Abstract
Ciliate and yeast telomerase possess a nucleolytic activity capable of removing DNA from the 3′ end of a single-stranded oligonucleotide substrate. The nuclease activity is thought to assist in enzyme proofreading and/or processivity. Herein, we report a previously uncharacterized human telomerase-associated nuclease activity that shares several properties with ciliate and yeast telomerases. Partially purified human telomerase, either from cell extracts or recombinantly produced, demonstrated an ability to remove 3′ nontelomeric nucleotides from a substrate containing 5′ telomeric DNA, followed by extension of the newly exposed telomeric sequence. This cleavage/extension activity was apparent at more than one position within the telomeric DNA and was influenced by sequences 5′ to the telomeric/nontelomeric boundary and by substitution with a methylphosphonate moiety at the telomeric/nontelomeric DNA boundary. Our data suggest that human telomerase is associated with an evolutionarily conserved nucleolytic activity and support a model in which telomerase-substrate interactions can occur distal from the 3′ primer end.
INTRODUCTION
Telomeres comprise the repetitive regions of DNA and associated proteins found at chromosomal termini and are essential for genome stability in many eukaryotic organisms (for review, see Blackburn, 2001). In most eukaryotes, telomeres are maintained by telomerase, a specialized polymerase that uses an internal RNA component and a reverse transcriptase to copy telomeric sequence, one nucleotide at a time, onto single-stranded telomeric DNA (for review, see Nugent and Lundblad, 1998). In humans, ciliates, and yeast, the telomerase RNA (TER) subunit and a telomerase reverse transcriptase (TERT) may be minimally sufficient for in vitro elongation of telomeric sequence (Lingner et al., 1997; Weinrich et al., 1997; Beattie et al., 1998; Collins and Gandhi, 1998; Nakayama et al., 1998; Bachand and Autexier, 1999; Masutomi et al., 2000; Wenz et al., 2001).
In addition to a telomere polymerization activity, ciliate and yeast telomerases also possess an associated nuclease activity (Collins and Greider, 1993; Autexier and Greider, 1994; Cohn and Blackburn, 1995; Melek et al., 1996; Bednenko et al., 1997; Bhattacharyya and Blackburn, 1997; Prescott and Blackburn, 1997; Greene et al., 1998; Lue and Peng, 1998; Niu et al., 2000). The nucleolytic activity of ciliate telomerase removes nontelomeric DNA that is 3′ of telomeric sequence, or it may remove a single nucleotide of telomeric DNA aligned at the 5′ end of the telomerase RNA template (Collins and Greider, 1993; Melek et al., 1996). The yeast telomerase-associated nuclease is stimulated by primer-template mismatches or noncanonical yeast telomere repeat sequences (Prescott and Blackburn, 1997; Niu et al., 2000). The nucleolytic activity may be inherent to the telomerase complex because it copurifies with ciliate and yeast telomerase over several steps (Melek et al., 1996; Greene et al., 1998; Niu et al., 2000). Moreover, the nucleolytic activity associated with Tetrahymena thermophila telomerase can be reconstituted in rabbit reticulocyte lysate (RRL) containing recombinant telomerase RNA and tTERT (Collins and Gandhi, 1998). Unpurified recombinant human telomerase also generated products shorter-than-input primer length when incubated with a telomeric oligonucleotide (Huard et al., 2003). The proposed functions of the telomerase nucleolytic activity include proofreading and/or reinitiation of a stalled polymerization complex (Collins and Greider, 1993; Melek et al., 1996). The telomerase-associated nuclease does not perform the resection of newly synthesized G strand overhangs that occurs at ciliate telomeres, because Tetrahymena cells depleted for telomerase still undergo 3′ end processing (Jacob et al., 2003).
In addition to the aforementioned findings, several studies suggest that sequences upstream of the 3′ primer end can influence substrate utilization and that telomerase-substrate contacts can occur distal to the 3′ primer end (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993; Lee and Blackburn, 1993; Melek et al., 1996; Bednenko et al., 1997; Wang and Blackburn, 1997; Bottius et al., 1998; Lue and Peng, 1998; Fitzgerald et al., 2001). One compelling piece of evidence for multiple contacts between telomerase and its substrate is the ability to cross-link Euplotes aediculatus telomerase to two sites within a telomeric oligonucleotide: one at the 3′ primer end and at a second site 20 nucleotides upstream (Hammond et al., 1997). These observations are consistent with a two-site recognition model in which telomerase contacts a substrate at the 3′ end and at a region upstream of the terminus, at a proposed anchor site. The anchor site may promote the association of telomerase with its substrate during translocation, that is, the reiterative translocation of the telomerase RNA template relative to the DNA substrate during processive elongation of the DNA substrate (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993). Experiments in E. aediculatus indicate that the intervening DNA sequence between the anchor site and the 3′ terminus may be looped out (Hammond et al., 1997). It is not known whether the putative anchor site within telomerase contributes to the recognition and cleavage of nontelomeric DNA from substrates.
We have used partially purified human telomerase to determine whether a telomerase-associated nucleolytic activity is conserved in higher eukaryotes. Specifically, we have examined our preparations for an activity that removes 3′ nontelomeric DNA from 5′ telomeric sequence. We used chimeric oligonucleotide substrates composed of a 5′ telomeric region and a 3′ nontelomeric region to examine the effect of primer length and sequence on cleavage and extension. In addition, the position of cleavage was probed with oligonucleotides containing a nuclease-resistant modification. Finally, we examined the influence of primer sequence on the mode of extension, either by direct elongation of the primer 3′ end or by cleavage and subsequent elongation.
MATERIALS AND METHODS
Oligonucleotides
Unless otherwise stated, unmodified oligonucleotides were synthesized and cartridge purified by ACGT (Toronto, Ontario, Canada). The methylphosphonate-containing oligonucleotides were synthesized either by TriLink Biotechnologies (San Diego, CA) or by Gene Link (Hawthorne, NY) and gel purified as described previously (Melek et al., 1996). Primers containing a 3′ dideoxynucleotide were end-labeled with Tdt (Niu et al., 2000) and gel purified (Melek et al., 1996) as described previously. Gel-purified oligonucleotides were electrophoresed on a 15% (wt/vol) denaturing gel (19:1, acrylamide: bisacrylamide), excised and eluted in TE buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA, pH 8.0) overnight. The eluate was then purified over a C18 reverse phase column (Waters, Milford, MA). The column was activated with 100% methanol and preequilibrated with TE. After loading the oligonucleotide, the column was washed with water several times, and the oligonucleotide was eluted with 50% methanol. The oligonucleotide was then lyophilized in a speed vacuum (Savant Instruments, Holbrook, NY) for several hours. The primers were 5′ end-labeled with [γ-32P]ATP (3000 Ci/mmol, 10 mCi/ml; PerkinElmer Life and Analytical Sciences, Boston, MA) and T4 Kinase (Invitrogen, Carlsbad, MA) according to the latter manufacturer's instructions. Approximately 0.2 fmol of end-labeled oligonucleotide was loaded for detection on a denaturing polyacrylamide gel.
Cell Culture and Lysate Preparation
Raji cells were grown in RPMI 1640 media [supplemented with 10% (vol/vol), fetal bovine serum and 1× l-glutamine] in a 100-liter reactor at Amgen (Thousand Oaks, CA), harvested in mid-log phase, washed in phosphate-buffered saline, and frozen in liquid nitrogen. Each frozen pellet of ∼2 × 1010 cells was thawed and resuspended in an equal volume of 2.3× hypo buffer (23 mM HEPES, 7 mM KCl, 2.3 mM MgCl2) supplemented with protease inhibitor cocktail tablets (Roche Diagnostics, Indianapolis, IN), RNase inhibitors (20 U/ml; Roche Diagnostics); and β-mercaptoethanol (2 mM). The mixture was subjected to ∼90 strokes in a Dounce homogenizer by using a loose pestle. Mechanical lysis was performed on ice at 4°C. The lysate was then subjected to centrifugation at 100,000 × g in a SW50.1 rotor (Beckman Coulter, Fullerton, CA) at 4°C for 1 h. The supernatant was removed, adjusted to between 0.1 and 0.15 M NaCl and 20% (vol/vol) glycerol, and snap frozen in liquid nitrogen.
In Vitro Transcription of hTER
hTER was synthesized using a MEGAscript T7 in vitro transcription kit (Ambion) as per manufacturer's instructions (Beattie et al., 1998). The telomerase RNA was transcribed from the linearized, gel-purified cDNA in the presence of RNase inhibitor (40 U/20-μl reaction; Roche Diagnostics) for 4 h at 37°C. The in vitro transcription products were treated with DNase I (2 U/20-μl reaction) for 15 min at 37°C. The reaction mixture was then phenol/chloroform extracted and precipitated at -20°C. Full-length hTER was obtained by gel purification: the reaction products were electrophoresed on a 4% (wt/vol) polyacrylamide denaturing gel (29:1, acrylamide/bisacrylamide), and UV shadowed at a wavelength of 254 nm. Full-length hTER was excised, eluted in water (1 h at 65°C), and reprecipitated at -20°C. After centrifugation, the pellet was washed with 70% (vol/vol) ethanol and then resuspended in water.
In Vitro Reconstitution of Human Telomerase
Telomerase was reconstituted in rabbit reticulocyte lysates (RRLs) as described previously (Beattie et al., 1998). Recombinant hTERT (accession no. NM_003219) was FLAG-tagged at both the amino and carboxyl termini. Full-length FLAG-tagged hTERT was synthesized (2 h at 30°C) in the presence of gel-purified, full-length hTER (0.5 μg/50-μl reaction) by using a TNT T7-coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. After reconstitution, the RRL extract was adjusted to 20% (vol/vol) glycerol, aliquoted, frozen on dry ice, and stored at -70°C.
Anion Exchange Chromatography
One milliliter of Raji cell lysate was thawed and clarified by centrifugation in a Microfuge for 5 min at 4°C. The supernatant was adjusted to 0.1 M NaCl (if necessary) by dilution with 2.3× hypo buffer (23 mM HEPES, 7 mM KCl, 2.3 mM MgCl2) containing 20% (vol/vol) glycerol, 2 mM β-mercaptoethanol, protease inhibitor cocktail tablets (Roche Diagnostics), and RNase inhibitors at 10 U/ml. All subsequent steps were performed at 4°C. A 400-μl DEAE column (Bio-Rad, Hercules, CA) was pre-equilibrated with 2.3× hypo buffer adjusted to 0.1 M NaCl and supplemented with RNase and protease inhibitors, glycerol, and β-mercaptoethanol as described above. The Raji lysate was passed over the resin three times (∼0.1 ml/min) followed by a wash with 5 ml of equilibration buffer. Telomerase was eluted with 2.3× hypo buffer adjusted to 0.3 M NaCl (supplemented as described above). Fractions (400 μl) were collected and tested for protein content with a Bradford assay (Bio-Rad). The fraction containing peak amounts of telomerase activity was used in subsequent experiments.
Immunopurification of Recombinant Human Telomerase
Anti-FLAG M2-agarose affinity gel (600 μl; Sigma-Aldrich, St. Louis, MO) was equilibrated with 2.3× hypo buffer containing 0.15 M NaCl, 0.1% (wt/vol) CHAPS, 20% (vol/vol) glycerol, 2 mM β-mercaptoethanol, protease inhibitor cocktail tablets (Roche Diagnostics), and RNase inhibitors at 10 U/ml. Three milliliters of rabbit reticulocyte lysate containing reconstituted human telomerase was passed over the M2 anti-FLAG resin three times at 4°C, at 0.3 ml/min. The anti-FLAG M2-agarose affinity gel was washed with 15 ml of equilibration buffer containing 0.3 M NaCl, washed again with equilibration buffer (at 0.15 M NaCl), and an aliquot of beads (200 μl) was removed from the column. The wash step was repeated with 15 ml of equilibration buffer containing 0.6 M NaCl. The resin was removed from the column and suspended in equilibration buffer adjusted to ∼0.15 M NaCl.
Standard Elongation Assay/Conventional Telomerase Assay
In a 40-μl reaction, 1× SEA buffer (2 mM dATP, 2 mM TTP, 1 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol, 50 mM KOAc, 50 mM Tris acetate, pH 8.5), 200 pmol of primer, and 40 μCi of [α-32P]dGTP (800 Ci/mmol, 10 mCi/ml; PerkinElmer Life and Analytical Sciences). According to the source of telomerase, the reaction also included one of the following: 20 μl of anion exchange purified telomerase from Raji cells (∼100 μg of protein), 20 μl of rabbit reticulocyte lysate containing reconstituted telomerase, or 10 μl of recombinant telomerase captured on anti-FLAG resin. For those samples pretreated with RNase A, 50 μg of DNase-free RNase A (Sigma-Aldrich) was added to the sample and incubated at room temperature for 15 min before adding oligonucleotide, buffer, and isotope. For all reactions, the remaining volume was adjusted to 40 μl with 2.3× hypo buffer (23 mM HEPES, 7 mM KCl, 2.3 mM MgCl2). The telomerase assays were carried out for 2 h in a 30°C water bath as described previously (Morin, 1989) with the following exceptions: samples containing rabbit reticulocyte lysate were phenol/chloroform extracted twice, and 1 μl of GenElute LPA (Sigma-Aldrich) was added to all samples immediately before precipitation (in place of carrier RNA). After precipitation and removal of the supernatant, the pellets were resuspended in 7 μl of load dye [80% (vol/vol) formamide, 0.1% (wt/vol) xylene cyanol, 0.6× Tris borate-EDTA (TBE), 10 mM EDTA]. The samples were placed in a heating block at 100°C for 5 min, quick spun and loaded onto a 10 or 12% (wt/vol) denaturing gel (29:1, acrylamide/bisacrylamide, 7 M urea, 0.6× TBE), which was prerun for at least 30 min. The gel was electrophoresed at 1000-1500 V in 0.6× TBE for ∼2 h (or until the bromphenol blue in the load dye in an adjacent lane was two-thirds of the way down the gel). The gel was dried for 1 h at 80°C and placed on a phosphor screen (Amersham Biosciences, Piscataway, NJ).
SDS-PAGE
Duplicate samples containing either 2.5 μl of RRL (with hTER/hTERT or no DNA) or 10 μl of immunopurified telomerase captured onto M2 anti-FLAG resin (Sigma-Aldrich) were suspended in SDS-PAGE load dye and resolved on 4-12% (wt/vol) gradient SDS-PAGE gels (Invitrogen). One gel was transferred to Immobilon P membrane (Millipore, Billerica, MA) and probed with 0.1 μg/ml polyclonal anti-TERT antibody. Another similar gel was silver-stained according to a previously described procedure (Wray et al., 1981).
Antibodies
The anti-hTERT polyclonal rabbit antibody was raised against a HIS-tagged hTERT fragment, spanning amino acids 560-956, purified from Escherichia coli. The antisera were affinity purified by coupling the same hTERT fragment to Affigel (Bio-Rad) according to the manufacturer's instructions.
RESULTS
A Human Nucleolytic Activity Removes Nontelomeric DNA from the 3′ End of Substrates Containing 5′ Telomeric Sequence
In the “standard elongation” or conventional human telomerase assay, telomerase elongation products are visualized by the incorporation of [α-32P]dGTP, in the presence of TTP and dATP, onto the 3′ terminus of a single-stranded DNA oligonucleotide (Figure 1A, lane 2) (Greider and Blackburn, 1985; Morin, 1989). This assay possesses several advantages over the more sensitive, polymerase chain reaction-based telomere repeat amplification protocol (Kim et al., 1994) in its ability to directly visualize telomerase extension products. In the standard elongation assay, the six-base periodicity observed in the telomerase extension products reflects product accumulation at the translocation site within the RNA template, which in humans corresponds to the first G in the telomeric repeat TTAGGG (Morin, 1989). To avoid elongation patterns due to contamination of less than full-length oligonucleotides, all substrates were purified after oligonucleotide synthesis (see MATERIALS AND METHODS). Similar results were obtained with cartridge, high-performance liquid chromatography, or gel-purified oligonucleotides.
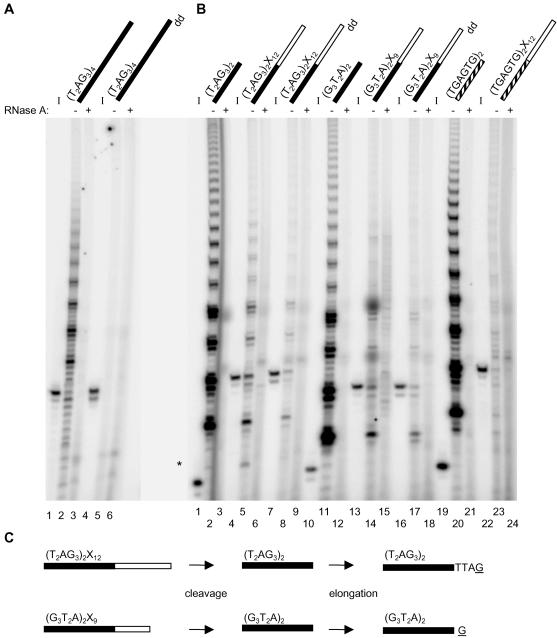
Figure 1.
(A) Telomerase elongation products generated by incubation with telomeric substrates. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and an oligonucleotide corresponding to four human telomeric repeats. Products of the standard elongation assay were electrophoresed on a 10% polyacrylamide denaturing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4). Oligonucleotides containing a dideoxynucleotide modification at the 3′ terminus are indicated by dd (lanes 4-6). Those samples pretreated with RNase A are indicated above the lane corresponding to their respective telomerase elongation reactions (+; lanes 3 and 6). (B) Telomerase elongation products generated by incubation with substrates containing 5′ telomeric DNA and 3′ nontelomeric DNA. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and the indicated oligonucleotide. Primer sequence composition is indicated by color code: black for telomeric DNA, white for nontelomeric DNA and striped for G-rich DNA. See Table 1 for oligonucleotide sequences. Products from the standard elongation assay were resolved on a 10% polyacrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, 10, 13, 16, 19, and 22). Oligonucleotides containing a dideoxynucleotide modification at the 3′ terminus are indicated by dd (lanes 7-9, 16-18). Perhaps due to an additional gel purification step, the dideoxynucleotide modified oligonucleotides generated slightly less intense elongation products relative to unmodified substrates (lanes 8 and 17, our unpublished results). Those samples pretreated with RNase A are indicated above the lane corresponding to their respective telomerase elongation reactions (+; lanes 3, 6, 9, 12, 15, 18, 21, and 24). An asterisk to the left indicates the position of the shortest product in lane 5. (C) Model for the cleavage and subsequent elongation of (T2AG3)2X12 and (G3T2A)2X9. Primer sequence composition is indicated by color: black for telomeric DNA and white for nontelomeric DNA. Primers are cleaved to expose telomeric DNA that becomes a substrate for elongation by telomerase (where G represents the incorporation of [α-32P]dGTP by telomerase). Note that regardless of whether cleavage occurs 5′ of or at the telomeric/nontelomeric DNA boundary, the elongation products will be similarly offset due to the permutation of telomeric DNA.
Human telomerase was partially purified from Raji cell extracts by anion exchange chromatography. Using this preparation, we examined the telomerase extension products upon incubation with the telomeric substrate (T2AG3)4 (Figure 1A, lane 2). In a separate reaction, the primer was also 5′ end-labeled with 32P to indicate the approximate position of the input oligonucleotide (“I”; Figure 1A, lane 1). As expected, incubation of telomerase with the telomeric substrate (T2AG3)4 generated elongation products (“P”) that initiated above the position of the full-length oligonucleotide, denoted P>I (Figure 1A, compare lanes 1 and 2), and possessed a periodicity that was consistent with the addition of TTAG, etc. (Morin, 1989). Products shorter than the input primer (P<I) also were generated, although these do not possess the periodicity typical of telomerase elongation products (Huard et al., 2003). All products were sensitive to treatment of the extract with RNase A before incubation with the substrate, confirming that their synthesis was RNA dependent (Figure 1A, lane 3). The elongation products were ablated by altering the 3′-terminal nucleotide to ddGTP (Figure 1A, lane 5), confirming that the preferential mode of elongation of this telomeric substrate is by extension of the 3′ primer end (Morin, 1989). Chimeric oligonucleotides were designed containing telomeric DNA at the 5′ end and nontelomeric sequence at the 3′ end. For example, the chimeric oligonucleotide (T2AG3)2X12 contains 12 nucleotides of telomeric DNA at the 5′ primer end, denoted (T2AG3)2, and 12 nucleotides of fixed nontelomeric sequence at the 3′ primer end, denoted X12 (see Table 1 for primer sequence). On incubation with the partially purified human telomerase preparation, the (T2AG3)2X12 primer generated products that migrated below the full-length oligonucleotide (denoted P<I) and possessed a six-base periodicity (Figure 1B, compare lanes 4 and 5). The extension products of (T2AG3)2X12 comigrated with the telomerase extension products generated by incubation with the 5′ telomeric sequence (T2AG3)2 (Figure 1B, compare lanes 2 and 5). These observations suggested that the (T2AG3)2X12 oligonucleotide was cleaved to (T2AG3)2 before extension by telomerase. Note that a less intense, shorter product was also apparent after incubation with (T2AG3)2X12, suggesting that a minority of oligonucleotide is cleaved at an alternate 5′ position within the telomeric region of the substrate (Figure 1B, lane 5, see asterisk). The relatively weaker intensity of the shortest product could be due to one of the following: infrequent utilization of a cleavage site 5′ to the telomeric/nontelomeric boundary, a reduction in precipitation efficiency or a low dissociation rate of short elongation products (Collins and Greider, 1993). All products, P>I or P<I, were sensitive to treatment of the extract with RNase A before incubation with substrate, confirming that their synthesis was RNA dependent (Figure 1B, lane 6).
Table 1.
Sequences of chimeric oligonucleotides
| Primer | Sequencea |
|---|---|
| (T2AG3)2X12 | (T2AG3)2CTACGCGATACT |
| (G3T2A)2X9 | (G3T2A)2CGCGATACT |
| (TGAGTG)2X12 | (TGAGTG)2CTACGCGATACT |
| (T2AG3)2X36 | (T2AG3)2CTACGCGA TCATAGCCACTATCG ACTACGCGATCAA |
| TA(G3T2A)2X9 | TA(G3T2A)2CGCGATACT |
| (G3T2A)3X9 | (G3T2A)3CGCGATACT |
| (G3T2A)2X5 · X4 | (G3T2A)2CGCGA · TACT |
| (G3T2A)2 · X9 | (G3T2A)2 · CGCGATACT |
| (T2AG3)2X6 · X6 | (T2AG3)2CTACGC · GATACT |
| (T2AG3)2 · X12 | (T2AG3)2 · CTACGCGATACT |
| −12/−25 | GGAGGTGGGTGGGG |
| −3/−25 | GGAGGTGGGTGGGGCGAGCAGAG |
| TA(G3T2A)2 −3/−11 | TA(G3T2A)2CGAGCAGAG |
| −12/−25X9 | GGAGGTGGGTGGGGCGCGATACT |
Where “·” indicates a methylphosphonate linkage.
To determine whether the observed elongation products initiated from a position internal to the input primer 3′ end, the partially purified telomerase preparation was incubated with an identical chimeric oligonucleotide that contained a 3′ terminal dideoxynucleotide residue. Replacement of the 3′ terminal nucleotide in (T2AG3)2X12 with a dideoxynucleotide (ddTTP) did not ablate formation of P<I products (Figure 1B, lane 8). These findings indicate that, unlike extension of (T2AG3)4, 3′ terminal sequence is removed from (T2AG3)2X12 before elongation by telomerase.
We tested another chimeric oligonucleotide, (G3T2A)2X9, that contained a permuted telomeric repeat at the 5′ end and a shorter 3′ nontelomeric end. The (G3T2A)2X9 primer also generated P<I products (Figure 1B, lane 14) that were sensitive to pretreatment of the extract with RNase A (Figure 1B, lane 15). As with (T2AG3)2X12, the P<I products were unperturbed by replacement of the 3′-terminal nucleotide in (G3T2A)2X9 with a dideoxynucleotide residue (ddTTP); again, consistent with the notion that the P<I products are generated by cleavage before extension by telomerase (Figure 1B, lane 17). The (G3T2A)2X9 P<I extension products comigrated with the P>I products of the telomeric sequence (G3T2A)2 indicating that cleavage occurred at or near the telomeric/nontelomeric boundary of (G3T2A)2X9 (Figure 1B, compare lanes 11 and 14).
In further support of the notion that both of the chimeric oligonucleotides were cleaved at or near the junction between telomeric and nontelomeric DNA, the major products generated by (T2AG3)2X12 and (G3T2A)2X9 are offset from each other as predicted based on the permutation of the telomeric sequence at the primer 5′ end (Figure 1B, compare lanes 5 and 14; see Figure 1C for schematic).
We next queried the sequence specificity of the nuclease cleavage activity by examining whether a scrambled 5′ telomeric sequence would lead to the generation of P<I products. The chimeric oligonucleotide, (TGAGTG)2X12, generated only faint P<I products (Figure 1B, lane 23) despite the efficient elongation of the scrambled telomeric sequence (TGAGTG)2 by telomerase (Figure 1B, lane 20). This result suggests that the P<I elongation products are more pronounced when a chimeric oligonucleotide contains telomeric sequence upstream of the 3′ primer end.
Immunoaffinity-purified Recombinant Telomerase Is Associated with Nucleolytic Activity
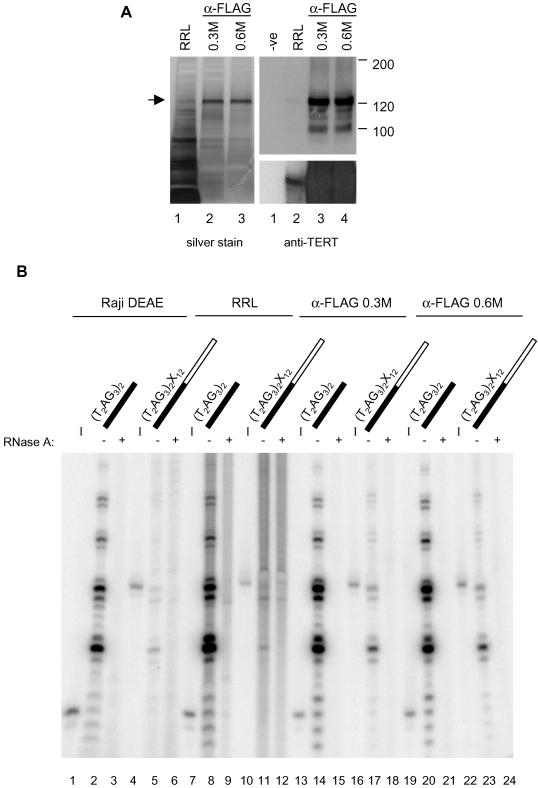
The aforementioned data suggest that partially purified human telomerase is associated with an activity that removes nontelomeric DNA from the 3′ end of a chimeric substrate. As a further demonstration that the observed nucleolytic activity copurifies with telomerase, recombinant hTER and FLAG-tagged hTERT were assembled in rabbit reticulocyte lysates (Weinrich et al., 1997; Beattie et al., 1998). Recombinant telomerase was captured on anti-FLAG affinity resin and washed with equilibration buffer containing either 0.3 M NaCl or 0.6 M NaCl. Immunopurification of recombinant telomerase enriched for a protein of ∼120 kDa that comigrated with hTERT by Western analysis (Figure 2A). This species was absent from mock anti-FLAG purifications with rabbit reticulocyte lysate alone (our unpublished data). The activity of reconstituted human telomerase was examined in the standard elongation assay (using both rabbit reticulocyte lysates and the immunopurified material). Rabbit reticulocyte lysates containing FLAG-tagged hTERT and hTER demonstrated elongation of the telomeric oligonucleotide (T2AG3)2 (Figure 2B, lane 8) (Weinrich et al., 1997; Beattie et al., 1998). Incubation of recombinantly produced human telomerase with the chimeric oligonucleotide (T2AG3)2X12 resulted in RNase-sensitive P<I products that comigrated with those generated by partially purified telomerase from Raji cells (Figure 2B, compare lanes 5 and 11). Purified recombinant telomerase (washed in 0.3 M NaCl) was also active when incubated with the telomeric oligonucleotide (T2AG3)2 and exhibited P<I products when incubated with the chimeric substrate (T2AG3)2X12 (Figure 2B, lanes 14 and 17). The P<I products observed with (T2AG3)2X12 comigrated with those generated by partially purified telomerase from Raji cells and were RNase-sensitive (Figure 2B, compare lanes 5 with 17 and lanes 17 with 18). Higher stringency washing (at 0.6 M NaCl) did not alter or reduce the P<I products generated by incubation of affinity-purified recombinant telomerase with (T2AG3)2X12 (Figure 2B, lane 23). These findings indicate that human telomerase synthesized in rabbit reticulocyte lysates is associated with a nucleolytic activity.
Figure 2.
Elongation products generated upon incubation with immunoaffinity-purified recombinant human telomerase. (A) Silver stain of an SDS-PAGE gel containing samples loaded as follows: 2.5 μl of rabbit reticulocyte lysate containing hTER and FLAG-tagged hTERT (RRL, lane 1), 10 μl of immunopurified recombinant telomerase washed with buffer containing 0.3 M NaCl (α-FLAG 0.3M, lane 2), or 0.6 M NaCl (α-FLAG 0.6M, lane 3). The protein enriched by immunopurification is indicated to the left with an arrow. Duplicate samples were electrophoresed on an SDS-PAGE gel, transferred to a membrane, and probed with anti-hTERT antibody. The order of samples is as follows: 2.5 μl of rabbit reticulocyte lysate containing no DNA (-ve, lane 1) or hTER and FLAG-tagged hTERT (RRL, lane 2), 10 μl of immunopurified recombinant telomerase washed with buffer containing 0.3 M NaCl (α-FLAG 0.3M, lane 3), or 0.6 M NaCl (α-FLAG 0.6 M, lane 4). The molecular mass (in kilodaltons) of protein markers is indicated at right. A longer exposure was performed to allow detection of hTERT synthesized in rabbit reticulocyte lysates (bottom; see lane 2). (B) Standard elongation assays were conducted with one of the following: 20 μl of partially purified telomerase from Raji cells (Raji DEAE, lanes 2, 3, 5, and 6), 20 μl of rabbit reticulocyte lysate containing hTER and FLAG-tagged hTERT (RRL lysate, lanes 8, 9, 11, and 12), 10 μl of immunopurified recombinant telomerase washed with buffer containing 0.3 M NaCl (α-FLAG 0.3M, lanes 14, 15, 17, and 18), or 0.6 M NaCl (α-FLAG 0.6M, lanes 20, 21, 23, and 24). Primer sequence is indicated above the corresponding lane: telomeric DNA is represented in black and nontelomeric DNA in white. The products resulting from incubation with (T2AG3)2 are shown in lanes 2, 3, 8, 9, 14, 15, 20, and 21. The products arising from incubation with (T2AG3)2X12 are shown in lanes 5, 6, 11, 12, 17, 18, 23, and 24. See Table 1 for oligonucleotide sequence. The products were resolved on a 12% polyacrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of the corresponding telomerase elongation reactions (I; lanes 1, 4, 7, 10, 13, 16, 19, and 22). Pretreatment with RNase A is indicated (+) for the samples corresponding to lanes 3, 6, 9, 12, 15, 18, 21, and 24.
As a demonstration that the nuclease activity was associated with human telomerase under alternate purification conditions, endogenous telomerase was isolated by gel filtration chromatography or antisense affinity selection (Schnapp et al., 1998; our unpublished data). Similar P<I products were obtained upon incubation of the chimeric oligonucleotide (T2AG3)2X12 with telomerase purified by anion exchange chromatography, gel filtration chromatography, or antisense affinity selection (our unpublished data).
Length Modulation of the Telomeric and Nontelomeric Regions within Chimeric Oligonucleotides
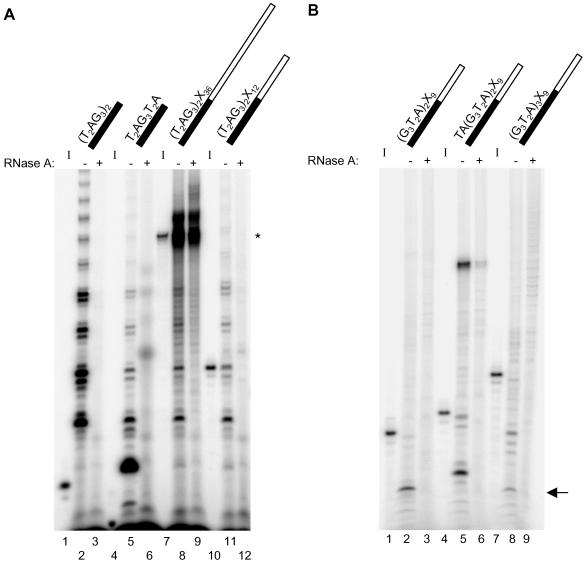
To further characterize the P<I products from the chimeric oligonucleotides, the length of the telomeric or nontelomeric region was varied within the substrate (while keeping the length and sequence of the other region constant). The chimeric oligonucleotides (T2AG3)2X36 and (T2AG3)2X12 possess the same 5′ telomeric DNA but differ in length by 24 nucleotides at the nontelomeric 3′ end (see Table 1 for primer sequence). Thus, if cleavage occurred in both substrates at or near the junction of telomeric and nontelomeric DNA, the resultant P<I elongation products should exhibit the same elongation pattern despite the difference in length and sequence of the cleaved 3′ fragments. Indeed, this prediction was borne out (Figure 3A, compare lanes 8 and 11). The P<I products generated by both (T2AG3)2X12 and (T2AG3)2X36 comigrated with the elongation products generated by incubation with (T2AG3)2 (Figure 3A, lane 2). The length of the 5′ telomeric region also was varied. The oligonucleotides (G3T2A)2X9 and TA(G3T2A)2X9 differ in their 5′ telomeric sequence by two nucleotides but possess a nontelomeric region of the same length and sequence (see Table 1 for primer sequence). The P<I products generated upon incubation with the primer TA(G3T2A)2X9 initiated at a position that differed by approximately two nucleotides relative to those of (G3T2A)2X9 (Figure 3B, compare lanes 2 and 5). In comparison, the oligonucleotide (G3T2A)3X9 contains an additional six nucleotides of telomeric sequence at the 5′ end relative to (G3T2A)2X9. Interestingly, the P<I products generated by (G3T2A)3X9 initiated at the same position as those generated by (G3T2A)2X9, despite a length difference in the 5′ telomeric region of the oligonucleotides (Figure 3B, compare lanes 2 and 8, see arrow at right), which supports the notion that cleavage can occur 5′ of the telomeric/nontelomeric DNA boundary. Identical results to those shown in Figure 3 also were obtained with immunoaffinity-purified recombinant human telomerase (our unpublished data).
Figure 3.
Telomerase elongation products generated by incubation with chimeric oligonucleotides containing different lengths of 3′ nontelomeric or 5′ telomeric DNA. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and the oligonucleotide indicated above each lane: black for telomeric DNA, white for nontelomeric DNA. Refer to Table 1 for oligonucleotide sequences. (A) Utilization of chimeric oligonucleotides with varying lengths of 3′ nontelomeric DNA. The standard elongation assay products were resolved on a 10% polyacrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, and 10). RNase A pretreatment is indicated by + above the corresponding lane (lanes 3, 6, 9, and 12). Note that the products in the upper region of lane 8 were not RNase A sensitive (lane 9) and are therefore not telomerase dependent (see asterisk beside this region in the gel). (B) Utilization of chimeric oligonucleotides with varying lengths of 5′ telomeric DNA. Products generated in the standard elongation assay were electrophoresed on a 12% polyacrylamide denaturing gel. 32P-5′ end-labeled primers are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, and 7). RNase A pretreatment is indicated by + above the corresponding lane (lanes 3, 6, and 9). The shortest major product generated by incubation with (G3T2A)2X9 and (G3T2A)3X9 is indicated by an arrow at right (lanes 2 and 8). Note that the products in the upper section of lanes 2, 5, and 8 were not sensitive to treatment with RNase A (lanes 3, 6, and 9, respectively) and are therefore not telomerase dependent.
A Nuclease-resistant Methylphosphonate Substitution Alters Substrate Utilization in a Position-specific Manner
Methylphosphonate modification was used to further examine the location of cleavage within the chimeric oligonucleotide substrates. By replacing the nonbridging oxygen of a phosphodiester bond with a methyl group, the linkage is rendered nuclease resistant (Stein and Cohen, 1988). If the modified site corresponds to a unique cleavage site, the reaction products will be altered relative to those from an unmodified oligonucleotide. For example, a chimeric oligonucleotide containing a methylphosphonate substitution at the boundary between telomeric and nontelomeric DNA inhibits the generation of P<I products upon incubation with Euplotes crassus telomerase (Melek et al., 1996; Bednenko et al., 1997).
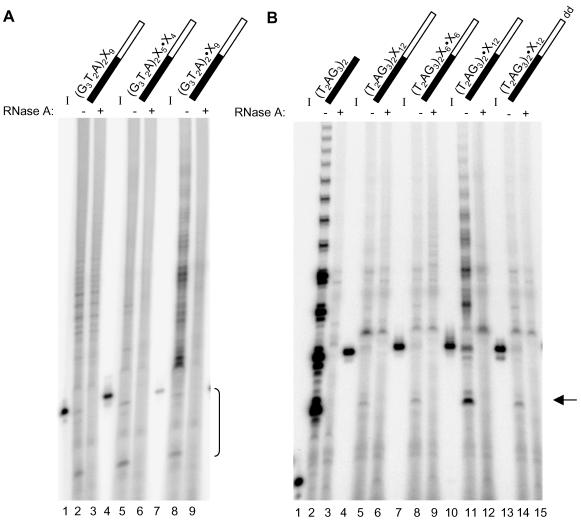
After incubation with partially purified telomerase from cells, (G3T2A)2X9 generated P<I products as observed previously (Figure 4A, lane 2; see bracket at right). (Note that the oligonucleotides used in Figure 4 were gel purified and generated fewer products in the standard elongation assay relative to cartridge-purified oligonucleotides used in other experiments). The chimeric substrate (G3T2A)2X9 was modified within the nontelomeric region generating the oligonucleotide (G3T2A)2X5·X4 (where · indicates a methylphosphonate linkage; see Table 1 for primer sequence). Incubation with (G3T2A)2X5·X4 resulted in P<I products similar to those generated with the unmodified oligonucleotide (G3T2A)2X9 (Figure 4A, compare lanes 2 and 5). This result indicates that methylphosphonate modification of the phosphodiester bond 4 nucleotides from the primer 3′ end does not prevent substrate binding or elongation. Because methylphosphonate substitution in the nontelomeric region would be expected to prevent formation of P<I products generated by an exonuclease, we speculate that an endonuclease is responsible for the cleavage activity, as described for the E. crassus telomerase-associated nuclease (Melek et al., 1996). Next, the effect of modification at the boundary between 5′ telomeric and 3′ nontelomeric DNA was examined with the oligonucleotide (G3T2A)2·X9. In contrast with (G3T2A)2X5·X4, incubation with (G3T2A)2·X9 resulted in alteration but not inhibition of product formation relative to the unmodified oligonucleotide (G3T2A)2X9 (Figure 4A, compare lanes 2, 5, with 8). Incubation with (G3T2A)2·X9 resulted in formation of both P<I and P>I products that were sensitive to pretreatment with RNase A (Figure 4A, compare lanes 8 and 9).
Figure 4.
Effect of methylphosphonate modification on the elongation of chimeric substrates. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and an oligonucleotide as indicated above each lane; black for telomeric DNA and white for nontelomeric DNA, where * indicates the position of a methylphosphonate linkage. Oligonucleotide sequences are listed in Table 1. Products from the standard elongation assay were resolved on a 10% polyacrylamide sequencing gel. All oligonucleotides used in these experiments were subjected to gel purification. (A) Elongation of the primer (G3T2A)2X9 containing methylphosphonate substitutions. Lanes 1-3, (G3T2A)2X9; lanes 4-6, (G3T2A)2X5·X4 primer modified with a methylphosphonate linkage within the 3′ nontelomeric DNA; lanes 7-9, (G3T2A)2·X9 primer modified with a methylphosphonate linkage at the telomeric/nontelomeric DNA boundary. The P<I products generated by the chimeric oligonucleotides are indicated by a bracket at right. Note that the products in the upper region of lane 2 were not RNase A sensitive (compare with lane 3) and did not occur in all experiments. However, the P>I products in lane 8 were sensitive to RNase A treatment (compare with lane 9). Input oligonucleotides were 32P-5′ end-labeled for reference (I; lanes 1, 4, and 7). Samples pretreated with RNase A are designated above by + (lanes 3, 6, and 9). (B) Elongation of the primer (T2AG3)2X12 containing methylphosphonate substitutions. Lanes 1-3, (T2AG3)2; lanes 4-6, (T2AG3)2X12; lanes 7-9, (T2AG3)2X6·X6 primer modified with a methylphosphonate linkage in the 3′ nontelomeric DNA; lanes 10-12, (T2AG3)2·X12 primer modified with a methylphosphonate residue at the telomeric/nontelomeric DNA boundary; lanes 13-15, (T2AG3)2·X12 containing a methylphosphonate residue at the telomeric/nontelomeric DNA boundary and a terminal dideoxy TTP. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, 10, and 13). Sample pretreatment with RNase A is represented above the respective lanes (+; lanes 3, 6, 9, 12, and 15). An arrow to the right indicates the position of the P<I product generated by the chimeric oligonucleotides. RNase A-insensitive products are observed in lanes 5, 6, 8, 9, 11, and 12. RNase A-sensitive P>I products were generated by incubation with (T2AG3)2·X12 (lane 11).
Similar results were obtained with modified versions of the chimeric substrate (T2AG3)2X12. A primer containing a methylphosphonate substitution in the nontelomeric region, (T2AG3)2X6·X6, did not inhibit or alter product formation relative to the unmodified oligonucleotide (T2AG3)2X12 (Figure 4B, compare lanes 5 and 8, see arrow at right; see Table 1 for primer sequence). The effect of modification at the boundary between the 5′ telomeric and the 3′ nontelomeric DNA was examined with the oligonucleotide (T2AG3)2·X12. Methylphosphonate modification at the telomeric/nontelomeric DNA junction altered but did not ablate product formation by human telomerase relative to the unmodified oligonucleotide (T2AG3)2X12 (Figure 4B, compare lanes 5 and 11); in fact, both P<I and P>I reaction products seemed more abundant relative to those generated by (T2AG3)2X12 or (T2AG3)2 X6·X6 (Figure 4B, compare lanes 5 with 11 and 8 with 11). Replacement of the 3′ terminal nucleotide in (T2AG3)2·X12 with a ddTTP residue did not ablate formation of P<I products (Figure 4B, lane 14). The reduction in overall efficiency of product formation may be due to an additional gel-purification step required to produce the dideoxy-modified version of (T2AG3)2·X12 (Figure 4B, compare lanes 11 and 14). Therefore, we could not conclude whether the P>I products resulting from incubation with (T2AG3)2·X12 are generated by extension of a 5′ cleavage product or by direct elongation of the nontelomeric 3′ end.
The persistence of P<I products upon introduction of a methylphosphonate linkage at the telomeric/nontelomeric junction suggests that cleavage occurred elsewhere in the oligonucleotides (G3T2A)2·X9 and (T2AG3)2·X12 (i.e., 5′ of the boundary) or via an alternate nucleolytic mechanism that was not inhibited by methylphosphonate modification. Methylphosphonate substitutions 5′ of the telomeric/nontelomeric border were not examined because such modifications can impair elongation when in proximity to a 3′ terminus capable of direct extension by ciliate telomerase (Shippen, personal communication). In addition to cleavage within the telomeric region, cleavage also may occur at the boundary between telomeric and nontelomeric DNA. The appearance of P>I products after incubation with (G3T2A)2·X9 and (T2AG3)2·X12 (and not their counterparts modified in the 3′ nontelomeric region) suggests that methylphosphonate modification at the telomeric/nontelomeric boundary does affect substrate utilization. Therefore, cleavage of (G3T2A)2·X9 and (T2AG3)2·X12 likely occurs at various positions, including sites 5′ of or at the junction between telomeric and nontelomeric DNA.
Primer Sequence Composition Influences the Mode of Substrate Utilization
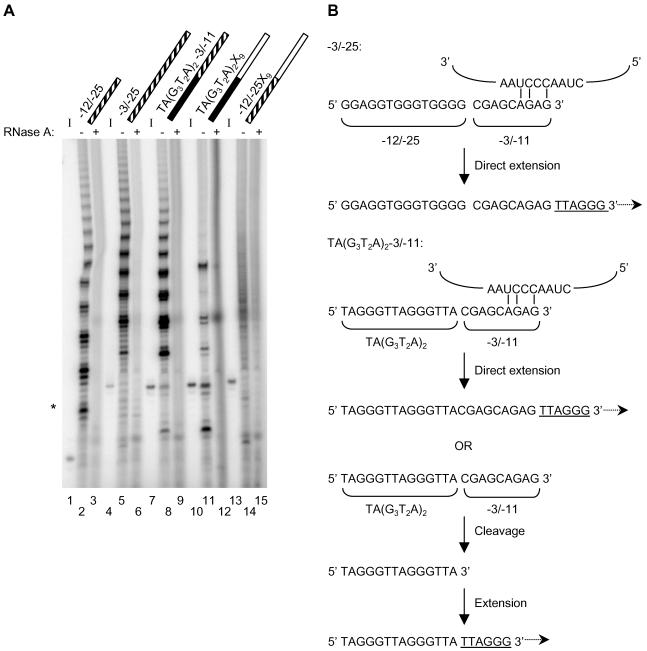
We next examined the contribution of the 3′ nontelomeric end to the cleavage and elongation of the chimeric DNA substrates. We tested the elongation properties of a chimeric oligonucleotide possessing complementarity to the telomerase RNA template at the 3′ primer end, to determine whether such a substrate would promote direct elongation of the primer 3′ end and perhaps interfere with the generation of P<I products. We used the chromosome 16 breakpoint sequence associated with the (αα)TI determinant of α-thalassemia (Wilkie et al., 1990). The terminally truncated chromosome is “healed” in vivo by the addition of telomeric sequence (Wilkie et al., 1990). Furthermore, the breakpoint sequence was shown to be a substrate for direct addition (i.e., direct extension of the primer 3′ end) by human and ciliate telomerases in vitro (Harrington and Greider, 1991; Morin, 1991). The -3/-25 primer encodes the sequence immediately upstream of one possible breakpoint junction (at the -3 position) and is a substrate for human telomerase in the standard elongation assay (Morin, 1991) (see also Figure 5A, lane 5; refer to Figure 5B for schematic). Two features promote utilization of the -3/-25 oligonucleotide by telomerase: a G-rich region at the 5′ end similar to telomeric DNA (designated -12/-25) and a 3′ region (-3/-11) able to pair with two to four nucleotides of the telomerase RNA template (Morin, 1991). The G-rich region at the 5′ end is thought to associate with the telomerase anchor site, whereas the nucleotides at the 3′ end allow minimal pairing to occur with the RNA template, thus fulfilling the “two-site” criteria for telomerase binding (Harrington and Greider, 1991; Morin, 1991).
Figure 5.
Telomerase elongation products generated by oligonucleotides containing 3′ sequence that is partially complementary to the telomerase RNA template. (A) Standard elongation assay was conducted using partially purified human telomerase from Raji cells and the oligonucleotide indicated above each lane: black for telomeric DNA, white for nontelomeric DNA, and striped for G-rich DNA. Refer to Table 1 for oligonucleotide sequences. Standard elongation assay products were resolved on a 10% acrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, 10, and 13). Those samples pretreated with RNase A are indicated above the corresponding lane (+; lanes 3, 6, 9, 12, and 15). RNase A-insensitive products are indicated by an asterisk alongside the gel (lanes 5 and 6). (B) Schematic for the direct elongation of -3/-25 and the proposed utilization of TA(G3T2A)2-3/-11 by human telomerase. The -3/-25 primer is composed of a G-rich 5′ region (denoted -12/-25) and a 3′ region capable of limited pairing with the telomerase RNA template (denoted -3/-11) (Morin, 1991). As previously shown, the -3/-25 primer can be directly extended by human telomerase (Morin, 1991). Potential base pairing with the telomerase RNA template is indicated above the primer sequence and the newly synthesized telomeric DNA is underlined. Both -3/-25 and TA(G3T2A)2-3/-11 possess the same 3′ sequence (designated -3/-11). Therefore, TA(G3T2A)2-3/-11 may participate in 3′ primer-template base-pairing interactions as with -3/-25. The P>I extension products generated by TA(G3T2A)2-3/-11 may result from the direct extension of this oligonucleotide by telomerase. The telomeric sequence at the 5′ end of TA(G3T2A)2-3/-11 may promote cleavage and subsequent extension by telomerase, consistent with the P<I products observed with this primer. The 5′ product of cleavage at the telomeric/nontelomeric boundary in TA(G3T2A)2-3/-11 is shown (although cleavage may also occur 5′ of the boundary).
In comparison with the -3/-25 oligonucleotide, the chimeric substrate TA(G3T2A)2X9 generated distinct P<I products, indicating that it was cleaved before extension by telomerase (Figure 5A, lane 11; see also Figure 3B, lane 5). We replaced the nine nontelomeric residues of TA(G3T2A)2X9 with nine nucleotides of 3′ sequence from the -3/-25 oligonucleotide. Since the resultant oligonucleotide, TA(G3T2A)2-3/-11, contains 3′ sequence capable of limited pairing with the telomerase RNA template, we asked whether this modified substrate would still support the generation of P<I products (see Figure 5B for primer sequence and schematic). We found that TA(G3T2A)2-3/-11 generated both P<I and P>I products (Figure 5A, compare lanes 7 and 8). Notably, TA(G3T2A)2-3/-11 and TA(G3T2A)2X9, which share the same 5′ telomeric sequence, produced comigrating P<I elongation products (Figure 5, compare lanes 8 and 11). Furthermore, oligonucleotides TA(G3T2A)2-3/-11 and -3/-25, which share the same 3′ sequence, generated similar P>I products (Figure 5, compare lanes 5 and 8). The dual pattern of products resulting from incubation with TA(G3T2A)2-3/-11 suggests that this chimeric oligonucleotide can be elongated either after removal of 3′ sequence or by direct extension of the 3′ terminus (Figure 5B).
Interestingly, when the 5′ telomeric sequence of TA(G3T2A)2X9 was replaced with a G-rich sequence corresponding to the 5′ region of -3/-25, the resulting primer (-12/-25X9) generated faint, albeit RNase A-sensitive P<I products (Figure 5A, compare lanes 14 and 15; see Table 1 for primer sequence). As observed previously with an oligonucleotide containing a G-rich (rather than telomeric) sequence at the 5′ end of the chimeric oligonucleotide, elongation of -12/-25X9 resulted in less distinct products (Figure 1B, lane 23). Furthermore, the P<I products generated upon incubation with -12/-25X9 did not comigrate with the P>I products from extension of the G-rich 5′ sequence -12/-25 (Figure 5A, compare lanes 2 and 14). These data suggest that -12/-25X9 is not cleaved at the boundary between the -12/-25 sequence and the X9 sequence, but rather elsewhere within the primer.
DISCUSSION
We have described a nucleolytic activity associated with partially purified endogenous and recombinant human telomerase. This study extends previous reports of a telomerase-associated nucleolytic activity in ciliates and yeast (Melek et al., 1996; Greene et al., 1998; Niu et al., 2000) by demonstrating that the nuclease activity robustly purifies with recombinant telomerase components (Figure 2B). Earlier reports indicated that a nucleolytic activity is present in unpurified rabbit reticulocyte preparations containing reconstituted telomerase (Collins and Gandhi, 1998; Huard et al., 2003). In the present study, detection of a nucleolytic activity relied on elongation of a cleaved chimeric substrate by telomerase. Thus far, we have been unable to develop a nucleolytic cleavage assay that is independent of elongation activity. However, the copurification of human telomerase with the nucleolytic activity argues in favor of a coupled cleavage/elongation process, rather than one in which telomerase extends nonspecifically degraded oligonucleotides in the reaction mixture.
Characteristics of the Human Telomerase-associated Nuclease Activity
The human telomerase-associated nuclease activity removes nontelomeric DNA from the 3′ terminus of substrates containing upstream G-rich (or telomeric) DNA. P<I products consistent with the removal (before elongation) of up to 36 nucleotides of nontelomeric DNA from the 3′ primer end were observed (Figure 3A, lane 8). Similarly, E. crassus telomerase can eliminate at least 29 nucleotides of nontelomeric DNA from the 3′ terminus of a chimeric oligonucleotide (Greene et al., 1998). The resulting 5′ primer fragment is the substrate for subsequent extension by human telomerase because a dideoxynucleotide modification at the 3′ end of a chimeric oligonucleotide did not block the formation of P<I products, also consistent with the properties of E. crassus telomerase (Melek et al., 1996; this study). In keeping with this notion, P<I product formation was sensitive to changes in telomeric register of the 5′ telomeric DNA (Figure 1B).
The cleavage and extension of chimeric substrates also was influenced by divergence from canonical telomeric sequence at the 5′ primer end. Replacement of 5′ telomeric sequence with G-rich sequence or noncanonical telomeric repeats, resulted in less pronounced P<I products (Figures 1B and 5). The indistinct pattern of products generated by (TGAGTG)2X12 and -12/-25X9 may be due to inefficient or nonuniform elongation of G-rich and noncanonical telomeric sequences. Alternatively, resection of (TGAGTG)2X12 and -12/-25X9 may be less efficient if the nucleolytic activity is sequence specific. Sequence specificity may be conferred by the template region of the RNA subunit, which has been implicated in the cleavage activity associated with both yeast and ciliate telomerases (Collins and Greider, 1993; Prescott and Blackburn, 1997; Greene et al., 1998).
Length variation within the 5′ telomeric region by using (G3T2A)2X9 and (G3T2A)3X9 indicated that cleavage can occur 5′ of or at the telomeric/nontelomeric boundary. Flexibility of the cleavage position also was observed upon incubation with methylphosphonate-modified oligonucleotides. Elongation products generated by (G3T2A)2X9 and (T2AG3)2X12 were altered but not inhibited by position-specific methylphosphonate modification at the boundary between telomeric and nontelomeric sequence. Although position-specific methylphosphonate modification inhibited product formation by E. crassus telomerase (Melek et al., 1996; Bednenko et al., 1997), the results from ciliate and human telomerase may not be directly comparable because the methylphosphonate substitutions were placed at different positions within the species-specific telomeric register and the oligonucleotides contained different lengths of 5′ telomeric sequence. In fact, flexible cleavage site positioning has been observed in E. crassus telomerase by using unmodified chimeric oligonucleotides (Greene et al., 1998). Furthermore, Saccharomyces cerevisiae telomerase can use an alternate cleavage site when the preferred site is blocked by a methylphosphonate linkage (Niu et al., 2000).
Other data also suggest that telomerase may be capable of flexible substrate utilization. We found that alteration of the 3′ primer end to allow limited pairing with the telomerase RNA template (as with TA(G3T2A)2-3/-11) resulted in a dual pattern of P<I and P>I products suggestive of direct and cleavage-mediated extension (Figure 5). Ciliate telomerase also may directly elongate or cleave and extend telomeric primers containing a mismatched nucleotide at the 3′ end (Collins and Greider, 1993).
Sequences Distal from the Primer 3′ Terminus Can Affect Substrate Utilization
Our findings indicate that sequences upstream from the 3′ terminus can influence substrate utilization by telomerase. Furthermore, the ability of telomerase to cleave an oligonucleotide at an upstream site does not depend on recognition of the 3′ terminus of the DNA substrate. Substrate recognition by telomerase both at the 3′ primer end and at an upstream site has been reported for telomerases from various organisms, including humans (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993; Lee and Blackburn, 1993; Melek et al., 1996; Bednenko et al., 1997; Hammond et al., 1997; Wang and Blackburn, 1997; Bottius et al., 1998; Lue and Peng, 1998; Fitzgerald et al., 2001). The 5′ recognition site may be positioned at varying distances from the 3′ terminus. Regardless of whether the 3′ sequences are extended directly or cleaved before elongation, telomerase can use substrates that contain 29-36 nucleotides of nontelomeric DNA at the primer 3′ end (Harrington and Greider, 1991; Melek et al., 1996; Greene et al., 1998; this study). An upper limit for the length of intervening nontelomeric DNA has not been determined. Telomerase might contact its substrate at the 3′ terminus and at a spatially distant upstream site by looping out the intervening DNA (Harrington and Greider, 1991; Morin, 1991). The loop-out model is supported by cross-linking analysis in E. aediculatus. Contact between E. aediculatus telomerase and a telomeric primer occurs at two sites spaced eight nucleotides apart on the telomerase RNA subunit and 20 nucleotides apart on the oligonucleotide substrate (Hammond et al., 1997).
Possible Roles for the Nucleolytic Activity of Human Telomerase
Detection of a telomerase-associated nuclease in ciliates, yeast, and humans suggests that this activity has an evolutionarily conserved function rather than a species-specific role related to developmentally programmed chromosome fragmentation (in ciliates) or a nonprocessive, nondissociative telomerase activity (in budding yeast). We speculate, as have others, that the telomerase-associated nuclease activity may serve a proofreading function and/or prevent stalling of the telomerase complex (Collins and Greider, 1993; Melek et al., 1996; Greene et al., 1998; Niu et al., 2000). By analogy, RNA polymerase has an intrinsic endonuclease activity thought to be located at or near the enzyme active site (Fish and Kane, 2002). The nucleolytic activity of RNA polymerase is stimulated by the cofactors GreA and GreB in prokaryotes and TFIIS in eukaryotes (Fish and Kane, 2002). This complex can relieve an arrested polymerase by cleaving back the nascent transcript. Stalling of RNA polymerase can occur during transcription of a particular sequence or due to nucleotide misincorporation. The fidelity of human telomerase is critical since alteration of the template region of hTER by a single nucleotide correlates with decreased cell viability (Marusic et al., 1997). Similarly, substitution of a single nucleotide within the human telomeric repeat prevents association with the telomere binding protein, TRF1 (Zhong et al., 1992). Removal of nontelomeric DNA from the 3′ end of a telomeric tract might thus prevent incorporation of incorrect sequence at the growing telomere. The error rate of human telomerase is estimated at 2 × 10-3 per nucleotide (Kreiter et al., 1995). The telomerase-associated nucleolytic activity is unlikely to be the sole determinant of telomere repeat fidelity because human and ciliate telomerase can directly extend primers that contain 3′ nontelomeric residues (Morin, 1989; Harrington and Greider, 1991; Collins and Greider, 1993; this study).
The nuclease activity also may facilitate reinitiation of telomerase elongation, for example, during translocation when the 3′ end of the DNA substrate is repositioned with respect to the template region of the telomerase RNA. In T. thermophila, removal of a single nucleotide of telomeric sequence has been observed when primer alignment extends to the extreme 5′ end of the RNA template, where translocation must occur before another round of template-mediated telomere addition (Collins and Greider, 1993). Further studies will be required to determine whether the nuclease activity associated with human telomerase also can remove a single nucleotide from the 3′ terminus of a telomeric primer that aligns with the 5′ end of the hTER template.
Note added in proof. Huard and Autexier have also recently reported that human telomerase is associated with a nucleolytic cleavage activity, and that 3′ terminal nucleotides can be removed from a telomeric primer (Huard, S., and Autexier, C. [2004]. Human telomerase catalyzes nucleolytic primer cleavage. Nucl. Acids Res. 32, 2171-2180.)
Acknowledgments
We thank members of the Harrington laboratory, Peter Cheung, Gregg Morin, Carolyn Price, Dorothy Shippen and the anonymous reviewers for critical comments and discussion. We thank Claudia Baikalov, Emil Brisan, Nicole Le, Zane Saremi, Geri Trail, Aaron Winter, and Amgen (Thousand Oaks, CA) for performing the large-scale growth of Raji cells. We also acknowledge assistance and reagents provided by Stephanie Hartsel and Bill Marshall (Amgen, Boulder, CO). This work was supported by a grant from the Canadian Institutes of Health Research to L.H. (MT14340) and a Canadian Institutes of Health Research Studentship to R.O. (MST-34566).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0178. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0178.
References
- Autexier, C., and Greider, C.W. (1994). Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev. 8, 563-575. [DOI] [PubMed] [Google Scholar]
- Bachand, F., and Autexier, C. (1999). Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274, 38027-38031. [DOI] [PubMed] [Google Scholar]
- Beattie, T.L., Zhou, W., Robinson, M.O., and Harrington, L. (1998). Reconstitution of human telomerase activity in vitro. Curr. Biol. 8, 177-180. [DOI] [PubMed] [Google Scholar]
- Bednenko, J., Melek, M., Greene, E.C., and Shippen, D.E. (1997). Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 16, 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, A., and Blackburn, E.H. (1997). A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc. Natl. Acad. Sci. USA 94, 2823-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- Bottius, E., Bakhsis, N., and Scherf, A. (1998). Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol. 18, 919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, M., and Blackburn, E.H. (1995). Telomerase in yeast. Science 269, 396-400. [DOI] [PubMed] [Google Scholar]
- Collins, K., and Gandhi, L. (1998). The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 95, 8485-8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K., and Greider, C.W. (1993). Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 7, 1364-1376. [DOI] [PubMed] [Google Scholar]
- Fish, R.N., and Kane, C.M. (2002). Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577, 287-307. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.S., Shakirov, E.V., Hood, E.E., McKnight, T.D., and Shippen, D.E. (2001). Different modes of de novo telomere formation by plant telomerases. Plant J. 26, 77-87. [DOI] [PubMed] [Google Scholar]
- Greene, E.C., Bednenko, J., and Shippen, D.E. (1998). Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol. Cell. Biol. 18, 1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C.W., and Blackburn, E.H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405-413. [DOI] [PubMed] [Google Scholar]
- Hammond, P.W., Lively, T.N., and Cech, T.R. (1997). The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 17, 296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.A., and Greider, C.W. (1991). Telomerase primer specificity and chromosome healing. Nature 353, 451-454. [DOI] [PubMed] [Google Scholar]
- Huard, S., Moriarty, T.J., and Autexier, C. (2003). The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31, 4059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, N.K., Kirk, K.E., and Price, C.M. (2003). Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell 11, 1021-1032. [DOI] [PubMed] [Google Scholar]
- Kim, N.W., Piatyszek, M.A., Prowse, K.R., Harley, C.B., West, M.D., Ho, P.L., Coviello, G.M., Wright, W.E., Weinrich, S.L., and Shay, J.W. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011-2015. [DOI] [PubMed] [Google Scholar]
- Kreiter, M., Irion, V., Ward, J., and Morin, G. (1995). The fidelity of human telomerase. Nucleic Acids Symp. Ser. 33, 137-139. [PubMed] [Google Scholar]
- Lee, M.S., and Blackburn, E.H. (1993). Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13, 6586-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Cech, T.R., Hughes, T.R., and Lundblad, V. (1997). Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94, 11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue, N.F., and Peng, Y. (1998). Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 26, 1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusic, L., Anton, M., Tidy, A., Wang, P., Villeponteau, B., and Bacchetti, S. (1997). Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol. Cell. Biol. 17, 6394-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutomi, K., Kaneko, S., Hayashi, N., Yamashita, T., Shirota, Y., Kobayashi, K., and Murakami, S. (2000). Telomerase activity reconstituted in vitro with purified human telomerase reverse transcriptase and human telomerase RNA component. J. Biol. Chem. 275, 22568-22573. [DOI] [PubMed] [Google Scholar]
- Melek, M., Greene, E.C., and Shippen, D.E. (1996). Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol. 16, 3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, G.B. (1989). The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521-529. [DOI] [PubMed] [Google Scholar]
- Morin, G.B. (1991). Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature 353, 454-456. [DOI] [PubMed] [Google Scholar]
- Nakayama, J., Tahara, H., Tahara, E., Saito, M., Ito, K., Nakamura, H., Nakanishi, T., Ide, T., and Ishikawa, F. (1998). Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 18, 65-68. [DOI] [PubMed] [Google Scholar]
- Niu, H., Xia, J., and Lue, N.F. (2000). Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol. Cell. Biol. 20, 6806-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C.I., and Lundblad, V. (1998). The telomerase reverse transcriptase: components and regulation. Genes Dev. 12, 1073-1085. [DOI] [PubMed] [Google Scholar]
- Prescott, J., and Blackburn, E.H. (1997). Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11, 528-540. [DOI] [PubMed] [Google Scholar]
- Schnapp, G., Rodi, H.P., Rettig, W.J., Schnapp, A., and Damm, K. (1998). One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 26, 3311-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, C.A., and Cohen, J.S. (1988). Oligodeoxynucleotides as inhibitors of gene expression: a review. Cancer Res. 48, 2659-2668. [PubMed] [Google Scholar]
- Wang, H., and Blackburn, E.H. (1997). De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 16, 866-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich, S.L., et al. (1997). Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17, 498-502. [DOI] [PubMed] [Google Scholar]
- Wenz, C., Enenkel, B., Amacker, M., Kelleher, C., Damm, K., and Lingner, J. (2001). Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20, 3526-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, A.O., Lamb, J., Harris, P.C., Finney, R.D., and Higgs, D.R. (1990). A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature 346, 868-871. [DOI] [PubMed] [Google Scholar]
- Wray, W., Boulikas, T., Wray, V.P., and Hancock, R. (1981). Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 118, 197-203. [DOI] [PubMed] [Google Scholar]
- Zhong, Z., Shiue, L., Kaplan, S., and de Lange, T. (1992). A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 12, 4834-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]