Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) is the most common form of malignancy in pancreatic carcinoma. Here we report our discovery on the correlations between transcriptional alternative splicing (AS) of NUMB, APP, VEGFA and PDAC in patients. Methods: The expression of NUMB, APP, VEGFA from patient samples was determined by qRT-PCR. AS of these genes was examined through laser induced fluorescence capillary electrophoresis. Correlation between the AS of the genes and results from clinical laboratory examinations were analyzed. Expression of NOTHC1 and NOTCH4 as downstream target genes was examined by qRT-PCR and Western blot. Results: Quantitative results indicated that expression of NUMB was significantly lower in tumor tissues (TT) than in para-tumor tissues (TP) (P<0.05), while APP (P<0.01) and VEGFA (P<0.05) were significantly higher. AS transcript percentage of NUMB PRRS was lower in TT than TP (P<0.05). AS transcript percentage of VEGFA (105+185) was significantly lower in TT than TP (P<0.05) compared to higher expression of VEGFA (206+338) (P<0.05). Regression analysis indicated that AS transcript of NUMB PRRL correlated with tumor size (P<0.01), while AS transcripts of APP and VEGFA correlated with results of laboratory examinations. To reveal the correlation between AS and its downstream targets, NOTCH1 and NOTCH4 were selected as NUMB gene targets and detected to be significantly higher in TT than TP (P<0.05). Conclusion: Alternative splicing of APP, VEGFA and NUMB may play an important role in pathogenesis of pancreatic ductal adenocarcinoma. Among the 3 genes, PRRL form of NUMB gene is highly expressed in TT and positively correlated with tumor size, while PRRS is lacking in TT and negatively correlated with NOTCH expression suggesting that PRRS might be protective in tumorogenesis and shows NOTCH pathway down regulation ability.
Keywords: Alternative splicing, NUMB, pancreatic ductal carcinoma
Introduction
Pancreatic ductal adenocarcinoma is the most common form of malignancy in pancreatic carcinoma, which accounts for more than 80% of total cases in clinic [1]. Researches have been undertaken to elucidate the mechanisms of this malignancy including alternative splicing (AS) in specific gene transcription. Newly synthesized transcripts (pre-mRNA) containing both exons and introns, which are coding regions and non-coding intervening regions, are subjected to post-transcriptional editing in eukaryotes. These process include 5’ capping, intron exclusion and 3’ cleavage and polyadenylation [2,3]. Pre-mRNA splicing is a crucial step that removes intronic sequences and joins exons together for efficient gene expression. Exons are joined and introns are removed through splicing, which are performed by the splicesome as a large ribonucleoprotein complex consisting five subcomponent small nuclear ribonucleoprotein particles (snRNP), U1, U2, U4, U5 and U6. Briefly, U1 snRNP recognize 5’ of intronic sequence and U2 complex binds to 3’, followed by binding of U4/U5/U6 tri-snRNP. Finally, intron sequences are excised and adjacent exons are joined [4].
Aberrations in this process can perturb normal gene coding or even generate antagonistic functional protein causing alternative splicing (AS) [5]. Alternative splicing is an activity to produce multiple isoforms of a gene via acting at alternative splice sites. Known isoforms of alternative splice have been revealed for their correlations with some cancers and other human diseases [6-8]. For example, alternative splicing of Caspase 8 was originally found responsible for apoptosis in leukemia. Similar situations were later found for androgen receptor and estrogen receptor variants in breast carcinoma [9-11].
Alternative splicing (AS) of vascular endothelial growth factor A (VEGFA), amyloid beta (A4) precursor protein (APP), and the Numb (Drosophila melanogaster) homolog (NUMB) was previously found to participate in non-small cell lung cancer (NSCLC), breast cancer or colonic cancer from a high-throughput mRNA AS screening study [12]. Variants of gene transcripts participating in cancer characteristic have been reported in the publications, which shed light on study of pancreatic malignancy. However, there are only two critical issues existing in the present study: 1) Without the knowledge from a broad investigation because there are only two alternative splicing cases about cholecystokinin-B/gastrin (CCK-B) receptor and secretin receptor were reported in pancreatic ductal adenocarcinoma [13,14]; 2) Studies were limited on cell lines rather than patient samples that were not able to provide a direct relation between PDAC and AS.
In this study, we tested 10 pairs of tumor and para-tumor tissue samples from PDAC patients. Total expressions of NUMB, VEGFA and APP were analyzed by qRT-PCR. We used a method for quantization of alternative splicing transcripts, which was well improved from any previous ones by Christine M. Misquitta-Ali et al. [x]. Laser induced fluorescence capillary electrophoresis (LIFCE) analysis instead of traditional RT-PCR/Southern blotting semi-quantitative approach was used to precisely calculate the tumor relating AS in tissues. Correlation between forms of AS and results Clinical or laboratory examinations were obtained from the analysis by linear regression methods. In order to more deeply clarify the mechanism of correlation between AS and PDAC, NOTCH as the downstream targets of NUMB was analyzed in our study. The gene expression of NOTCH was up regulated in tumor tissue, which was responsible for cancer growth and proliferation. Remarkably, NUMB has been reported to work against NOTCH pathway through endocytosis of NOTCH receptors and the other NUMB regulated pathways involving in some ubiquitin network [15]. The expression of NOTCH1 and NOTCH4 was analyzed by qRT-PCR on transcriptional level and Western blotting on translational level. Furthermore, correlation between PRRS and NOTCH1/4 was analyzed in our study.
Materials and methods
Patients and sampling
10 pancreatic ductal adenocarcinoma patients were enrolled in this study. The diagnoses were confirmed based on clinical manifestation, pathological and serological examinations. Pregnant patients and patients with other chronic diseases or cancer were not included. The cancer classification assessment was based on NCCN guideline [16]. Tumor tissues were named TT in short and Para-tumor Tissues were abbreviated as TP in this article. Tissues were excised during operation and was preserved in Anti-degradation Buffer (SLNco, Cinoasia ltd, China) in -80°C refrigerator followed the manufacturer’s instructions. All donors were well informed, and the methods of processing were approved by Ethics Committee of Changhai Hospital, School of Medicine, Second Military Medical University, Shanghai, China.
Quantitative real-time PCR
Total RNA from tumor tissues and para-tumor tissues was extracted by RNA Extraction Kit (SLNco, Cinoasia, China), and cDNA was synthesized using PrimeScript RT reagent Kit (TaKaRa Biotechnology, Japan). The primers for general NUMB, APP and VEGFA quantization and NOTHC1, NOTCH4 were described below. In Table 1, Primers were designed with PRIMER 5.0 (ABI, Foster City, CA, USA) and synthesized by Generay (Shanghai, China). General products of NUMB, APP and VEGFA after Reverse transcription were amplified from Exon 5~6, Exon 5~6 and Exon 1~3 as templates respectively as those were reported with exons constantly included regions (Figure 1). Reverse transcription was performed at 37°C for 15 minutes and at 85°C for 5 seconds using the ReverTra Ace® qPCR RT Kit (FSQ-101, TOYOBO, Japan) following the manufacturer’s protocol. PCRs were performed 5 min at 95°C, followed by 45 cycles of 15 s at 95°C, and 1 min at 60°C. The processed were conducted on a Real-time Thermo Cycler (FTC3000, Funglyn, Canada) with SYBR Green Real-time PCR Master Mix (QPK-201, TOYOBO, Japan). The specificity of real-time PCR was confirmed by melting-curve analysis. Relative expressions were determined by normalizing expression of each Ct value to GAPDH Ct value. Data were analyzed according to 2-ΔΔCt formula.
Table 1.
Quantitative Real-time PCR primers for general GAPDH, NUMB, APP, VEGFA, NOTHC1 and NOTCH4 quantization
| Gene name | Primer sequence (5’ to 3’) | Amplicon size |
|---|---|---|
| h-gapdh-F | TGAAGGTCGGAGTCAACGGA | 225 bp |
| h-gapdh-R | CCTGGAAGATGGTGATGGGAT | |
| h-numb-F | GCTGGATCTGTCACTGCTTCAT | 118 bp |
| h-numb-R | CCACATTCCTTCTCCCGCTTC | |
| h-app-F | GGCGGAGCAGACACAGACTA | 131 bp |
| h-app-R | ACCTCATCACCATCCTCATCGT | |
| h-vegfa-F | TTGCTGCTCTACCTCCACCAT | 270 bp |
| h-vegfa-R | GGTGATGTTGGACTCCTCAGTG | |
| h-notch1-F | ACATCAACGAGTGTGCCAGTG | 128 bp |
| h-notch1-R | GCAGTCAGGCGTGTTGTTCT | |
| h-notch4-F | ACCTGCTCAACGGCTTCCA | 193 bp |
| h-notch4-R | AGGCACTCATCCACCTCTGTT |
Figure 1.
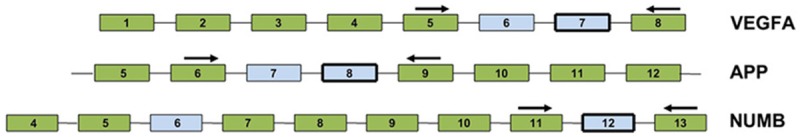
Black arrows stand for forward and reverse primers selected to bridge variable exons for NUMB, APP and VEGFA alternative transcripts.
FAM labeled alternative splicing PCR
PCR amplifications of NUMB APP and VEGFA were performed with 5’-6-carboxyfluorescein (FAM)-labeled forward primers. Primers were designed to produce PCR products covering with or without exons of each transcript (Table 2; Figure 1). Reverse transcription was performed as above described using the ReverTra Ace® qPCR RT Kit (FSQ-101, TOYOBO, Japan) following the manufacturer’s protocol. PCRs were performed 5 min at 95°C, followed by 45 cycles of 15 s at 95°C, and 1 min at 60°C. The processed were conducted on a Real-time Thermo Cycler (FTC3000, Funglyn, Canada) with SYBR Green Real-time PCR Master Mix (QPK-201, TOYOBO, Japan).
Table 2.
FAM labeled primers for GAPDH, NUMB, APP and VEGFA gene alternative splicing quantization
| Gene name | Primer sequence (5’ to 3’) | Amplicon size | Primer Starts/Ends |
|---|---|---|---|
| h-gapdh-F | FAM-TGAAGGTCGGAGTCAACGGA | 225 bp | |
| h-gapdh-R | CCTGGAAGATGGTGATGGGAT | ||
| h-numb-F | FAM-TGCTCCGATGACCAAACCAG | 157 bp (PRRS)/301 bp (PRRL) | EXON 11 |
| h-numb-R | CACCTCTTCTAACCATCGGTC | EXON 13 | |
| h-app-F | FAM-CCTACGAAGAAGCCACAGAG | 162 bp/330 bp/387 bp | EXON 6 |
| h-app-R | GGGCATGTTCATTCTCATCC | EXON 9 | |
| h-vegfa-F | FAM-TGAGCTTCCTACAGCACAAC | 105 bp/185 bp/205 bp/338 bp | EXON 5 |
| h-vegfa-R | TCGATGGTGATGGTGTGGTG | EXON 8 |
Laser induced fluorescence capillary electrophoresis analysis (LIFCE)
The FAM-labeled PCR products were diluted (to 1/10) and 1 μl was supplemented with 10 μl formamide (Sangon, China) and 0.5 μl of GeneScan ROX 500 (Applied Biosystems, USA). The mixture was denatured at 95°C for 3 min, and subsequently cooled on ice, followed by separation on an ABI 3730 capillary sequencer. Data were collected and analyzed with the GeneMarker software. The amount of each isoform in per PCR reaction was calculated by the area under the curve. Subsequent comparisons of isoform were normalized to GAPDH curve and proportions were made by standardized calculation of percentage. All values are given as mean of triplicate PCRs including standard deviation.
Western blotting
Total protein from tumor and para-tumor tissues was extracted using nucleic/plasma protein extraction kit (SLNco, Cinoasia, China). Protein samples were then loaded to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (Biorad, USA). Samples were separated in 10% SDS-PAGE and transferred onto a nitrocellulose membrane for 2 hours (Millipore, USA) at 4°C, 200 mA. Then the membrane is blocked for 1 hour at room temperature in 3% non-fat milk. Incubation with the NOTCH1, NOTCH4 and GAPDH primary antibodies are applied (ab8925, ab134831, ab8245, 1:500, Abcam, UK) with continuous gentle agitation overnight at 4°C. The membrane is incubated for 1 hour with HRP-conjugated secondary antibodies (1:1000, Beyondtime, China) at room temperature, and finally developed by Pierce ECL system (Thermo Pierce, USA).
Laboratory examination
Clinical and laboratory examinations were carried out by Changhai Hospital, Second Military Medical University when patients were admitted. Tumor size, corpuscular hemoglobin concentration, platelet distribution, small round cell number, serum sodium and serum chloride. Values were compared with AS expression level and calculated by linear regression methods.
Statistics
Results were presented as mean ± SEM. GraphPad Prism 5.0 (GraphPad Software, San Diego CA, USA) was used in statistics procedure. Correlation between AS transcripts and examinations were made by linear regression. P<0.05 was considered significant in unpaired t-test. (*stands for P<0.05; **P<0.01; ***P<0.001).
Results
Total expression levels for transcriptions of NUMB, APP and VEGFA
Results of total expression levels for the three transcripts revealed that NUMB showed a significantly lower expression in TT than that in TP (P<0.05), while APP (P<0.01) and VEGFA (P<0.05) were significantly higher in TP than that in TT (Figure 2).
Figure 2.
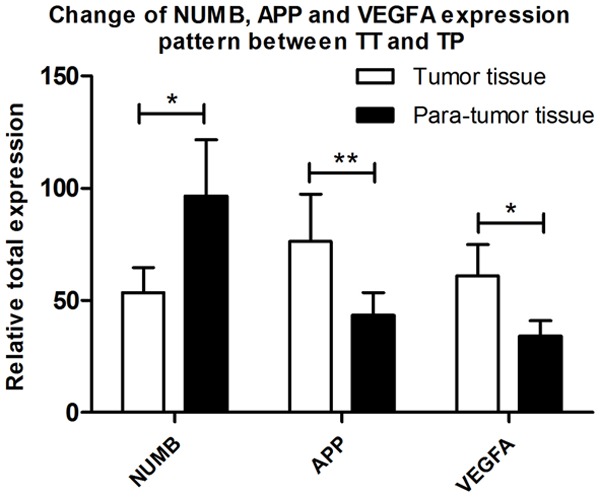
Overall expression pattern of NUMB, APP and VEGFA between Tumor Tissue and Para-tumor Tissue samples.
Alternative splicing changes in tumor tissues vs. para-tumor tissues
Results showed NUMB Exon12 exclusion AS (PRRS) had lower expression level in TT than that in TP (P<0.05). On the other hand, Exon12 inclusion of NUMB PRRL was significantly richer in TT than that in TP (P<0.05) (Figure 3).
Figure 3.
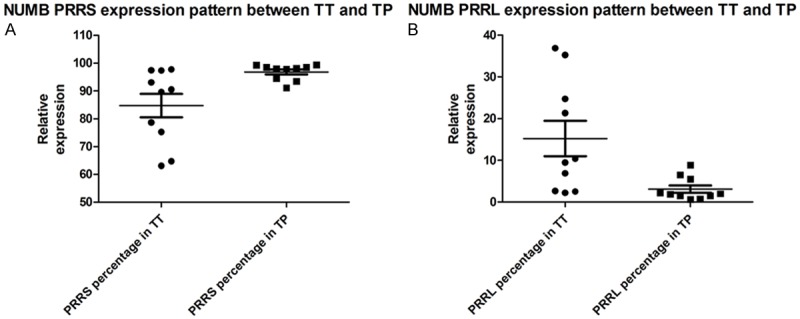
Alternative splicing changes of NUMB transcripts in TT samples vs. TT tissues by LIFCE. A and B show comparison of PRRL and PRRS quantization level of NUMB between TT and TP. Results reveal that PRRL (P<0.05)expresses much higher in TT while PRRS (P<0.05) is on the contrary.
Alternative splicing changes of APP transcripts in TT samples vs. TP tissues were detected by LIFCE. Panels of (a) (b) and (c) showed the comparisons of quantization percentage levels for APP 162, APP 330 and APP 387 between in TT and in TP. Results showed that there was no difference of expression pattern of these transcripts between TT and TP. The panels of (d) (e) (f) and (g) showed the comparisons of quantization percentage levels for VEGFA 105, VEGFA 185, VEGFA 206 and VEGFA 338 between TT and TP. Results showed that there was also no difference of expression pattern difference the transcripts between TT and TP. However, Panel (h) showed percentage of VEGFA 105+185 as a shorter form of VEGFA was present to be significantly lower in TT than in TP (P<0.05) (Figure 4).
Figure 4.
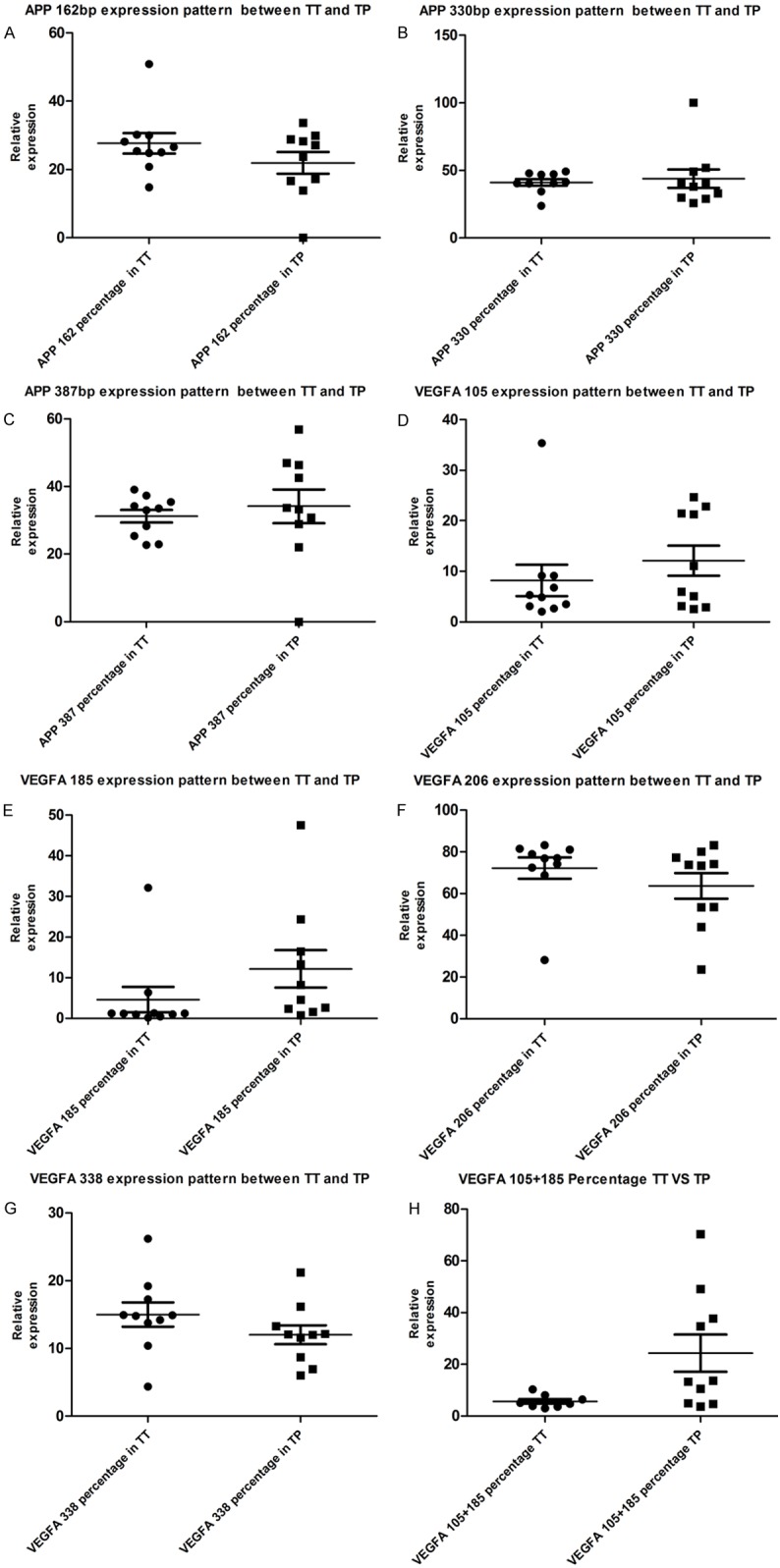
Alternative splicing changes of APP transcripts in TT samples vs. TP tissues by LIFCE. A-C. Show comparison of APP 162, APP 330 and APP 387 quantization percentage level of APP between TT and TP. Results show no expression pattern difference between TT and TP in these transcripts. D-G. Show comparison of VEGFA 105, VEGFA 185, VEGFA 206 and VEGFA 338 quantization percentage level of VEGFA between TT and TP. Results show no expression pattern difference between TT and TP in these transcripts. H. However VEGFA 105+185 percentage, a shorter form of VEGFA is present to be significantly lower in TT and TP (P<0.05).
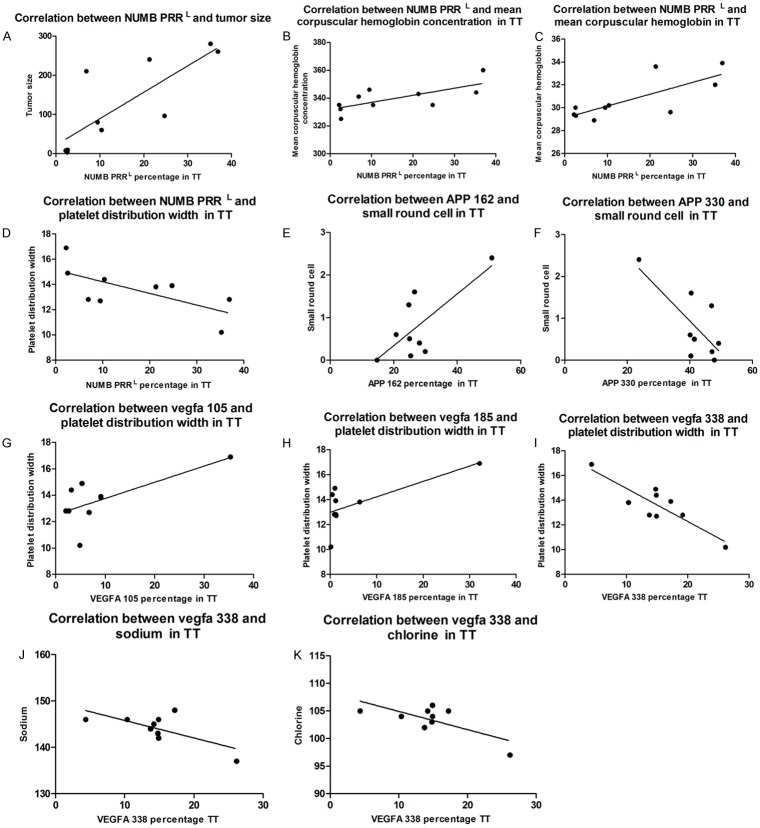
Correlation between as transcripts of NUMB, APP, VEGFA and morphological results of clinical or laboratory examinations
Linear regression results showed percentage of Numb PRRS transcript in TT positively correlated with platelet distribution width (r2=0.46, *P<0.05), but negatively correlated with tumor size (r2=0.65, **P<0.01), mean corpuscular hemoglobin (r2=0.59, ***P<0.001), mean corpuscular hemoglobin concentration (r2=0.51, *P<0.05) (Figure 5).
Figure 5.
Linear regression results show PRRL transcript of Numb percentage in TT is positively correlated with tumor size (r2=0.65, P<0.01), mean corpuscular hemoglobin (r2=0.59, P<0.001), mean corpuscular hemoglobin concentration (r2=0.51, P<0.05), and negatively correlated with platelet distribution width (r2=0.46, P<0.05). APP162 expression percentage in tumor tissue is correlated with small round cell count and negatively correlated with Lymphocyte count (r2=0.5, P<0.05). APP 330 is correlated with small round cell count but negatively correlated with platelet distribution width.
Quantitation of the expressions for NOTCH1 and NOTCH4
Results of QRT-PCR showed that both NOTCH1 and NOTCH4 were expressed significantly higher in TT than those in TP (P<0.05). However, results of western blotting indicated the gentle contrast between each other. Results also indicated that proteins of NOTCH1 and NOTCH4 expressed more in TT than in TP. PRRS is negatively correlated with NOTCH1 expression (Figure 6).
Figure 6.
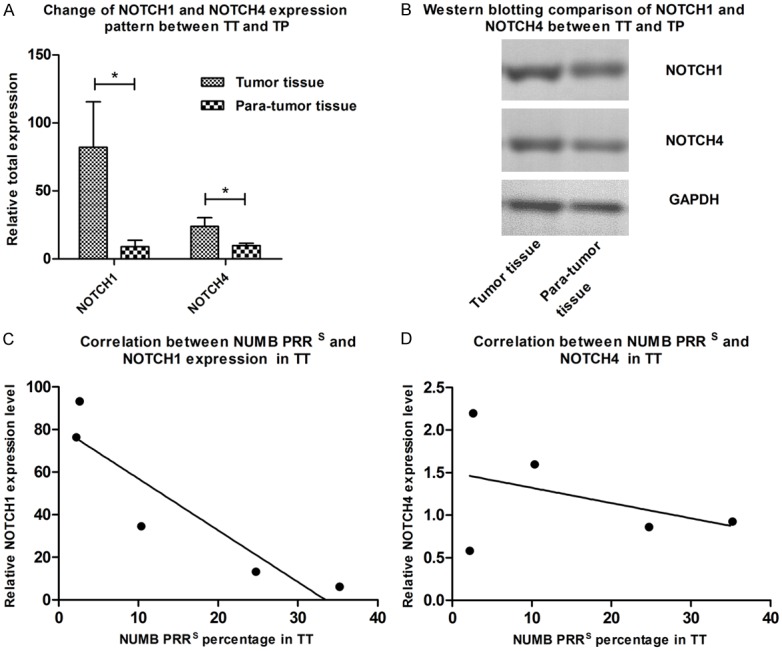
A. NOTCH1 and NOTCH4 were expressed significantly higher in TT compared to that in TP (P<0.05). B. Western blotting results however presented a more gentle contrast between each other but also indicating that TT expressed more NOTCH1 and NOTCH4 protein than TP. C and D. Show that expression of NOTCH1 is negatively correlated with PRRS in TT with significance, while NOTCH4 is not.
Discussion
Our results clearly indicated the correlation between NUMB gene expression and PDAC, which suggested that the reduced level of NUMB gene expression might play an important role in tumor suppressive function. Previously, NUMB has been reported to work against NOTCH pathway through endocytosis of NOTCH receptors and the involvement in ubiquitin network through which NUMB regulates signaling pathways [15]. Therefore, down regulation of NOTCH transcripts and proteins were verified by assays of qRT-PCR and western blot. The high level of overall APP gene expression in TT found in our results might act as the critical enhancers for PDAC proliferation and oncogenesis. In addition, quantization result of overall VEGFA level in TT was in agreement with the results in a previously report that the silencing VEGF gene could directly decrease cell viability [17].
Next, our research was focused on the correlation between AS expression levels and PDAC Tumor Tissue. Our finding indicated the NUMB with exclusion of exon 12, PRRS showed a reduced expression level in TT, which implied that this PRRS transcript, instead of overall NUMB, might be generally important in cancer suppression for all cell types. On the other hand, VEGFA and APP with exon exclusions (VEGFA with E6 and E7 exclusion; APP with E7 and E8 exclusion) had the increased expression in tumor tissues in comparison with those expressions in para-tumor tissues, suggesting that these genes may take part in pathological activities of PDAC or they may be induced or regulated in tumor tissues by PDAC metabolism and progression.
Overall, NUMB, APP and VEGFA have already been found in pancreas development, physiology and cancer progression [18,19]. Hereby our research focused on NUMB gene and its downstream targets furthermore. Previous researches revealed that endocrine/exocrine lineage developments were regulated by differential activation of Notch1-mediated signaling. Mammalian NUMB is an intracellular inhibitor of Notch signaling, including four isoforms. In our study, NOTCH1 and NOTCH4 were up-regulated in both transcription and translational levels. Furthermore, NOTCH1 is negatively associated with PRRS, which means that the PRRS transcript might play more important role in regulating NOTCH1 than that of PRRL. In sum, PRRL form of NUMB gene is highly expressed in TT and positively correlated with tumor size, while PRRS is lacking in TT and negatively correlated with tumor size and NOTCH expression suggesting that PRRS might be protective in tumorogenesis as well as NOTCH pathway down regulative ability. Spatiotemporal expression patterns of these NUMB transcripts affect NOTCH family expression and pancreatic development. Similarly, we also presume that NUMB isoforms could also play a very important role in PDAC for overall biological activities instead of just overall NUMB transcripts [20]. Here, we found alternative splicing phenomena of three genes in PDAC and revealed that the specific AS genes are related to PDAC. Therefore, our new interpretation about PDAC progression could go to the deeper understanding though the revealed correlations between AS (for all NUMB, APP and VEGFA) and PDAC, rather than just between their overall transcripts expression level, affecting their downstream effectors such as NOTCH family and PDAC. As traditional cancer gene oriented quantization might show different results from the same disease and the same gene among samples such as P53 [21]. Therefore, further studies are needed to identify whether AS causes PDAC tumorogenesis or AS is just enrolled in or the result of tumorogenesis.
Acknowledgements
This work is supported by 2013 Science Foundation of Shanghai Health and Family Planning Commission for Young Scholar (NO. 20134Y139), SHSFC (14ZR1408700).
Disclosure of conflict of interest
None.
References
- 1.Alexakis N, Halloran C, Raraty M, Ghaneh P, Sutton R, Neoptolemos JP. Current standards of surgery for pancreatic cancer. Br J Surg. 2004;91:1410–27. doi: 10.1002/bjs.4794. [DOI] [PubMed] [Google Scholar]
- 2.Kaida D, Schneider-Poetsch T, Yoshida M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 2012;103:1611–6. doi: 10.1111/j.1349-7006.2012.02356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–59. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 6.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–93. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 7.Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, Zhen M, Ciruna B, Blencowe BJ. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–93. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugahara K, Hayashi T, Dateki N, Hirakata Y, Harasawa H, Tomonaga M, Yamada Y, Kamihira S. Possible attenuation of fas-mediated signaling by dominant expression of caspase-8 aberrant isoform in adult T-cell leukemia cells. Int J Hematol. 2002;76:50–4. doi: 10.1007/BF02982718. [DOI] [PubMed] [Google Scholar]
- 10.van Dijk MA, Hart AA, van‘t Veer LJ. Differences in estrogen receptor alpha variant messenger RNAs between normal human breast tissue and primary breast carcinomas. Cancer Res. 2000;60:530–3. [PubMed] [Google Scholar]
- 11.Zhu X, Daffada AA, Chan CM, Dowsett M. Identification of an exon 3 deletion splice variant androgen receptor mRNA in human breast cancer. Int J Cancer. 1997;72:574–80. doi: 10.1002/(sici)1097-0215(19970807)72:4<574::aid-ijc4>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 12.Misquitta-Ali CM, Cheng E, O’Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011;31:138–50. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding WQ, Cheng ZJ, McElhiney J, Kuntz SM, Miller LJ. Silencing of secretin receptor function by dimerization with a misspliced variant secretin receptor in ductal pancreatic adenocarcinoma. Cancer Res. 2002;62:5223–9. [PubMed] [Google Scholar]
- 14.Ding WQ, Kuntz SM, Miller LJ. A misspliced form of the cholecystokinin-B/gastrin receptor in pancreatic carcinoma: role of reduced sellular U2AF35 and a suboptimal 3’-splicing site leading to retention of the fourth intron. Cancer Res. 2002;62:947–52. [PubMed] [Google Scholar]
- 15.Pece S, Confalonieri S, R Romano P, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 16. http://www.nccn.org/professionals/physician_gls/f_guidelines.asp#pancreatic.
- 17.Song J, Cao L, Li Y. RNA interferencemediated inhibition of survivin and VEGF in pancreatic cancer cells in vitro. Mol Med Rep. 2013;7:1651–5. doi: 10.3892/mmr.2013.1361. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, Guo P, Matsuda K, Truong N, Lee A, Chun C, Cheng SY, Korc M. Pancreatic cancer cell-derived vascular endothelial growth factor is biologically active in vitro and enhances tumorigenicity in vivo. Int J Cancer. 2001;92:361–9. doi: 10.1002/ijc.1202. [DOI] [PubMed] [Google Scholar]
- 19.Woods NK, Padmanabhan J. Inhibition of amyloid precursor protein processing enhances gemcitabine-mediated cytotoxicity in pancreatic cancer cells. J Biol Chem. 2013;288:30114–24. doi: 10.1074/jbc.M113.459255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida T, Tokunaga A, Nakao K, Okano H. Distinct expression patterns of splicing isoforms of mNumb in the endocrine lineage of developing pancreas. Differentiation. 2003;71:486–95. doi: 10.1046/j.1432-0436.2003.7108006.x. [DOI] [PubMed] [Google Scholar]
- 21.Allende-Vega N, Dayal S, Agarwala U, Sparks A, Bourdon JC, Saville MK. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene. 2013;32:1–14. doi: 10.1038/onc.2012.38. [DOI] [PubMed] [Google Scholar]