grown cells is visible. (C) Promotion of opaque to white phenotypic switching. The opaque form of strain Red3/6 was transformed with pBT-145 (PCK1p-EFH1) or with control vector pBT-44 (PCK1p) and transformants were grown at 25°C on PCK1p-repressing S4D or -inducing SCAA medium containing phloxine B. Note the speckled appearance of pBT-145 transformants on SCAA plates consisting mainly of the white form but containing red sectors indicative of the opaque form.
We showed previously that EFG1 overexpression blocks the formation of true hyphae (Tebarth et al., 2003). Interestingly, EFH1-overexpressing cells generated the same phenotype: pseudohyphal cells incapable of forming true hyphae in response to serum (Figure 3B). In contrast, cells expressing EFH1 at low levels did respond to serum, forming strongly aggregating germ tubes and true hyphae. Hypha production was still blocked in efg1 cells overexpressing EFH1, indicating that the requirement for Efg1p cannot be bypassed by EFH1 overexpression.
EFG1 overexpression causes rapid phenotypic switching from opaque to white cells (Sonneborn et al., 1999b). To test whether EFH1 overexpression affects this process, we examined the behavior of opaque Red3/6 cells carrying a PCK1p-EFH1 fusion (pBT-145) on media containing phloxine B (which stains opaque colonies pink, whereas white-form colonies are white). All of the cells formed pink colonies on PCK1p-repressing S4D-medium indicating that the opaque form was stable at low EFH1 expression levels (Figure 3C). Control transformants grown on SCAA medium remained purely opaque. In contrast, EFH1 overexpression on SCAA medium generated mostly white colonies, although some pink spots indicative of opaque cells did remain (Figure 3C). Thus, EFH1 overexpression was able to induce the opaque to white conversion, whereas it did not affect the reverse switching process (white-opaque) (our unpublished data). To test whether forced opaque to white conversion was dependent on Efg1p, we overexpressed EFH1 in the MTLa efg1- homozygous strain (HLC67[pDB35]a), which on phloxine plates formed opaque-like pink colonies (Sonneborn et al., 1999b). In this case, overexpression of EFH1 did not give rise to white form colonies. We conclude that opaque to white switching, which is triggered by EFH1 overexpression, is dependent on Efg1p.
Transcriptomes of Strains Lacking APSES Proteins
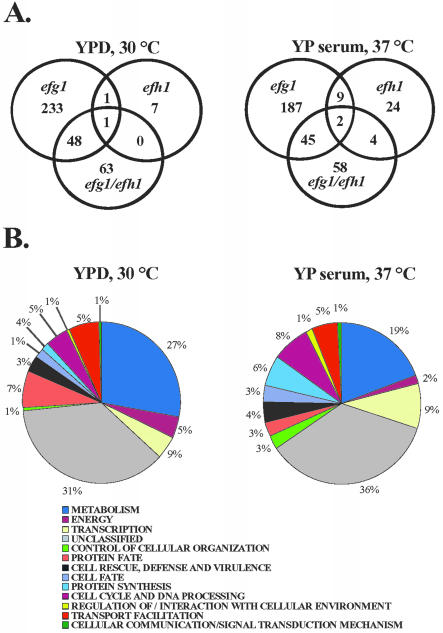
Transcript profiling was used to examine further the roles of the APSES proteins in C. albicans. DNA microarrays carrying an almost complete set of C. albicans ORFs were used for these experiments, in which the transcriptomes of efg1 and efh1 mutants were compared with wild-type cells under identical growth conditions. Genes that displayed reproducible and statistically significant changes in expression relative to wild-type cells were identified after exposure to hypha-inducing or -noninducing growth conditions (Figure 4A). Lists of all genes that displayed significant regulation by Efg1p and Efh1p are provided as supplemental data (Supplemental Data, Tables 1-6).
Figure 4.
Transcript profiles of C. albicans mutants lacking the APSES proteins Efg1p or Efh1p. (A) Venn diagrams of genes regulated by Efg1p and Efh1p. The wild-type strain CAF2-1 was used as reference for the efg1 mutant (HLC52), the efh1 mutant (C4/d6-3) and the efg1 efh1 double mutant (H/1.22), which were all grown in identical conditions: YPD/30°C (non-inducing) or for 30 min in hypha-inducing YP serum, 37°C. (B) Functional categories of genes regulated by Efg1p in the indicated conditions.
To explore the functions of the APSES proteins under conditions that promote growth in the yeast form, we compared the transcriptomes of wild-type and mutant strains in YPD medium at 30°C. The deletion of EFG1 altered the expression of 283 genes by >1.5-fold, with 100 genes being up-regulated and 183 being down-regulated. Only nine genes were affected by the deletion of EFH1 (8 genes were up- and 1 was down-regulated) (Figure 4A). Only two genes of unknown function (IPF4696 and IPF4328) were commonly affected in both single mutants. The simultaneous deletion of EFG1 and EFH1 showed a transcriptional pattern that overlapped with that of efg1 single mutants (49 genes). However, a new subset of 63 genes was affected in the efg1 efh1 double mutant that was not affected in the efg1 single mutant. This is consistent with the existence of synthetic interactions between EFG1 and EFH1. These results indicate that under standard growth conditions Efg1p (but not Efh1p) has an important role in gene regulation.
Categorization of the regulated genes according to their functional categories of the MIPS database (http://mips.gsf.de/genre/proj/yeast/searchCatalogFirstAction.do?db=CYGD) revealed that deletion of EFG1 alone or in combination with an EFH1 deletion had a striking effect on carbon metabolism (Figures 4B and 5; Supplemental Data, Table 2). Transcript levels for almost all glycolytic genes (e.g., FBA1, PFK1, and ENO1) and for genes involved in the metabolism of reserve sugars trehalose and glycogen (e.g., TPS2, TPS3, and GSY1) were reduced in an efg1 mutant by a factor of 2 or 3. In addition, transcript levels for several citric acid cycle enzymes (e.g., FUM12 and KGD1), enzymes of the mitochondrial ATPase complex, and glycine and formate catabolic enzymes were increased (Figure 6). In contrast, the inactivation of Efh1p affected few glycolytic genes only slightly (Figure 5 and Supplemental Data, Table 2).
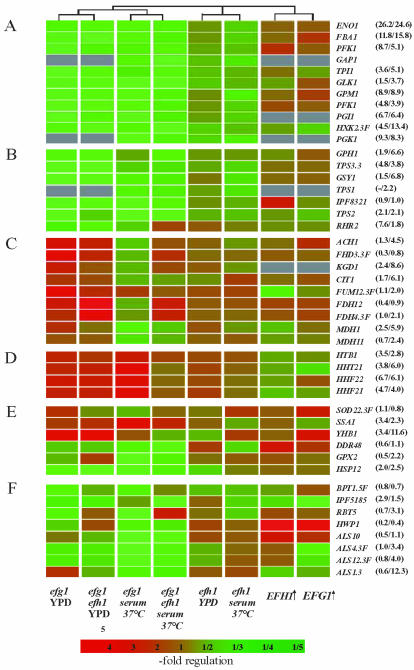
Figure 5.
Transcriptional regulation of individual genes by Efg1p or Efh1p. Representative genes responsible for specific cellular functions are grouped and their regulation in efg1 and efh1 single and double mutants and strains overexpressing EFG1 or EFH1 relative to the CAF2-1 wild-type strain is listed. Genes involved in glycolysis (A), reserve sugar metabolism (B), citric acid cycle (C), histone biosynthesis (D), stress response (E), and cell wall biosynthesis (F) are compared. Numbers next to gene designations indicate the relative transcript levels of such genes (transcript level of individual gene relative to the average level of all genes) in the wild-type strain CAF2-1, grown in YPD (30°C) and YP serum (37°C), respectively.
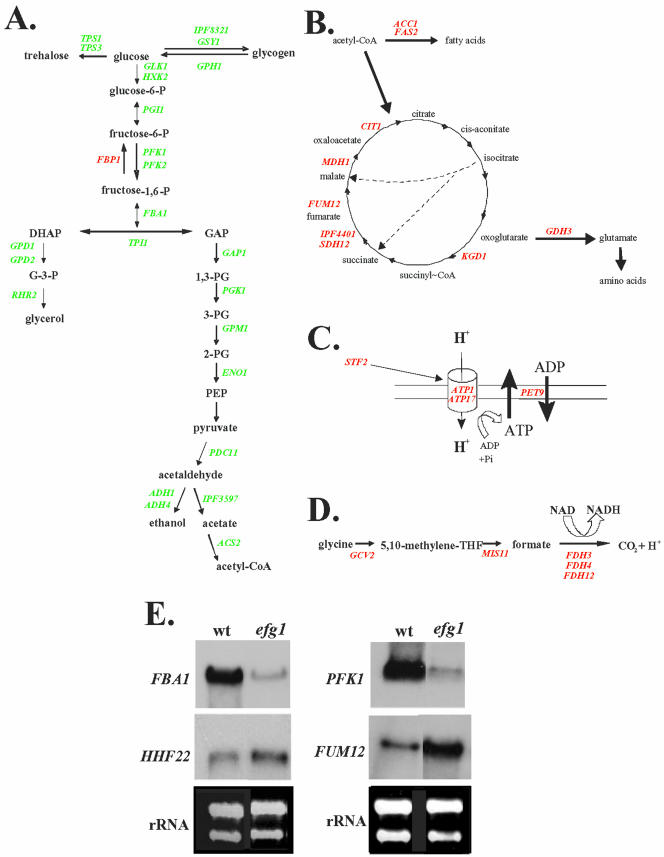
Figure 6.
Regulation of genes involved in carbon metabolism by Efg1p. Genes regulated by Efg1p are involved in glycolysis (A), citric acid cycle (B), ATP biosynthesis (C), and glycin degradation (D) (genes up-regulated by Efg1p, green; genes down-regulated by Efg1p, red). (E) Northern blots on 30 μg of RNA of strain CAF2-1 (wt) and strain HLC52 (efg1), by using probes for the indicated genes. Equal loading was verified by ethidium-bromide staining of rRNA (rRNA).
To confirm the transcriptional profiling data, we performed Northern analysis on a selection of Efg1p-regulated genes (Figure 6). The results confirmed the reduced levels of two glycolytic transcripts in the efg1 mutant (FBA1 and PFK1) and the increased levels of the HHF22 and FUM12 transcripts in the efg1 mutant (Figure 6E).
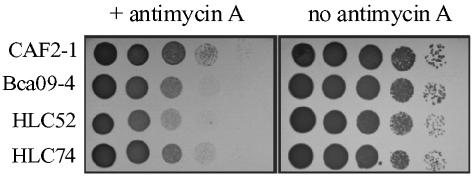
To verify the biological consequences of a presumed reduced glycolytic flux in efg1 strains, we compared its growth to a wild-type strain in the presence of antimycin A, which blocks ATP production by the respiratory chain. We indeed observed that two independently constructed efg1 mutants (strains Bca09-4 and HLC52) grew similar to the wild-type in medium without antimycin A, but in the presence of antimycin A were retarded in growth (Figure 7). Insertion of a single wild-type copy of EFG1 into the mutant background of strain HLC52 (strain HLC74) partially increased antimycin A-resistance as expected from a reduced EFG1 gene dosage (Fu et al., 2002). To confirm the enhanced antimycin A-sensitivity of the efg1 mutants, we also performed a mixing experiment, in which the wild-type CAF2-1 and the efg1 mutant HLC52 were grown in the same YPD culture. At the starting OD600 nm of 0.05 efg1 mutant cells amounted to 60% of cells in the mixed culture and slightly decreased to 40% after 12 h of growth (final OD600 nm = 12). In contrast, growth in the presence of 100 μM antimycin A for 12 h led to an strong decrease of efg1 mutant cells to 10% (final OD600 nm = 5). These results are consistent with reduced glycolytic ATP-production in efg1 mutants, suggesting a dual function of Efg1p in morphogenesis and metabolism.
Figure 7.
Supersensitivity of efg1 mutants to antimycin A. Wild-type strain CAF2-1 and efg1 mutants HLC52 and Bca09-4 (efg1), as well as an efg1 mutant reconstituted with EFG1 (HLC74) were grown in YPD and a series of tenfold dilutions was spotted on YPD agar containing 10 μg/ml antimycin A and grown at 30°C for 3 d.
Other genes regulated by Efg1p under noninducing conditions include genes encoding histones, stress response-proteins membrane transporters, and putative cell wall proteins (Figure 5). Another interesting group of genes that seems to be repressed by Efg1p encode for putative transcriptional activators (CTA21, CTA24, and CTA26), raising the possibility that Efg1 is a global regulator of gene expression. The highest Efg1p-dependent regulation (30-fold up-regulation) was observed for a putative multidrug resistance protein (IPF14540) (Supplemental Data, Table 1).
Transcriptomes during Morphogenesis
In a previous study, the transcriptomes of wild-type and efg1 strains were compared after growth in hypha-inducing conditions for several generations (Nantel et al., 2002). In this study, we focused upon the identification of genes whose expression depends on the APSES proteins during an early stage of hyphal development. We generated transcriptional profiles for efg1 and efh1 cells exposed to serum for only 30 min at 37°C, by using a wild-type strain (CAF2-1) as reference. At this time point ∼50% of wild-type cells showed short protrusions, which developed into germ tubes and ultimately formed hyphae during prolonged incubation. Under these hypha-inducing conditions, the deletion of EFG1 caused major changes in the transcriptome. The expression of 243 genes was affected by a factor of >1.5 in efg1 cells compared with wild-type cells. In contrast, the deletion of EFH1 had a relatively small effect, affecting the expression of 39 genes (Figure 4A). However, 58 new genes were affected by the efg1 efh1 double mutant, supporting the idea that EFG1 and EFH1 interact synthetically to regulate the expression of some C. albicans genes.
Again, glycolytic genes were expressed at lower levels in cells lacking Efg1p (Figures 4B and 5). However, in contrast to the analysis under yeast growth conditions, genes involved in oxidative metabolism were not upregulated in efg1 cells during hyphal development (e.g., CIT1, MDH1, KGD1, FDH3.3F, FDH4.3F, and FDH12) (Figure 5C). Thus, Efg1p seems to repress these genes under noninducing conditions, but not during hyphal development. This pattern might be due to the lack of glucose in the inducing medium used. Therefore, Efg1p-dependent repression of genes involved in oxidative metabolism might require the presence of glucose.
Transcript Profiling of EFG1- or EFH1-Overexpression
To identify genes that are regulated by high levels of APSES proteins, we performed transcript profiling of C. albicans cells overexpressing EFG1 (pRC2312P-H) or EFH1 (pDB35) from the PCK1 promoter. Cells containing the empty expression vector (pBI-1) were used as reference. The transcript levels of 85 and 81 genes were affected by elevated EFG1 or EFH1 expression, respectively (Figure 8; Supplemental Data, Tables 7 and 8).
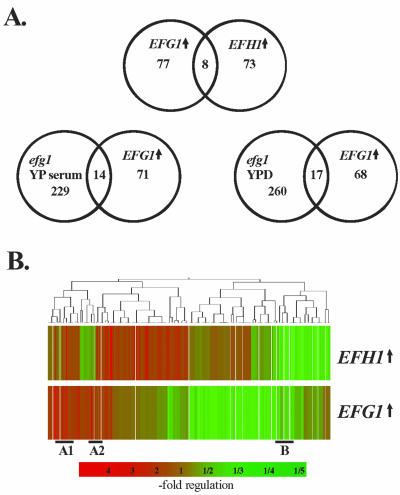
Figure 8.
Genes regulated by EFG1 or EFH1 overexpression. (A) Venn diagram of 129 genes regulated by EFG1- (EFG↑) or EFH1 (EFH↑) overexpression. Expression ratios of genes regulated in strain CAI4[pRC2312P-H] overexpressing EFG1 or in strain CAI4[pDB35] overexpressing EFH1 relative to a control transformant (CAI4[pBI-1]) grown identically were determined. Strains were grown in SCAA medium at 30°C. (B) Clustering of genes regulated by EFG1/EFH1 overexpression. Genes in sections designated A1, A2, and B are shown in more detail in Supplemental Data, Figure 4.
Among the 85 genes regulated by EFG1 overexpression, 53 genes were down-regulated (1.5- to 3.5-fold) and 32 genes were up-regulated (1.5- to 19-fold). The up-regulated genes encode cell wall proteins such as Hwp1p, Als10p, Rbt5p, Ece1p, and Phr1p, which are known to be associated with filamentous growth in C. albicans (Birse et al., 1993; Bailey et al., 1996; Fonzi, 1999; Sharkey et al., 1999; Braun and Johnson, 2000; Braun et al., 2000; Lane et al., 2001) (Figure 5F). The DDR48 gene, encoding a stress protein, and IPF1222 (SOD5), encoding a hypha-specific superoxide dismutase (Nantel et al., 2002), also were up-regulated. Most of these genes make up the overlap of 14 genes reduced by the absence of Efg1p in inducing conditions, and induced by EFG1 overexpression (Figure 8A). Whereas EFG1 down-regulated a majority of genes, EFH1 overexpression up-regulated most genes (53 up-regulated and 28 down-regulated genes). In agreement with the related phenotypes generated by EFG1 and EFH1 overexpression, the corresponding transcriptional profiles were similar (Figure 8B). Especially filament-specific genes such as HWP1 and ECE1 but also metabolic genes such as ADH5 and MET6 were up-regulated by EFG1 and EFH1 overexpression (Supplemental Data, Figure 4). These experiments also revealed that in general, a different set of genes is regulated during the formation of true hyphae (dependent on Efg1p) compared with pseudohyphal formation (triggered by EFG1 overexpression). Clearly, significant molecular differences exist between these morphological states.
Table 2.
Regulation of genes encoding metabolic enzymes
| Accession no. |
Fold regulation |
||||
|---|---|---|---|---|---|
| Gene | Description (CandidaDB) | efg1 | efg1-efh1 | efh1 | |
| Glycolysis | |||||
| ENO1 | CA3874 | Enolase I | 0.66 | 0.60 | 0.96 |
| FBA1 | CA5180 | Fructose-bisphosphate aldolase | 0.37 | 0.46 | 0.99 |
| GLK1 | CA0263 | Aldohexose specific glucokinase | 0.40 | 0.54 | 0.80 |
| GPM1 | CA4671 | Phosphoglycerate mutase | 0.40 | 0.56 | 0.99 |
| PFK1 | CA1834 | Phosphofructokinase, alpha subunit | 0.46 | 0.53 | 0.97 |
| PFK2 | CA3112 | Phosphofructokinase, beta subunit | 0.41 | 0.65 | 0.93 |
| PGI1 | CA3559 | Glucose-6-phosphate isomerase | 0.50 | 0.64 | 0.96 |
| TPI1 | CA5950 | Triose phosphate isomerase | 0.44 | 0.58 | 0.82 |
| HXK2.3F | CA0127 | Hexokinase II | 0.36 | 0.49 | 0.76 |
| Reserve | |||||
| carbohydrates | |||||
| TPS2 | CA5066 | Threalose-6-phosphate phosphatase | 0.45 | 0.83 | 0.89 |
| TPS3.3 | CA5505 | Trehalose-phosphate synthase, regulatory subunit | 0.41 | 0.49 | 0.92 |
| IPF8321 | CA2938 | Initiator of glycogen synthesis | 0.41 | 0.53 | 1.01 |
| GSY1 | CA5467 | Glycogen synthase | 0.45 | 0.51 | 1.04 |
| GPH1 | CA5206 | Glycogen phosphorylase | 0.35 | 0.57 | 0.98 |
| RHR2 | CA5788 | DL-glycerol phosphatase | 0.23 | 0.60 | 1.16 |
| Citric acid | |||||
| cycle | |||||
| ACC1 | CA5816 | Acetyl-coenzyme-A carboxylase | 1.5 | 1.42 | 1.08 |
| ACH1 | CA0345 | Acetyl-coenzyme-A hydrolase | 2.3 | 1.82 | 1.10 |
| FUM12.3F | CA4351 | Fumarate hydratase, 3-prime end | 3.1 | 1.60 | 1.10 |
| FUM12.5F | CA4349 | Fumarate hydratase, 5-prime end | 2.1 | 1.57 | 1.05 |
| GDH3 | CA1579 | NADP-glutamate dehydrogenase | 2.5 | 0.79 | 1.07 |
| IPF4401 | CA0903 | Succinate dehydrogenase | 1.7 | 0.99 | 1.24 |
| KGD1 | CA3149 | 2-oxoglutarate dehydrogenase | 2 | 1.22 | 1.15 |
| MDH1 | CA5164 | Mitochondrial malate dehydrogenase precursor | 1.6 | 1.21 | 1.20 |
| SDH12 | CA2470 | Succinate dehydrogenase | 1.5 | 1.08 | 1.09 |
| ACS2 | CA2858 | Acetyl-coenzyme-A synthetase | 1.51 | 1.5 | 1.02 |
Expression of genes in an efg1 mutant (HLC52), an efh1 mutant (C4/d6-3) and an efg1 efh1 mutant (H/1.22) relative to the wild-type CAF2-1. Cells were grown in YPD medium at 30°C prior to transcriptional profiling.
Activity of Efg1p and Efh1p in One- and Two-Hybrid Assays
Efg1p is assumed by some to act as a transcriptional activator on the basis that the expression of hypha-specific genes depends upon Efg1p. However, other data suggest that Efg1 acts as a repressor. Therefore, we performed one- and two-hybrid assays to test directly whether the APSES proteins act as transcriptional activators or repressors.
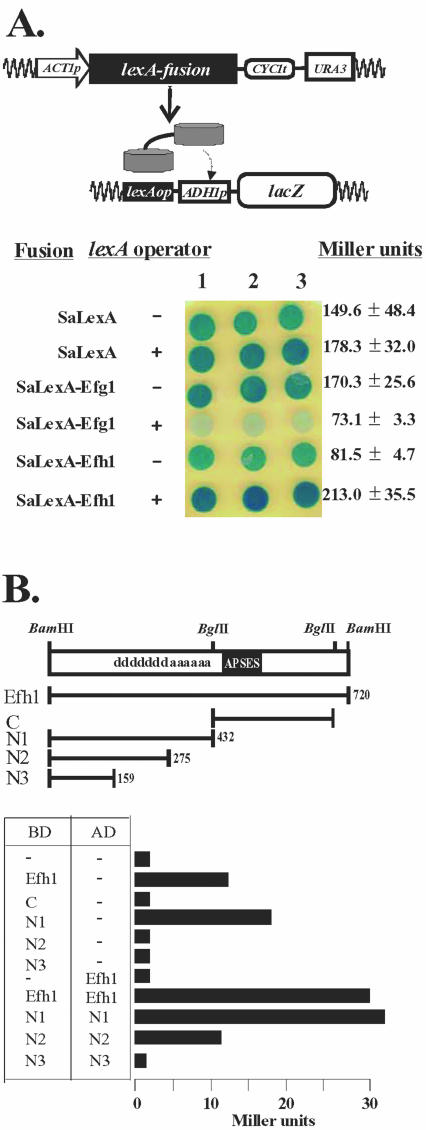
In a first set of experiments, we used a recently developed one-hybrid assay, which allows an assessment of transcriptional regulatory activity in C. albicans (Russell and Brown, unpublished data). In these experiments LexA fusions to Efg1p or Efh1p were tested for their ability to repress or activate a lacZ reporter gene. This reporter was transcribed by a basal ADH1 promoter fused to a lexA operator (Figure 9A, top). Transformants expressing lexA alone showed a basal level of β-galactosidase activity as indicated by blue staining on X-gal plates (Figure 9A, bottom). In contrast, β-galactosidase activity was significantly reduced in cells producing a LexA-Efg1 fusion. This reduction did not occur in a reporter lacking the lexA operator. Hence, Efg1p seems to act as a transcriptional repressor in C. albicans. Further experiments showed that the repressing function of the LexA-Efg1p was not affected by hypha-inducing stimuli, such as 10% serum or growth in microaerophilic conditions. In contrast to Efg1p, a LexA-Efh1p fusion significantly activated lacZ expression (Figure 9A, bottom), suggesting that Efh1 acts as a transcriptional activator.
Figure 9.
Transcriptional activities of Efg1p and Efh1p in one- or two-hybrid analyses. (A) One-hybrid assay in C. albicans. Expression plasmids encoding S. aureus LexA (SaLexA) or LexA-fusions to Efg1p (SaLexAEfg1) or Efh1p (SaLexA-Efh1) were transformed into strain CRC104 (105, 106) carrying a lacZ reporter gene under control of a lexA operator (+). As controls strains CRC101 (102, 103), which carry lacZ without a lexA operator (-), were used as hosts. LacZ activity of cells grown on SD medium was determined using X-Gal indicator. (B) Two-hybrid assay in S. cerevisiae. Genes encoding full-length and truncated versions of Efh1p were inserted into pGBD- or GAD-vectors to express fusions to the Gal4p-DNA binding (BD) or activation domain (AD). Pairs of BD- or AD-plasmids were transformed into S. cerevisiae PJ69-4A; β-galactosidase levels of transformants, encoded by a GAL7-lacZ fusion in this strain, were determined in a liquid assay and expressed as Miller units. The localization of the APSES-domain in Efh1p, as well as presumed activation (a) and dimerization (d) domains, are indicated.
In a second set of experiments, we performed standard two-hybrid assays in S. cerevisiae to examine the regulation and possible interactions of APSES proteins. For this purpose, we tested fusions of Efg1p or Efh1p to the Gal4p DNA-binding (BD) or activating domains (AD). In contrast to the one-hybrid assays in C. albicans, we found that BD-Efg1p-fusions did not influence lacZ reporter expression in S. cerevisiae. This suggests a requirement for C. albicans- specific components in the repressor function of Efg1p. In contrast, Efh1p-BD-fusions activated the lacZ reporter significantly (Figure 9B), which was consistent with the abovementioned one-hybrid data obtained with C. albicans.
A C-terminally truncated Efh1p comprising 432 residues (N1) also activated the reporter gene in S. cerevisiae. However, further truncated versions retaining 275 or 159 residues (N2 and N3) were inactive (Figure 9B). These results suggested that Efh1p contains an activation domain between residues 275 and 432. An AD-Efh1p fusion did not activate the reporter, presumably because binding of the fusion protein to the promoter did not occur. However, a strain containing both the BD-Efh1p fusion and the AD-Efh1p fusion contained twice the activity compared with a strain only containing a BD-Efh1p, suggesting homodimerization of Efh1p. This effect also was observed for the N1- and N2-proteins, but not the N3-truncated forms suggesting that N2 proteins are still able to dimerize and that a putative dimerization domain may be located between residues 159 and 275 of Efh1p. Similar experiments for the Efg1 protein did not reveal a homodimerizing activity for this transcriptional regulator nor heterodimerization with Efh1p.
From these experiments, we conclude that Efg1p and Efh1p have different regulatory potential. Efh1p functions as a transcriptional activator, as demonstrated in both C. albicans and S. cerevisiae. In contrast, Efg1p functions as a repressor, a function that for unknown reasons is only detected in the homologous host C. albicans. Furthermore, Efh1p but not Efg1p is able to form homodimers. As expected, the activation and dimerization domains of Efh1p are in regions that lack significant homology between these APSES proteins.
DISCUSSION
The availability of the complete C. albicans genomic sequence has allowed us to identify the complete set of APSES proteins in this organism, consisting of Efg1p and Efh1p. As a result, we were able to perform a comprehensive phenotypic and transcriptional analysis of APSES proteins, and in particular we were able to assess the relative contributions of both proteins to growth and morphogenesis. Furthermore, we examined functional interactions between both proteins and their cellular roles in C. albicans. This has allowed us to compare their functions to the APSES proteins of S. cerevisiae, which represent the only other known complete set of APSES protein in a fungal species.
Overexpression of both EFG1 and EFH1 induces pseudohyphal growth and represses the formation of true hyphae (Stoldt et al., 1997; Tebarth et al., 2003). At normal expression levels, both proteins cooperate to repress an alternative pathway of true hypha formation, which only seems to be operative in embedded or microaerophilic conditions. The function of the alternative pathway of filamentation in C. albicans is yet unknown, but it could promote morphogenesis in deep tissues or in phagocytic vacuoles when oxygen becomes limiting and/or cells are surrounded by an extracellular matrix. Our data suggest that the APSES proteins down-regulate this alternative morphogenetic pathway, thereby preventing filamentation under these conditions. On the other hand Efg1p is positively required for maintenance of the yeast growth form. In the absence of Efg1, cells change to a rod-like (opaque-like) form (Sonneborn et al., 1999b; Srikantha et al., 2000). Furthermore, Efg1p is required for the competence of yeast cells to undergo hyphal morphogenesis in standard (aerobic) conditions (Stoldt et al., 1997; Lo et al., 1997). Thus, Efg1p may have both activator and repressor functions in C. albicans, unlike APSES proteins in S. cerevisiae. Our one- and two-hybrid analyses confirmed a repressor function for Efg1p and an activator function for Efh1p, which had the ability to form homodimers in these tests. As expected, activation and homodimerization domains of Efh1p were mapped to regions outside of the conserved APSES domain.
Sequence comparisons reveal that Efh1p is more similar to the activator Phd1p, whereas Efg1p is more similar to the repressor Sok2p. For example, a potential PKA phosphorylation site is present in the APSES domains of Efg1p and Sok2 (RVT), but it is absent in Efh1p and Phd1 (KVA448 and RVI, respectively) (Bockmühl and Ernst, 2001). A direct role of Efg1p as a repressor was shown recently by its role as a negative autoregulator at the EFG1 promoter (Tebarth et al., 2003). The apparent repressor function of Efh1p in embedded/hypoxic conditions may in fact reflect Efh1p-dependent activation of Efg1p repressor activity, or of EFG1 expression under these conditions. Such an indirect role for Efh1p is also suggested by the findings that EFH1 overexpression phenotypes including pseudohypha- and opaque-white-induction, require Efg1p, and by the synthetic effects of Efh1 and Efg1 upon the C. albicans transcriptome. Thus, our data suggest that in C. albicans, the relative importance of the APSES proteins is skewed toward the main regulator Efg1p, whose role is facilitated under some conditions by the auxiliary factor Efh1p. In contrast, the S. cerevisiae APSES proteins seem to have opposing functions. A schematic overview of APSES protein functions in C. albicans is shown in Figure 10.
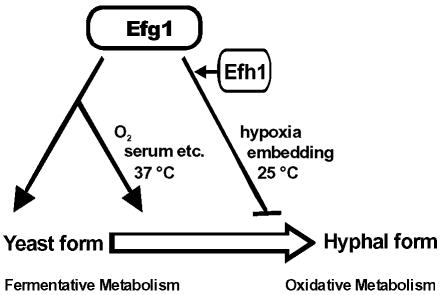
Figure 10.
Model of APSES protein functions in C. albicans. Efg1p favors the yeast growth form and a fermentative mode of metabolism. It also is required for the transition from the yeast to the hyphal growth form, which is initiated by inducers including serum. In embedded and hypoxic conditions, at lower temperatures, Efg1p represses rather than induces hypha formation. Efg1p could act directly, as a direct transcriptional activator, or indirectly, by repressing yet undefined repressors of morphogenesis and metabolism (as suggested by Efg1p-repressor function in one-hybrid experiments). Efh1p enhances the activation and repression functions of Efg1p in metabolism and morphogenesis, presumably by activation of EFG1 expression; Efh1p may also have yet unknown Efg1p-independent functions.
The influence of an efg1 mutation upon expression of a limited set of genes, and more recently upon the C. albicans transcriptome, has been described previously (Lane et al., 2001; Nantel et al., 2002; Sohn et al., 2003). Our genome-wide transcript profiling extends these results significantly by 1) including the second APSES-encoding gene, EFH1; 2) examining the effects of overexpressing the APSES proteins; 3) analyzing the importance of APSES proteins at an early stage of hypha induction; and 4) establishing the role of APSES proteins in metabolism in addition to morphogenesis. Nantel et al. (2002) mainly described differences between the transcript profiles of cells under noninducing and inducing conditions. In contrast, we have compared mutant and wild-type cells under the same (inducing or noninducing) medium to identify even relatively small but statistically significant changes in the transcript profiles mediated by the APSES proteins. Also, we used strain CAF2-1 as the wild-type strain, because it is more directly related to the genetic background of the mutants we examined.
The inactivation of Efg1p is known to exert strong phenotypic effects upon C. albicans (Lo et al., 1997; Stoldt et al., 1997). Hence, it is not surprising that transcript profiling revealed that numerous genes were affected by the absence of Efg1p. In contrast, deletion of EFH1 led to relatively few changes in the transcriptional profile. However, this was consistent with the minimal phenotypic effects of the efh1 deletion.
When the functions of Efg1-regulated genes were examined, almost every gene encoding an enzyme involved in glycolysis was found to be stimulated by Efg1p. In contrast, genes encoding oxidative enzymes of the citric acid cycle were repressed by Efg1p. These differences were due to the loss of Efg1 rather than growth effects, because we examined the transcriptomes of efg1 and wild-type cells under identical growth conditions. Thus, the presence of Efg1p favors fermentative and represses oxidative metabolism. A similar statement was made recently after comparisons of the transcriptomes of white and opaque cells. Lan et al. (2002) showed that the white (regular yeast) cells had enhanced expression of glycolytic genes compared with the opaque (rod-like) cells, which seemed to be in an state favoring oxidative metabolism. Intriguingly, EFG1 expression is insignificant in opaque cells, and efg1 mutants have an opaque-like appearance (Sonneborn et al., 1999b). Therefore, the overlaps between the transcriptomes of opaque and efg1 cells might be due, at least in part, to the absence of Efg1p. We show that the effects of Efg1p disruption upon glycolysis are of biological significance, because the growth of efg1 mutants is relatively sensitive to antimycin A, an inhibitor of respiration.
EFG1 and EFH1 overexpression affected the expression of several genes in common. This was to be expected because the overexpression of both genes causes similar phenotypes, including the induction of pseudohyphal growth and promotion of opaque to white phenotypic switching. The induction of pseudohyphal growth by EFH1 required the presence of EFG1, suggesting that Efh1p acts indirectly to stimulate EFG1 expression or Efg1p activity, which consequently would lead to (partially) overlapping transcriptional profiles. Genes commonly regulated by Efg1p- and Efh1p-over-production included ALS1, ALS10, and HWP1. This is probably closely correlated to the pseudohyphal or filamentous growth modes of these cell types (Braun and Johnson, 2000; Lane et al., 2001). Nevertheless, other genes were differentially regulated by Efg1p and Efh1p, probably reflecting the structural differences of both proteins. Interestingly, the set of genes identified in overexpression experiments did not overlap significantly with genes affected by the absence of APSES proteins in hypha-inducing conditions. Again, this highlights that significant differences exist between pseudohyphal and true hyphal modes of growth. Surprisingly, overexpression of EFG1 and EFH1 down-regulated IPF5185 (FLO1), encoding a putative yeast-specific cell wall protein, although in an efg1 mutant it was down-regulated as well (Nantel et al., 2002; Sohn et al., 2003). This can be explained by the suggestion that Efg1p acts as an activator as well as a repressor for this gene (Sohn et al., 2003).
To summarize, the results presented in this study suggest that APSES proteins are important regulators of metabolism and morphogenesis in C. albicans. Their effects upon morphogenesis include inducible dimorphism and spontaneous phenotypic switching. The typical yeast form (white) exploits a fermentative mode of metabolism, which is enhanced by the APSES protein Efg1p. This also renders these cells competent to form true hyphae in standard inducing conditions (lack of glucose, presence of inducers, 37°C). Homozygosity at the MTL locus, low temperatures (Miller and Johnson, 2002), or the down-regulation of EFG1/EFH1 (Sonneborn et al., 1999b) allows the generation of the opaque form, which exploits an oxidative mode of metabolism. This is refractory to standard inducers of hyphal growth.
We previously pointed out several structural and functional parallels between Efg1p and human myc proteins (Stoldt et al., 1997; Tebarth et al., 2003). Interestingly, it has been reported recently that, in addition to its role in cellular differentiation, myc induces fermentative metabolism (in particular glucose import and glycolysis: Osthus et al., 2000). Intriguingly, Myc-like proteins in Drosophila bind to ∼15% of all coding regions (Orian et al., 2003) suggesting a general role in gene expression. Collectively, the data suggest that APSES proteins play important roles in coordinating central metabolism with cellular differentiation.
Acknowledgments
We are grateful to B. Cormack, G. Fink, H. Liu, and D. Soll for strains and plasmids. We thank K. Lengeler for critical reading of the manuscript. Nucleotide sequence data for C. albicans were obtained from the Stanford Genome Technology Center Web site at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the National Institute of Dental and Craniofacial Research and the Burroughs Wellcome Fund. Annotations of C. albicans genes were obtained from the Candida database Web server (http://genolist.pasteur.fr/CandidaDB/). This study was supported by EU-project “Galar Fungail” (QLRT-1999-30795) and the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich SFB 590, Düsseldorf). We acknowledge the Alexander von Humbodt foundation for the support of S.K. C.R. was supported by a studentship from the BBSRC (01/B1/P/07007), and A.B. was supported by grants from the Biotechnology and Biological Sciences Research Council (1/P17124) and the Wellcome Trust (063204 and 068143).
Article published online ahead of print. Mol. Biol. Cell 10.1091/10.1091/mbc.E03-11-0782. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E03-11-0782.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Aramayo, R., Peleg, Y., Addison, R., and Metzenberg, R. (1996). Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144, 991-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, D.A., Feldmann, P.J., Bovey, M., Gow, N.A., and Brown, A.J. (1996). The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178, 5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse, C.E., Irwin, M.Y., Fonzi, W.A., and Sypherd, P.S. (1993). Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61, 3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmühl, D.P., and Ernst, J.F. (2001). A potential phosphorylation site for an A-kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157, 1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B.R., Head, W.S., Wang, M.X., and Johnson, A.D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B.R., and Johnson, A.D. (2000). TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155, 57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, Jr., D.H., Giusani, A.D., Chen, X., and Kumamoto, C.A. (1999). Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34, 651-662. [DOI] [PubMed] [Google Scholar]
- Delbrück, S., and Ernst, J.F. (1993). Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol. Microbiol. 10, 859-866. [DOI] [PubMed] [Google Scholar]
- Dutton, J.R., Johns, S., and Miller, B.L. (1997). StuA is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16, 5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W.A. (1999). PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 181, 7070-70779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W.A., and Irwin, M.Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, P.K., and Davis, R.W. (1994). Cloning vectors for the synthesis of epitope-tagged, truncated and chimeric proteins in Saccharomyces cerevisiae. Gene 144, 63-68. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Ibrahim, A.S., Sheppard, D.C., Chen, Y.-C., French, S.W., Cutler, J.E., Filler, S.G., and Edwards, Jr., J.E. (2002). Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44, 61-72. [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., and Fink, G.R. (1994). Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Biol. Cell 14, 2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusani, A.D., Vinces, M., and Kumamoto, C.A. (2002). Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160, 1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, C.-Y., Newport, G., Murillo, L.A., Jones, T., Scherer, S., Davis, R.W., and Agabian, N. (2002). Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99, 14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, S., Birse, C., Zhou, S., Matson, R., and Liu, H. (2001). DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276, 48988-48996. [DOI] [PubMed] [Google Scholar]
- Lee, K.L., Buckley, H.R., and Campbell, C.C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13, 148-153. [DOI] [PubMed] [Google Scholar]
- Liu, H., Köhler, J., and Fink, G.R. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266, 1723-1726 [DOI] [PubMed] [Google Scholar]; (Erratum: Science 267, 17).
- Leng, P., Lee, P.R., Wu, H., and Brown, A.J. (2001). Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 183, 4090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, H.J., Köhler, J.R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G.R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Losberger, C., and Ernst, J.F. (1989). Sequence and transcript analysis of the C. albicans gene (URA3) encoding orotidine-5′-phosphate decarboxylase. Curr. Genet. 16, 153-157. [DOI] [PubMed] [Google Scholar]
- Magee, B.B., and Magee, P.T. (2000). Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289, 310-313. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Miller, M.G., and Johnson, A.D. (2002). White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110, 293-302. [DOI] [PubMed] [Google Scholar]
- Morschhäuser, J., Michel, S., and Staib, P. (1999). Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 32, 547-556. [DOI] [PubMed] [Google Scholar]
- Nantel, A., et al. (2002). Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 13, 3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasi, S., Ciarapica, R., Jucker, R., Rosati, J., and Soucek, L. (2001). Making decisions through Myc. FEBS lett. 490, 153-162. [DOI] [PubMed] [Google Scholar]
- Orian, A., et al. (2003). Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Mol. Biol. Cell 17, 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus, R.C., Shim, H., Kim, S., Li, Q., Reddy, R., Mukherjee, M., Xu, Y., Wonseq, D., Lee, L.A., and Dang, C.V. (2000). Deregulation of glucose transporter 1 and glycolytic gene expression by c-myc. J. Biol. Chem. 275, 21797-21800. [DOI] [PubMed] [Google Scholar]
- Sharkey, L.L., McNemar, M.D., Saporito-Irwin, S.M., Sypherd, P.S., and Fonzi, W.A. (1999). HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBT1. J. Bacteriol. 181, 5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenhar, G., and Kassir, Y. (2001). A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G., and Hicks, J. (1986). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sohn, K., Urban, C. Brunner, H., and Rupp, S. (2003). EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47, 89-102. [DOI] [PubMed] [Google Scholar]
- Sonneborn, A., Bockmühl, D.P., and Ernst, J.F. (1999a). Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67, 5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn, A., Tebarth, B., and Ernst, J.F. (1999b). Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 67, 4655-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha, T., Tsai, L.K., Daniels, K., and Soll, D.R. (2000). EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 182, 1580-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt, V.R., Sonneborn, A., Leuker, C.E., and Ernst, J.F. (1997). Efg1p, an essential regulator of morphogenesis of the human fungal pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16, 1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebarth, B., Doedt, T., Krishnamurthy, S., Weide, M., Monterola, F., Dominguez, A., and Ernst, J.F. (2003). Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 328, 949-962. [DOI] [PubMed] [Google Scholar]
- Ward, M.P., Gimeno, C.J., Fink, G.R., and Garrett, S. (1996). SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15, 6854-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]