Abstract
The eukaryotic flagellum is a large structure into which specific constituent proteins must be targeted, transported and assembled after their synthesis in the cytoplasm. Using Trypanosoma brucei and a proteomic approach, we have identified and characterized a novel set of adenylate kinase proteins that are localized to the flagellum. These proteins represent unique isoforms that are targeted to the flagellum by an N-terminal extension to the protein and are incorporated into an extraaxonemal structure (the paraflagellar rod). We show that the N-terminal extension is both necessary for isoform location in the flagellum and sufficient for targeting of a green fluorescent protein reporter protein to the flagellum. Moreover, these N-terminal extension sequences are conserved in evolution and we find that they allow the identification of novel adenylate kinases in the genomes of humans and worms. Given the existence of specific isoforms of certain central metabolic enzymes, and targeting sequences for these isoforms, we suggest that these isoforms form part of a complex, “solid-phase” metabolic capability that is built into the eukaryotic flagellum.
INTRODUCTION
Motility is often central to cell feeding, survival, or reproduction in most eukaryotic organisms. Fast cell motility or locomotion of material over the cell surface is apparently best produced by the use of cilia or flagella. Although there are intriguing variations in some cell types and genera, most motile eukaryotic cilia and flagella have the canonical 9 + 2 microtubule structure termed the axoneme. This regular and evolutionarily conserved pattern of the nine doublet microtubules and two central pair singlet microtubules in the axoneme is intimately involved in the production of motility via the dynein arm bridges that form between adjacent outer doublets (Porter and Sale, 2000; King, 2003). The resulting microtubule doublet sliding is constrained by other connections so converting the applied force into a bending motion.
In a variety of microbial eukaryotes and in flagella of certain animal cell types (often sperm), a diversity of extraaxonemal structures occurs along with the microtubule axoneme. For example, in mammalian sperm tails, nine outer dense fibers run along the length of the flagellum, and in the principal piece a second extraaxonemal structure, the fibrous sheath, surrounds the outer dense fibers (Eddy et al., 2003). One of the most intriguing of these extraaxonemal structures is the elaborate paraflagellar rod (PFR) found in the kinetoplastid protozoa, the Euglenoids, and the dinoflagellates (Bastin et al., 1996b). In the African trypanosome Trypanosoma brucei, the PFR is present alongside the axoneme from its point of exit from the flagellar pocket. The T. brucei PFR has three zones, each of characteristic ultrastructure, and the whole structure is linked to axonemal microtubule doublets 4 through 7 (Gull, 1999).
There are a number of traditional views on the function of these diverse extraaxonemal structures, often involving suggestions that they may alter the mechanical properties of the flagellum in some advantageous manner. More modern molecular interrogations of such structures have revealed that they are important for motility. For example, in the mouse a knockout of the major component of the fibrous sheath prevented normal flagellum development and the resulting sperm were incapable of progressive motility (Miki et al., 2002). Likewise, in T. brucei RNA interference (RNAi) knockdown of the major paraflagellar rod component PFRA resulted in trypanosomes that lacked the major portion of the PFR structure, and although viable, were paralyzed (Bastin et al., 1998, 2000a).
Whereas we have some molecular knowledge of the major structural components in the PFR and fibrous sheath, there is a paucity of data on less abundant proteins, which may nonetheless contribute to important regulatory, signaling, or enzymatic functions in flagella. Here, we have used the PFR minus, paralyzed mutants of T. brucei to address issues of what biochemistry an extraaxonemal structure contributes to the eukaryotic flagellum. We have isolated flagella from the snl-2 mutant of T. brucei. This mutant is an inducible RNAi mutant. In the noninduced state, it presents a normal PFR and is actively motile (Bastin et al., 2000a). When switched to conditions where RNAi is induced, the trypanosomes lack a full PFR and are paralyzed, yet grow well. Isolated flagella were subjected to two-dimensional (2D) gel analysis and mass spectrometry to identify protein components present in the flagellum that exhibits a PFR, but which are then absent from flagella with no PFR. This proteomic comparison has allowed the identification of a novel set of adenylate kinase proteins that localize to the extraaxonemal structure and are targeted to the flagellum by an N-terminal extension to the protein. We show that the N-terminal extension is both necessary and sufficient for targeting of a GFP reporter protein to the flagellum. Moreover, these N-terminal extension sequences are conserved in evolution, and we find that they allow the identification of novel adenylate kinases in the genomes of humans and worms. These observations reveal a “solid-phase” enzymatic biochemistry for adenine nucleotide homeostasis that is built into the structure of a eukaryotic flagellum.
MATERIALS AND METHODS
Trypanosome Strains, Growth, and Transformation
Procyclic T. brucei was maintained at 27°C in SDM-79 medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum as described previously (Brun and Schonenberger, 1979). Proteomics and analysis of Ty-epitope-tagged TbADKA and TbADKA were investigated using the snl-2 mutant (Bastin et al., 2000a). RNAi experiments were carried out using the 29-13 cell line (Wirtz et al., 1999). Transformations were carried out essentially as described previously, by using ZPFM buffer and electroporation of 3 × 100-ms pulses at 1.5 kV with an Electro Square Porator (BTX) (Wickstead et al., 2003).
Trypanosome Fractionation
Cells were harvested by centrifugation (850 × g, 10 min, 20°C) and washed twice with phosphate-buffered saline. Cytoskeletons were isolated by extraction of 108 cells ml-1 in 1% (vol/vol) Nonidet-P40 in 0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, pH 6.9, and containing 5 μg ml-1 pepstatin A, 10 μg ml-1 leupeptin, and 10 μg ml-1 phenylmethylsulfonyl fluoride. After centrifugation (3400 × g, 10 min, 4°C), the detergent-insoluble cytoskeletal pellet was extracted twice in 0.1 M PIPES, 1 M NaCl, 2 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, pH 6.9, plus protease inhibitors (ice, 10 min). Depolymerization of the subpellicular microtubules was visualized by light microscopy, and flagella were subsequently isolated by centrifugation (16 000 × g, 15 min, 4°C). The flagella pellet was washed once (0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, pH 6.9) and resuspended in 20 mM Tris-HCl, 5 mM MgCl2, 1% (vol/vol) Nonidet-P40 and incubated on ice (15 min) with 2.5 μg of RNase, and 8.75 μg of DNaseI. Sixty milligrams of electrophoresis grade urea and 100 μl of a solution containing 2% (vol/vol) Nonidet-P40, 2% (vol/vol) ampholines, pH 3.5-10, 5% (vol/vol) β-mercaptoethanol, and 9.5 M urea were then added, and the samples stored at -80°C before analysis.
Electrophoresis and Mass Spectrometry of Flagellar Proteins
Samples were separated in the first dimension by isoelectric focusing and in the second dimension by SDS-PAGE, and proteins were visualized by silver staining as described previously (Burland et al., 1983). In-gel trypsin digestion of proteins (Shevchenko et al., 1996) was followed by peptide mass finger-printing, by using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (Voyager DE-STR; Applied Biosystems, Warrington, United Kingdom), and peptide sequencing by using quadrupole-time-of-flight tandem mass spectrometry (Q-ToF; Waters/Micromass, Manchester, United Kingdom). Peptide sequences obtained from tandem mass spectrometry were used to search the T. brucei genome database (http://www.genedb.org/genedb/tryp/index.jsp) and the identity of candidate proteins confirmed by matching the peptide masses obtained by matrix-assisted laser desorption/ionization-time of flight mass spectrometry with those obtained from in silico digestion of the candidate (PeptideMass, http://www.expasy.org).
Plasmid Construction
Construction of plasmids used for 1) expression of Ty-epitope-tagged TbADKA and TbADKB from their endogenous RNA polymerase II-transcribed loci, 2) inducible overexpression of TbADKA::Ty and TbADKAΔ(1-43)::Ty from a rDNA locus by using the RNA polymerase I-transcribed procyclin promoter, and 3) RNAi of flagellar adenylate kinases are all described previously (Pullen, 2002). Briefly, for expression of tagged proteins from their endogenous loci the tagged open reading frames were located between the endogenous 5′ untranslated region (UTR) and an aldolase 3′ UTR. In the overexpression experiments, tagged open reading frames were cloned into pHD430, in which trans-splicing and polyadenylation processing signals are provided by a procyclin 5′ UTR and aldolase 3′ UTR, respectively (Wirtz and Clayton, 1995). The parent vector for the RNAi experiments was pZJM (Wang et al., 2000). The expression of TbADKAΔ(55-260)-GFP was achieved after polymerase chain reaction amplification of sequence that encodes the first 55 amino acids of TbADKA by using the primer combination ADKAN5′ (5′-ATATTTGTGGTACTGCGACC-3′) and ADKAN3′ (5′-TGACATCCATGGCAATGAATATTTGCAAAGG-3′) and Herculase DNA polymerase (Stratagene, La Jolla, CA). The resulting polymerase chain reaction product was digested with NcoI (site italicised in ADKAN3′, and an additional site spans the start codon of ADKA) and cloned into pGAD8-VSG-G4 (Wickstead et al., 2003) that had been digested with NcoI across the start codon of the GFP gene. The resulting plasmid was digested with BamHI for stable integration into a minichromosomal locus.
Adenylate Kinase Assay
Detergent-insoluble cytoskeletons and flagella (5 × 107 cell equivalents) were assayed in a final volume of 1 ml in 0.03 M Tris-HCl, pH 7.75, 1.2 mM MgSO4, 5 mM glucose, 1 mM NADP+, 10 U of hexokinase, and 6 U of glucose-6-phosphate dehydrogenase. The rate of NADPH formation was followed at 340 nm.
Immunofluorescence and Immunoblotting
Immunofluorescence using BB2, which recognizes the Ty-epitope, and ROD-1 monoclonal antibodies, and immunoblotting by using BB2 were carried out using previously described methods (Bastin et al., 1996a,b).
RESULTS
Proteomic Analysis of the PFR
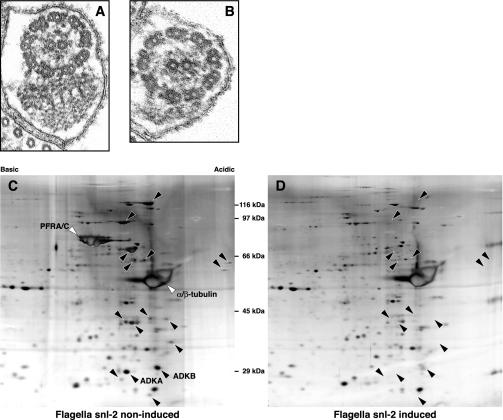
The snl-2 mutant of T. brucei is an inducible RNAi mutant that possesses a PFR and is motile when the RNAi is not induced, but becomes PFR minus and paralyzed when RNAi is induced (Figure 1, A and B) (Bastin et al., 2000a). Loss of the PFR structure does not affect viability, and populations of cells can therefore be grown in both states: with or without the PFR. The induction of RNAi ablates the expression of only one protein (PFRA), but this is sufficient to compromise the construction of the major portion of the PFR. We conjectured that many flagellar proteins will be synthesized but not assembled in the flagellum when the PFR is missing. This provided an opportunity to isolate the axoneme and PFR complex from the snl-2 mutant in both the RNAi induced and noninduced states, and use 2D gels to compare their protein profiles. We performed this analysis and detected a number of spots clearly missing from the flagellum of the paralyzed, induced RNAi population in comparison with the noninduced, motile population of the snl-2 strain (Figure 1, C and D). Four specific spots were subjected to mass spectrometry and sequences derived from the genome database. Two were trypanosome specific and are likely to represent PFR structural proteins. However, intriguingly, two sequences were putative adenylate kinases, which we termed TbADKA and TbADKB (Figure 2).
Figure 1.
Composition of the PFR can be determined by comparative proteomics of an inducible RNAi mutant. Transmission electron micrograph of thin sections through the flagellum show the wild-type organization of the axoneme and PFR in the noninduced snl-2 mutant (A) and the absence of the PFR in paralyzed, induced snl-2 (B). Proteomic analysis of flagella purified from induced and noninduced snl-2 indicates that the PFR is composed of at least 18 proteins (C and D). The adenylate kinases identified in this study are indicated as ADKA and ADKB in C.
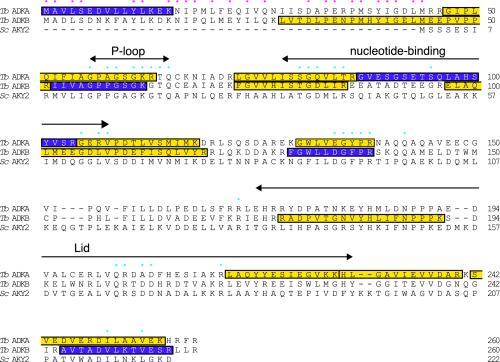
Figure 2.
Identification of two flagellar adenylate kinases. Sequences of the two adenylate kinases identified in this study, TbADKA and TbADKB, and S. cerevisiae adenylate kinase (accession no. AAA66319) were aligned using ClustalW. The gene identities for the T. brucei adenylate kinases are Tb927.2.5660 (TbADKA) and Tb10.70.7330 (TbADKB); there is 43% identity between the two trypanosome enzymes. Blue dots highlight conserved residues that are important for catalysis or enzyme structure (Schricker et al., 1995; Sanchez and Muller, 1998). Domains involved in the binding of ATP and AMP substrates, and a third characteristic region, known as the LID domain which changes conformation after substrate binding (Fukami-Kobayashi et al., 1996; Miura et al., 2001), are indicated by double-ended arrows. The red dots highlight residues that are identical in the N-terminal extensions of TbADKA and TbADKB. Purple shading indicates peptide fragments that were identified by tandem mass spectrometry sequencing; yellow shading indicates peptides identified on a basis of their mass only.
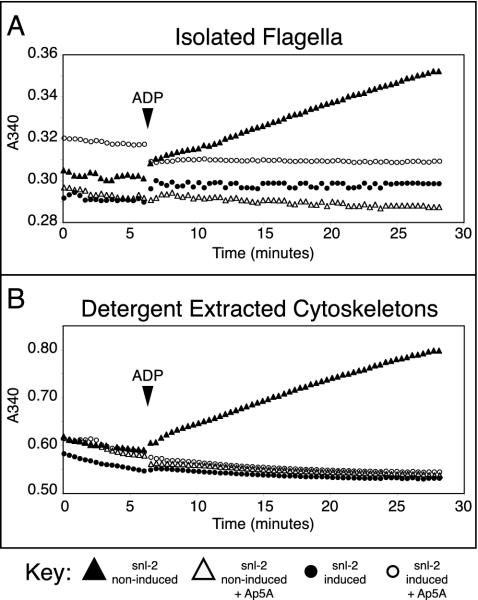
Given the role of adenylate kinases in adenine nucleotide homeostasis and the requirement for such adenine nucleotide balance for dynein motor protein activity in the flagellum (Yagi, 2000), the finding of two adenylate kinases in the assembled flagellum was intriguing. We therefore asked whether we could detect the corresponding biochemical activity in the flagellum. We used the classic approach of detergent extraction of whole cells to produce cytoskeletons (the subpellicular microtubule cortex and the flagellum), followed by salt extraction to depolymerize the subpellicular microtubule cortex, yielding purified flagella (Schneider et al., 1987; Robinson et al., 1991) and assayed this flagella preparation for adenylate kinase activity. Figure 3A shows that adenylate kinase activity could be detected in flagella preparations from noninduced snl-2, but not in paralyzed flagella isolated from RNAi-induced snl-2. This activity could be inhibited by diadenosine pentaphosphate (Ap5A) indicative of bona fide adenylate kinase activity (Figure 3A). As expected, adenylate kinase activity was also present in the cytoskeletal fraction of the noninduced snl-2 mutant. However, this activity was absent from cytoskeletons of the induced snl-2 mutant (Figure 3B). Because there is no difference between the cytoskeletons from these two induction states, other than the lack of PFR in the flagellum of induced snl-2, we can conclude that adenylate kinase activity is only associated with the flagellum, and not with other areas of the cytoskeleton. The difference in levels of activity between the cytoskeletal and flagellar preparations is likely to arise as a consequence of the salt extraction used to depolymerize the subpellicular microtubules. The use of stringent detergent and salt extractions shows that the adenylate kinase activity must be intimately associated with the flagellum. This demonstration of activity reflected precisely the results of our proteomic investigation.
Figure 3.
Adenylate kinase activity is associated with purified flagella (A) and cytoskeletons (the subpellicular microtubule and flagellar fraction (B). Activity occurs after the addition of ADP to the equilibrated assay mixture (solid arrowhead).
Localization of Adenylate Kinase along the PFR
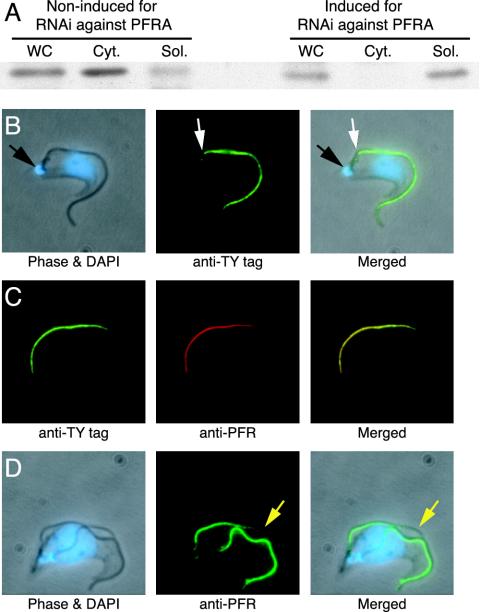
These results raised the question of where the adenylate kinases are located in the flagellum. We expressed an epitope-tagged version of TbADKA from its endogenous locus and used the tag to ascertain whether this adenylate kinase was associated with the detergent-insoluble cytoskeletal fraction. Figure 4A shows that this was indeed the case in snl-2 in its noninduced state. However, when PFRA RNAi was induced and the population displayed the paralyzed, PFR minus phenotype, the tagged TbADKA was located in the detergent-soluble fraction and was absent from the cytoskeletal fraction (Figure 4A). Immunofluorescence images of noninduced cells showed that the TbADKA protein was located specifically in the PFR and not in the axoneme (Figure 4, B-D). The axoneme extends from the flagellar basal bodies, which are physically connected to the kinetoplast (Ogbadoyi et al., 2003) to the end of the flagellum, but the PFR is only present from the point where the flagellum exits the cell body (Gull, 1999). Thus, PFR localization was confirmed by the gap in the immunofluorescence between the kinetoplast and the start of the PFR (Figure 4B). Moreover, the TbADKA immunofluorescence was entirely coincident with an antibody that specifically detects the PFR (Figure 4C). Finally, the TbADKA signal lags behind that of the axoneme as the latter extends, exactly as one would expect for a PFR protein (Figure 4D). The same results were observed for TbADKB (our unpublished data).
Figure 4.
TbADKA is a PFR-associated protein in motile parasites, but it is not present in the flagella of the induced snl-2 mutant. (A) Immunoblot analysis of TbADKA tagged with the Ty-epitope at its C terminus by using the monoclonal antibody BB2. Cell equivalents (107) were loaded per lane. ADKA, expressed from its endogenous locus, partitions into the detergent-insoluble cytoskeletal fraction (Cyt.), except under conditions where the PFR is not assembled (snl-2 induced mutant), when it is found only in the detergent-soluble cytoplasmic fraction (Sol.). WC, whole cell extract. (B) Immunolocalization of ADKA::Ty to the flagellum by using BB2. The immunofluorescence pattern indicates that ADKA is present from the point where the flagellum leaves the cell body (white arrow) to its distal tip, but not at the point where the axoneme is nucleated from the basal body that is attached to the mitochondrial genome (black arrow; Ogbadoyi et al., 2003). The positions of the nuclear and mitochondrial (kinetoplast) genomes are revealed by the blue 4′, 6-diamidino-2-phenyindole stain (DAPI). (C) Colocalization of ADKA::Ty with the monoclonal antibody ROD-1, which recognizes a large molecular weight component of the PFR. (D) Incorporation of ADKA into the new flagellum (yellow arrowhead) lags behind the elongation of the axoneme. Cells in C and D were processed as described for B.
Unusual N-Terminal Extensions Characteristic of the Flagellar Adenylate Kinases
Examination of the TbADKA and TbADKB sequences and comparison with many other known adenylate kinase sequences revealed the conservation of residues known to be important for catalysis and adenylate kinase structure in the trypanosome proteins (Figure 2; Schricker et al., 1995; Sanchez and Muller, 1998). A most unusual feature, however, was the presence of a 55-amino acid-long N-terminal extension, upstream of the conserved P-loop on both TbADKA and TbADKB (Figure 2). The N-terminal extensions of the two flagellar enzymes showed some sequence conservation. Interestingly, two lines of evidence indicated that neither extension was cleaved after translation of TbADKA or TbADKB. First, the predicted molecular weights and isoelectric points of the full-length proteins matched the values observed when each protein was resolved by 2D electrophoresis (Figure 1C). Second, and more directly, mass spectrometry of both proteins included peptide mass and sequence data in the N-terminal regions (Figure 2).
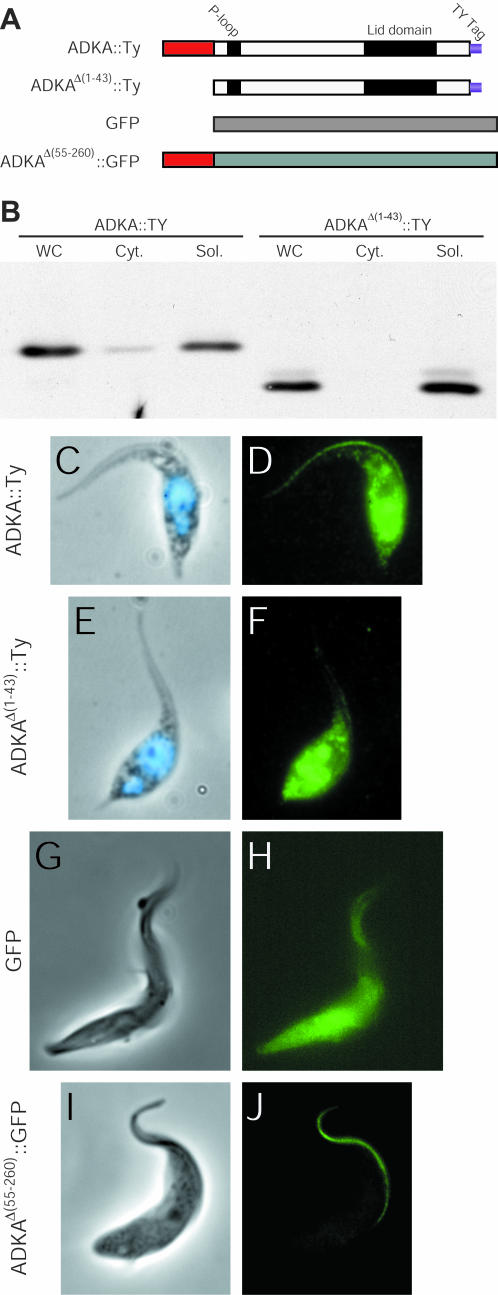
The presence of the N-terminal extensions clearly raised the issue of whether they are responsible for the unusual location of TbADKA and TbADKB in the flagellum. To address this issue, we constructed a deletion of the N-terminal region of TbADKA [ADKAΔ(1-43)::Ty] and produced cell lines expressing the epitope-tagged versions of either TbADKA or the truncated TbADKA from a strong promoter (Figure 5A). We subjected these cell lines to the same detergent extraction as used earlier and found that although a significant amount of soluble protein was produced, due to expression from a strong promoter (Figure 5, B-D), only full-length TbADKA, and not ADKAΔ(1-43), could be targeted to the flagellum (Figure 5D). The overexpression of these proteins emphasized that even though large amounts of the N-terminal-truncated protein were produced in the cytoplasm, the system was unable to concentrate such protein in the flagellum compartment. We also engineered cell lines that expressed either GFP, or GFP fused at its N terminus to the N-terminal 55-amino acids of TbADKA. Fluorescence analysis of these cell lines showed that GFP on its own localized to the cell body with insignificant amounts in the flagellum (Figure 5H), whereas GFP carrying the N-terminal adenylate kinase extension localized to the flagellum (Figure 5J). This signal was not seen in cytoskeletal preparations (detergent-extracted cells; our unpublished data), presumably because it was either in a soluble state within flagella before detergent extraction, or, if the extension also provides structural features contributing to PFR incorporation, was only loosely associated with the PFR. Overall, therefore, our data demonstrated that the N-terminal extension present on TbADKA, and presumably the extension present on TbADKB, too, are both necessary and sufficient for targeting proteins to the trypanosome flagellum. However, it also seems that once in the flagellum there are additional structural determinants that are needed to anchor the flagellar adenylate kinases within the PFR.
Figure 5.
N-terminal extension of the flagellar adenylate kinases is an essential targeting determinant. (A) Cartoon showing the structures of the N-terminal truncation [ADKAΔ(1-43)::Ty] and the chimeric GFP [ADKAΔ(55-260)::GFP] used to obtain the results shown in B-J. (B) Immunoblot analysis of overexpressed ADKA::Ty and ADKAΔ(1-43)::Ty with antibody BB2. Cell equivalents (2 × 106) were loaded per lane. Full-length ADKA is present in both detergent-insoluble and -soluble fractions, but the N-terminal truncated protein is present only in the soluble cytoplasmic fraction. (C-F) Immunofluorescence analysis of ADKA::Ty and ADKAΔ(1-43)::Ty. Methanol-fixed trypanosomes (phase and 4,6-diamidino-2-phenylindole shown in C and E) were used for immunofluorescence with BB2 (D and F). Even though an intense signal can be seen in the cytoplasm, overexpression of ADKAΔ(1-43)::Ty still does lead to any of protein being transported to the flagellum (F). (G-J) Fluorescence analysis of GFP and ADKAΔ(55-260)::GFP (H and J) in formaldehyde-fixed cells (phase, G and I).
The Unusual N-Terminal Extensions Identify Novel Conserved Adenylate Kinases in Nematode and Human Genomes
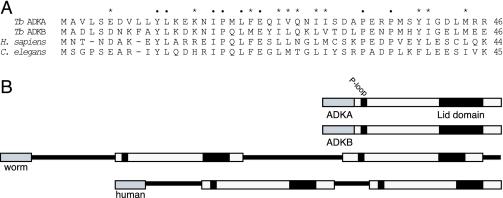
We considered the possibility that the N-terminal extensions of TbADKA and TbADKB, or motifs contained within these extensions, might be used to target other proteins to the axoneme or PFR. Whereas a bioinformatic search of the trypanosome genome database identified a number of proteins that share some sequence identity with the N-terminal regions of TbADKA and TbADKB, respectively, these do not include known flagellar proteins. Nor could we, by comparison with known flagellar components, identify motifs within the primary sequences of TbADKA and TbADKB that were suggestive of flagellar targeting sequences common to many flagellar components. However, when the bioinformatic analysis was extended to the genomes of other organisms we were surprised to find that specific blast analyses using only the N-terminal extensions of TbADKA or TbADKB identified similar sequences in the Caenorhabditis elegans and human genomes that showed unexpectedly high identity (Figure 6A). These human and C. elegans sequences turned out to be the N termini of predicted open reading frames that both contained two complete adenylate kinase domains in tandem (Figure 6B). Identification of conserved catalytic or structurally essential residues in these tandem adenylate kinases suggested that both domains would be able to catalyze phosphotransfer between a nucleotide triphosphate donor and nucleotide monophosphate acceptor.
Figure 6.
Identification of unusual adenylate kinases in C. elegans and Homo sapiens. (A) Sequence alignment of the first 46 amino acids of TbADKA and TbADKB with two sequences that were identified when these N-terminal regions were used in blast analysis of the genome databases available at National Center for Biotechnology Information. Positions of identity in all four sequences are highlighted using black dots; asterisks indicate positions where functional similarity is conserved. (B) When the two novel sequences were examined more closely (accession nos. CAA15625 [C. elegans] and NP_777283 [H. sapiens]) both proteins were found to be tandem adenylate kinases in which both putative catalytic domains were downstream of the conserved N-terminal region shown in A.
Phenotype Analysis in a Flagellar Adenylate Kinase RNAi Knock-Down Mutant
Given that induced snl-2 lacks a full PFR and is paralyzed, is it possible that removal of only adenylate kinase from the structure might be sufficient to phenocopy the original snl-2 paralyzed phenotype? We used an inducible RNAi system (Bastin et al., 2000a; Wang et al., 2000) to address this question, and we were able to successfully ablate the expression of both flagellar adenylate kinases simultaneously (as evidenced by proteomic analysis by 2D electrophoresis and a decrease of >80% in the adenylate kinase activity found in cytoskeletal extracts) using sequences specific to each gene to generate double-stranded RNA. However, when RNAi-induced and control cell lines were analyzed by videomicroscopy no differences in either speed or direction of movement, or waveform or beat pattern were detected between RNAi-induced and RNAi-noninduced cells. We also saw no structural alterations to the PFR by using electron microscopy.
DISCUSSION
Insights into Adenylate Kinase Function in the Flagellum
Motility mediated by the activity of axonemal dynein ATPases requires ATP hydrolysis, but this motility is also sensitive to the balance of ATP and ADP concentrations with recent work suggesting that some inner arm dyneins are strongly inhibited under ADP-free conditions (Yagi, 2000). Studies over the last 35 yr or so have indicated that enzymatic activities that could regulate adenine nucleotide concentrations can be detected in flagellar, ciliary, and axonemal preparations. These reports include accounts of adenylate kinase, nucleotide diphosphate kinase, and phosphagen (creatine and arginine) kinase activities in organisms as diverse as Tetrahymena, Chlamydomonas, sea uchin, and mammalian sperm (Gibbons, 1966; Watanabe and Flavin, 1976; Schoff et al., 1989; Wothe et al., 1990; Ogawa et al., 1996; Nakamura et al., 1999; Patel-King et al., 2002; Miranda-Vizuete et al., 2003; Sadek et al., 2003). An obvious difficulty, acknowledged by some authors, of biochemical fractionation studies is the definition of the specificity of location of, for example, adenylate kinase activity when this is such a ubiquitous enzyme in the cell (Schoff et al., 1989). Our present study now provides direct evidence that particular isoforms of adenylate kinase are associated with the eukaryotic flagellum, and moreover we provide the explanation for the flagellar specificity of these isoforms.
The recognition of the N-terminal extensions on these isotypes provides an explanation for the mechanisms by which unique isotypes of a protein family could be targeted to the flagellum. Adenylate kinases are historically recognized as enzymes of the cytoplasm or mitochondrion in microbes or animal cells. Our discovery of these isotypes has facilitated an initial description of a novel protein sequence necessary and sufficient to endow flagellar localization. This system is distinct from both the targeting signals located within the C-terminal region of the major PFR proteins (Bastin et al., 1999a), and the N-terminal sequences important for targeting of certain flagellar membrane proteins (Godsel and Engman, 1999; Bloodgood, 2000; Ignatushchenko et al., 2004). A further key point is the biological function of flagellar adenylate kinase activity, and our data provide three important contributions to this debate. The adenylate kinase activity is present in multiple isoforms, they are built into the cytoskeletal structure, and finally they are distributed in a uniform manner along essentially the length of the flagellum. Although multiple isoforms might indicate redundancy or specificity of function, their distributed location does not suggest a local function at either the entry point into the flagellum from the cytoplasm, or the distal tip where assembly of axonemal and PFR components is achieved (Bastin et al., 1999a). Also this distribution does not suggest a local function in the initiation of axonemal beating. However, this distribution would indicate a possible function of flagellar adenylate kinase in providing a phosphotransfer relay (Dzeja and Terzic, 2003) that would facilitate a more rapid and efficient movement of adenine nucleotides along the flagellum than would be achieved by diffusional exchange alone. RNAi knockdown of the two flagellar adenylate kinases that we have identified does not obviously affect flagellar motility. It is possible that adenylate kinase activity is required for flagellar motility and that the residual activity (<20% of normal) is sufficient to sustain flagellar motility. Alternatively, adenylate kinase activity is not required under normal culture conditions, or this adenylate kinase system can be bolstered by a redundancy involving yet more homeostatic mechanisms.
The notion of compensatory phenotypes for localized adenylate kinase deficiency may not be so surprising in the light of investigations into the roles played by individual adenylate kinase, and creatine kinase, isoforms in mammals. Transgenic mice deficient in the major cytosolic isoform of adenylate kinase display striking metabolic rearrangements in which wide-scale remodeling of glycolytic and mitochondrial pathway networks are sufficient to sustain high energy phosphotransfer in muscle tissue, except under conditions of metabolic stress (Janssen et al., 2000; Carrasco et al., 2001; Janssen et al., 2003a,b). These observations are relevant when one considers the metabolic plasticity of cultured trypanosomes (Bochud-Allemann and Schneider, 2002; van Weelden et al., 2003) and indicate the most direct way to assess whether flagellar adenylate kinases contribute to the economy of energy metabolism will be transmission of null mutants through the natural tsetse fly vector and so explored within the natural nutritional context. This experiment, however, could not be executed with the laboratory-adapted strains currently available.
Our studies clearly locate the flagellar adenylate kinases to the PFR in the trypanosome flagellum. Such extraaxonemal structures are important features of diverse eukaryotic flagella (Santrich et al., 1997; Bastin et al., 1998; Miki et al., 2002). Our data also show that the PFR of trypanosomes is not merely an architectural feature but rather acts as a matrix into which enzymes such as adenylate kinase and most likely other regulatory proteins can be built. This analysis parallels recent conclusions concerning the biochemical nature of another extraaxonemal structure, the fibrous sheath of mammalian spermatozoa (Miki et al., 2002; Eddy et al., 2003), and perhaps provides an intriguing example of convergent evolution between phylogenetically distant organisms. Major components of the fibrous sheath have been revealed to be AKAP4, AKAP3, and TAKAP-80, which provide anchoring opportunities for cAMP-dependent protein kinase, and specific isoforms of enzymes such as glyceraldehyde-3-phosphate dehydrogenase and hexokinase (Eddy et al., 2003). However, it is important to realize that there is a significant difference between the flagellum of a metazoan sperm and that of a eukaryotic microbe. In essence the sperm lacks cytoplasm, whereas the eukaryotic microbial cell possesses abundant cytoplasm with organelles. Thus, localization of metabolic functions in the flagellum of the latter is best viewed as an isotype differential localization problem, whereas in the former it maybe viewed as more of a cell type-dependent expression phenomenon.
Results from many systems, including trypanosomes, indicate that the localization of cAMP-dependent signaling pathways to flagella is ubiquitous, and at least in some cases is linked to motility (Inaba, 2003; King, 2003). Key enzymes in this process, adenylate cyclases and cAMP phosphodiesterases, have been localized to flagella in several organisms. Thus, adenine nucleotide homeostasis is likely to require extensive regulation in the flagellar compartment to take account of concurrent motility and signaling phenomena and the maintenance of a balance between them. Redundancy in homeostatic systems may therefore explain the absence of singular phenotypes after adenylate kinase RNAi, but we believe that the structural incorporation of enzymes into extraaxonemal structures is significant in explaining why complete removal (mouse knock-outs for AKAP4 and RNAi of PFR-A in trypanosomes, respectively) of the structure has such dramatic consequences (Bastin et al., 1998, 2000a; Miki et al., 2002). This provides an alternative to considering a merely physical reduction in the diameter of the flagellum as being influential in explaining the abnormal motility phenotype in trypanosomatid mutants that fail to assemble a PFR (Santrich et al., 1997). Although this biophysical explanation remains possible, it is important to note that the diameter of the PFR varies by 100% between Trypanosoma, Crithidia, and Leishmania species (150-300 nm) with no observable concomitant change in the motility of these organisms (Bastin et al., 1996b, 2000b). In addition, in trypanosomatid species such as C. oncopelti that were reported to lack a PFR there was no discernible difference in their motility in comparison with Crithidia fasciculata, which does assemble a normal PFR (Bastin et al., 1996b). Moreover, we have recently observed that in fact a very small PFR is present in Crithidia oncopelti (Gadelha and Gull, unpublished), so providing further support for our hypothesis that the PFR provides a critical platform into which regulatory or metabolic enzymes are anchored.
Insights into Adenylate Kinase Targeting into the Flagellum
Our analysis of the N-terminal extension of TbADKA provides the molecular explanation for targeting of these isoforms; the extension is both necessary and sufficient for locating proteins to the flagellum. The N-terminal sequences of TbADKA and TbADKB show a conservation of several motifs and are clearly related. However, although these adenylate kinases are built into the PFR structure, as defined by detergent and high salt treatment, N-terminal-tagged GFP is targeted to the flagellum but it is not stably incorporated into the PFR, such that it can withstand such treatments. Thus, some additional feature is likely to influence stable incorporation. Although we have previously described a C-terminal motif important in some proteins for targeting to the flagellum (Bastin et al., 1999a; Ersfeld and Gull, 2001) and there are at least two distinct mechanisms for targeting proteins to the flagellar membrane (Godsel and Engman, 1999; Bloodgood, 2000; Ignatushchenko et al., 2004), the present N-terminal extension sequences represent additional excellent illustrations of eukaryotic flagellar targeting sequences. Are these sequences widely seen in the T. brucei genome, and hence could they facilitate identification of other flagellar proteins? The answer here is that we could recognize a number of putative proteins containing sequences that shared identity with the N-terminal sequences described here. However, if there are certain common motifs that define a whole family of flagellar proteins further experimental work will be needed to confirm such candidates.
Targeting motifs are likely to be used by cargo proteins for transportation into the flagellum by intraflagellar transport (Scholey, 2003) or other systems (such as ones necessary for the targeting of membrane proteins (Godsel and Engman, 1999; Ignatushchenko et al., 2004). With this in mind, we were therefore intrigued to find predicted proteins in other genomes, such as human and C. elegans, that share identity at their respective N termini with the T. brucei adenylate kinases. In particular, a [YLxxxxxIPxLxE] sequence, followed by two conserved prolines was present in both of the examples shown in Figure 6. Moreover, both putative polypeptides contain two apparently complete, but non-identical, adenylate kinase units downstream of the conserved N-terminal region. Interestingly, we recognize that this tandem organization provides some echoes of the unusual triplet creatine kinase targeted to sperm flagella in sea urchins (Wothe et al., 1990) and the triplet nucleotide diphosphate kinases that have been localized to the axoneme in sea urchin and Chlamydomonas (Ogawa et al., 1996; Patel-King et al., 2002) and the fibrous sheath in mammalian sperm (Miranda-Vizuete et al., 2003). Given the existence of specific isoforms of certain central metabolic enzymes, and targeting sequences for these isoforms, we suggest that these isoforms form part of a conserved complex, cytoskeletal-anchored metabolic capability that is built into the eukaryotic flagellum.
Acknowledgments
We thank Flavia Moriera-Leite for the illustrative electron micrographs. Sequence data for the T. brucei genome were obtained from The Institute for Genome Research and The Sanger Institute; sequencing of the T. brucei genome was accomplished as part of the Trypanosoma Genome Network with support from The National Institute of Allergy and Infectious Diseases (National Institutes of Health) and The Wellcome Trust. This work was supported by grants from the Wellcome Trust and the Royal Society. T.J.P. was supported by a Biotechnology and Biological Sciences Research Council studentship. M.L.G. is a Royal Society University Research Fellow and K.G. is a Wellcome Trust Principal Research Fellow.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0217. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0217.
Abbreviations used: Ap5A, diadenosine pentaphosphate; PFR, paraflagellar rod; RNAi, RNA interference.
References
- Bastin, P., Bagherzadeh, Z., Matthews, K.R., and Gull, K. (1996a). A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235-239. [DOI] [PubMed] [Google Scholar]
- Bastin, P., Ellis, K., Kohl, L., and Gull, K. (2000a). Flagellum ontogeny in trypanosomes studied via an inherited and regulated RNA interference system. J. Cell Sci. 113, 3321-3328. [DOI] [PubMed] [Google Scholar]
- Bastin, P., MacRae, T.H., Francis, S.B., Matthews, K.R., and Gull, K. (1999a). Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 19, 8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin, P., Matthews, K.R., and Gull, K. (1996b). The paraflagellar rod of Kinetoplastida: solved and unsolved questions. Parasitol. Today 12, 302-307. [DOI] [PubMed] [Google Scholar]
- Bastin, P., Pullen, T.J., Moreira-Leite, F.F., and Gull, K. (2000b). Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes Infect. 2, 1865-1874. [DOI] [PubMed] [Google Scholar]
- Bastin, P., Pullen, T.J., Sherwin, T., and Gull, K. (1999b). Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 112, 3769-3777. [DOI] [PubMed] [Google Scholar]
- Bastin, P., Sherwin, T., and Gull, K. (1998). Paraflagellar rod is vital for trypanosome motility. Nature 391, 548. [DOI] [PubMed] [Google Scholar]
- Bloodgood, R.A. (2000). Protein targeting to flagella of trypanosomatid protozoa. Cell Biol. Int. 24, 857-862. [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann, N., and Schneider, A. (2002). Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849-32854. [DOI] [PubMed] [Google Scholar]
- Brun, R., and Schonenberger. (1979). Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36, 289-292. [PubMed] [Google Scholar]
- Burland, T.G., Gull, K., Schedl, T., Boston, R.S., and Dove, W.F. (1983). Cell type-dependent expression of tubulins in Physarum. J. Cell Biol. 97, 1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, A.J., et al. (2001). Adenylate kinase phosphotransfer communicates cellular energetic signals to ATP-sensitive potassium channels. Proc. Natl. Acad. Sci. USA 98, 7623-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja, P.P., and Terzic, A. (2003). Phosphotransfer networks and cellular energetics. J. Exp. Biol. 206, 2039-2047. [DOI] [PubMed] [Google Scholar]
- Eddy, E.M., Toshimori, K., and O'Brien, D.A. (2003). Fibrous sheath of mammalian spermatozoa. Microsc. Res. Tech. 61, 103-115. [DOI] [PubMed] [Google Scholar]
- Ersfeld, K., and Gull, K. (2001). Targeting of cytoskeletal proteins to the flagellum of Trypanosoma brucei. J. Cell Sci. 114, 141-148. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi, K., Nosaka, M., Nakazawa, A., and Go, M. (1996). Ancient divergence of long and short isoforms of adenylate kinase: molecular evolution of the nucleoside monophosphate kinase family. FEBS Lett. 385, 214-220. [DOI] [PubMed] [Google Scholar]
- Gibbons, I.R. (1966). Studies on the adenosine triphosphatase activity of 14 S and 30 S dynein from cilia of Tetrahymena. J. Biol. Chem. 241, 5590-5596. [PubMed] [Google Scholar]
- Godsel, L.M., and Engman, D.M. (1999). Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 18, 2057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull, K. (1999). The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53, 629-655. [DOI] [PubMed] [Google Scholar]
- Ignatushchenko, M., Nasser, A., and Landfear, S.M. (2004). Sequences required for the flagellar targeting of an integral membrane protein. Mol. Biochem. Parasitol. 135, 89-100. [DOI] [PubMed] [Google Scholar]
- Inaba, K. (2003). Molecular architecture of the sperm flagella: molecules for motility and signaling. Zool. Sci. 20, 1043-1056. [DOI] [PubMed] [Google Scholar]
- Janssen, E., de Groof, A., Wijers, M., Fransen, J., Dzeja, P.P., Terzic, A., and Wieringa, B. (2003a). Adenylate kinase 1 deficiency induces molecular and structural adaptations to support muscle energy metabolism. J. Biol. Chem. 278, 12937-12945. [DOI] [PubMed] [Google Scholar]
- Janssen, E., Dzeja, P.P., Oerlemans, F., Simonetti, A.W., Heerschap, A., de Haan, A., Rush, P.S., Terjung, R.R., Wieringa, B., and Terzic, A. (2000). Adenylate kinase 1 gene deletion disrupts muscle energetic economy despite metabolic rearrangement. EMBO J. 19, 6371-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, E., Terzic, A., Wieringa, B., and Dzeja, P.P. (2003b). Impaired intracellular energetic communication in muscles from creatine kinase and adenylate kinase (M-CK/AK1) double knock-out mice. J. Biol. Chem. 278, 30441-30449. [DOI] [PubMed] [Google Scholar]
- King, S.M. (2003). Organization and regulation of the dynein microtubule motor. Cell Biol. Int. 27, 213-215. [DOI] [PubMed] [Google Scholar]
- Miki, K., Willis, W.D., Brown, P.R., Goulding, E.H., Fulcher, K.D., and Eddy, E.M. (2002). Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev. Biol. 248, 331-342. [DOI] [PubMed] [Google Scholar]
- Miranda-Vizuete, A., Tsang, K., Yu, Y., Jimenez, A., Pelto-Huikko, M., Flickinger, C.J., Sutovsky, P., and Oko, R. (2003). Cloning and developmental analysis of murid spermatid-specific thioredoxin-2 (SPTRX-2), a novel sperm fibrous sheath protein and autoantigen. J. Biol. Chem. 278, 44874-44885. [DOI] [PubMed] [Google Scholar]
- Miura, K., et al. (2001). Cloning and characterization of adenylate kinase from Chlamydia pneumoniae. J. Biol. Chem. 276, 13490-13498. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Iitsuka, K., and Fujii, T. (1999). Adenylate kinase is tightly bound to axonemes of Tetrahymena cilia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 124, 195-199. [DOI] [PubMed] [Google Scholar]
- Ogawa, K., Takai, H., Ogiwara, A., Yokota, E., Shimizu, T., Inaba, K., and Mohri, H. (1996). Is outer arm dynein intermediate chain 1 multifunctional? Mol. Biol. Cell 7, 1895-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbadoyi, E.O., Robinson, D.R., and Gull, K. (2003). A high-order transmembrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar Basal bodies in trypanosomes. Mol. Biol. Cell 14, 1769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King, R.S., Benashski, S.E., and King, S.M. (2002). A bipartite Ca2+-regulated nucleoside-diphosphate kinase system within the Chlamydomonas flagellum. The regulatory subunit p72. J. Biol. Chem. 277, 34271-34279. [DOI] [PubMed] [Google Scholar]
- Porter, M.E., and Sale, W.S. (2000). The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151, F37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen, T.J. (2002). A proteomic investigation of the trypanosome flagellum. Ph.D. dissertation, University of Manchester, Manchester, United Kingdom.
- Robinson, D., Beattie, P., Sherwin, T., and Gull, K. (1991). Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 196, 285-299. [DOI] [PubMed] [Google Scholar]
- Sadek, C.M., et al. (2003). Characterization of human thioredoxin-like 2. A. novel microtubule-binding thioredoxin expressed predominantly in the cilia of lung airway epithelium and spermatid manchette and axoneme. J. Biol. Chem. 278, 13133-13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, L.B., and Muller, M. (1998). Cloning and heterologous expression of Entamoeba histolytica adenylate kinase and uridylate/cytidylate kinase. Gene 209, 219-228. [DOI] [PubMed] [Google Scholar]
- Santrich, C., Moore, L., Sherwin, T., Bastin, P., Brokaw, C., Gull, K., and LeBowitz, J.H. (1997). A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol. Biochem. Parasitol. 90, 95-109. [DOI] [PubMed] [Google Scholar]
- Schneider, A., Sherwin, T., Sasse, R., Russell, D.G., Gull, K., and Seebeck, T. (1987). Subpellicular and flagellar microtubules of Trypanosoma brucei brucei contain the same alpha-tubulin isoforms. J. Cell Biol. 104, 431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoff, P.K., Cheetham, J., and Lardy, H.A. (1989). Adenylate kinase activity in ejaculated bovine sperm flagella. J. Biol. Chem. 264, 6086-6091. [PubMed] [Google Scholar]
- Scholey, J.M. (2003). Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19, 423-443. [DOI] [PubMed] [Google Scholar]
- Schricker, R., Magdolen, V., Strobel, G., Bogengruber, E., Breitenbach, M., and Bandlow, W. (1995). Strain-dependent occurrence of functional GTP:AMP phosphotransferase (AK3) in Saccharomyces cerevisiae. J. Biol. Chem. 270, 31103-31110. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- van Weelden, S.W., Fast, B., Vogt, A., van der Meer, P., Saas, J., van Hellemond, J.J., Tielens, A.G., and Boshart, M. (2003). Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 278, 12854-12863. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Morris, J.C., Drew, M.E., and Englund, P.T. (2000). Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174-40179. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., and Flavin, M. (1976). Nucleotide-metabolizing enzymes in Chlamydomonas flagella. J. Biol. Chem. 251, 182-192. [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld, K., and Gull, K. (2003). The frequency of gene targeting in Trypanosoma brucei is independent of target site copy number. Nucleic Acids Res. 31, 3993-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz, E., and Clayton, C. (1995). Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268, 1179-1183. [DOI] [PubMed] [Google Scholar]
- Wirtz, E., Leal, S., Ochatt, C., and Cross, G.A. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89-101. [DOI] [PubMed] [Google Scholar]
- Wothe, D.D., Charbonneau, H., and Shapiro, B.M. (1990). The phosphocreatine shuttle of sea urchin sperm: flagellar creatine kinase resulted from a gene triplication. Proc. Natl. Acad. Sci. USA 87, 5203-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi, T. (2000). ADP-dependent microtubule translocation by flagellar inner-arm dyneins. Cell Struct. Funct. 25, 263-267. [DOI] [PubMed] [Google Scholar]