Abstract
Recently, an increasing number of reports have revealed that long non-coding RNAs (LncRNAs) play important roles in a variety of aspects of cell activity, and their aberrant expression is closely associated with multigenetic diseases, including carcinoma. In the present study, through microarray analysis, we screened out a new LncRNA (LncRNA-AP001631.9), which was regulated by FOXM1, a well-known carcinogenetic factor and aimed to reveal the functional roles of this novel LncRNA in gastric cancer development. The data from qRT-PCR confirmed that the expression level of LncRNA-AP001631.9 was positively correlated with that of FOXM1. The transwell and wound healing assays indicated that LncRNA-AP001631.9 was required for the migration of gastric cancer cells. The downregulation of LncRNA-AP001631.9 by small interference RNA suppressed the migratory ability of MGC803 and AGS cells, while the overexpression of LncRNA-AP001631.9 promoted the movement of BGC823 and SGC7901 cells. Furthermore, a tail vein injection was administered, and the obtained results suggested that LncRNA-AP001631.9 contributed to the distant metastasis of SGC7901 cells. Moreover, we also collected 36 paired samples of gastric cancer tissues to explore the expression levels of LncRNA-AP001631.9. The qRT-PCR data indicated that the LncRNA-AP001631.9 expression was frequently increased in gastric cancer tissues. Taken together, our findings established that LncRNA-AP001631.9 plays critical roles in gastric cancer progression and can serve as a potential new target for the treatment of gastric cancer.
Keywords: FoxM1, LncRNA, cell migration, gastric cancer
Introduction
Gastric cancer is one of the most common digestive malignancies [1]. Although the incidence of GC and the mortality of patients diagnosed with this disease have experienced a decline over the years, the 5-year overall survival rate (OS) is still lower than 25% [2,3]. As a result of the lack of specific tumor markers for early detection, most patients with GC are diagnosed at later stages and have a poor prognosis [4,5]. Apart from the application of conventional histological and molecular biomarkers, the identification of novel GC-specific molecular targets would contribute to a better understanding of the pathogenesis of gastric cancer [6,7].
Cancer is a complicated disease involving multiple phases and steps in the process of tumor progression. With the development of biomedical technology, a serious of protein-coding genes have been recognized or identified as cancer-specific markers. However, genome sequencing indicated that less than 2% of the total genes can be transcribed into protein-coding RNAs, and the others are always transcribed into non-coding RNAs (ncRNAs) [8]. The long non-coding RNAs (LncRNAs), a subcategory of these ncRNAs, are commonly composed of more than 200 nucleotides in length [9,10]. To date, many reports have documented that LncRNAs play important roles in a variety of aspects of cell activity, and their aberrant expression is closely associated with multigenetic diseases, including carcinoma [11-13].
The forkhead box protein M1 (FOXM1), a transcription factor which contains a 100-amino acid winged-helix DNA-binding domain, is a valued member of the forkhead protein family [14,15]. This protein is significantly upregulated in embryonic and fetal tissues [16,17] and exerts a central influence in cell proliferation and differentiation [18].
Existing data revealed that the expression of FOXM1 is frequently increased in multifarious malignancies, including hepatoma, cervical carcinoma, colorectal carcinoma, prostatic carcinoma, and lung carcinoma [19,20]. In addition, the findings of some reports have demonstrated that the FOXM1 level is significantly elevated in gastric cancer tissues and is correlated closely with tumorigenesis and lymph node metastases of GC [21].
Until now, only a limited number of studies concerning the correlation between FOXM1 and LncRNAs have been conducted. Lately, LncRNAs microarray screening was performed in MGC803 cells, which were stably transfected with LV-shNC or LV-shFOXM1. A serious of LncRNAs were decreased in MGC803 cells with shFOXM1. In the present study, we focused on one of them, designated as LncRNA-AP001631.9, which we previously identified as likely to be regulated by FOXM1. LncRNA-AP001631.9 is an LncRNA located on chromosome 17, with a length of 422 nucleotides. The qRT-PCR results confirmed that LncRNA-AP001631.9 was positively regulated by FOXM1, which was consistent with the microarray data. The evidence from several in vitro and in vivo functional analyses suggested that LncRNA-AP001631.9 exhibited significant roles in cell migration and metastasis. Furthermore, 36 paired GC tissue samples were collected to explore the expression of LncRNA-AP001631.9. The data also indicated that LncRNA-AP001631.9 was always increased in cancer tissues. Thus, our findings suggested that LncRNA-AP001631.9, as a FOXM1-regulated LncRNA, plays a central role in gastric cancer progression and may represent a valuable potential biomarker for early diagnosis of gastric cancer.
Materials and methods
Cell lines and culture conditions
A total of four human gastric cancer cell lines (AGS, MGC803, BGC823, and SGC7901) were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). They were cultured in MEM medium, supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 U/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Gastric cancer tissue samples
All gastric cancer samples were obtained from patients with gastric cancer who had undergone surgery at the Shanghai East Hospital, Tongji University. The tissue specimens were rapidly frozen in liquid nitrogen and stored at -80°C until use. No prior treatment of these patients had been conducted, and the tissue samples were examined and verified by pathologists. This study was approved by the Human Resources Ethics Committee of the Shanghai East Hospital affiliated to Tongji University. All patients had provided written informed consent.
Plasmid transfection and RNA interference
To elevate the expression level of LncRNA-AP001631.9, we amplified the full-length cDNA of LncRNA-AP001631.9 from human gastric cancer cell MGC803 and cloned it into the pCDH-CMV-EF1-copGFP expression vector to construct apCDH-LncRNA-AP001631.9 plasmid. The accuracy of reading frame insertion was confirmed by DNA sequencing. SiRNA for LncRNA-AP001631.9 and a negative control (siNC) were designed, whose sequences were as follows: siLncRNA-AP001631.9 (sense 5’-CAAGUAUACACCUCAGCCU-3’) and siNC (sense 5’-UUCUCCGAACGUGUCACGUdTdT-3’). The sequence of siRNA specifically targeting FOXM1 was designed as siFOXM1 (sense 5-CUCUUCUCCCUCAGAUAUAdTdT-3). All the siRNAs used in the investigation were obtained from GenePharma, Shanghai, China. The cell transfection of the plasmid or siRNA was conducted by Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions. The lentivirus overexpressing FOXM1 or LncRNA-AP001631.9 (LV-FOXM1, LV-LncRNA-AP001631.9) and decreasing FOXM1 or LncRNA-AP001631.9 (LV-shFOXM1, LV-shLncRNA-AP001631.9) were packaged by and purchased from GenePharma, Shanghai, China by using the corresponding sequences mentioned above. The efficiency of knockdown or overexpression was determined by quantitative real-time PCR.
Cell migration assay
Gastric cancer cells (3×104) were transfected with a plasmid or siRNAs in serum-free media and were placed into the upper part of a transwell chamber in a 24-well format with 8 mm diameters (Corning, USA). Aliquots (600 µl) of MEM medium with 10% FBS were added into the bottom chambers as a chemoattractant, and the chambers were moved into the 5% CO2 incubator and kept at 37°C for 48 hours. The cells on the upper surface of the chambers were removed by cotton-tipped swabs, while those that migrated through the membrane were stained with 0.05% crystal violet for about 2 hours. Then, the relocated cells were washed in PBS buffer, counted in 5 random fields under a microscope, and the average number of the cells in the observed 5 fields was calculated. All above assays were performed in triplicate and repeated three times.
Scratch wounding assay
Transfected cells (3.5×105) were seeded into a six-well plate to form a 90% confluent monolayer in a 10% FCS-containing medium. The confluent monolayer cells were scratched by a plastic tip and washed with PBS buffer to remove the cell debris. Aliquots of 0.5% FBS-containing MEM were then added to each well, and the scratched monolayer was incubated at 37°C in a 5% CO2 incubator for 48 h. The initial scratched gap breadth (0 h) and the residual scratched gap breadth (48 h) were measured by using the light microscope (Nikon, Japan).
Tail vein injections into nude mice
Four-week-old male athymic mice were provided by the Animal Center of the Chinese Academy of Science (Shanghai, China) and raised in laminar flow cabinets under specific pathogen-free conditions. MGC803 cells that were stably transfected with LncRNA-AP001631.9 or with an empty vector and SGC7901 cells stably transfected with LncRNA-AP001631.9 or with an empty vector were harvested and washed with PBS buffer. Then, the transfected cells were suspended at a density of 1.5×107 cells/ml. The mice were randomly divided into four groups at the age of 6 weeks and were injected with 100 µl cell suspension through the tail veins as mentioned previously. Six weeks after injection, the mice were euthanized. The lungs were removed and photographed, and the visible tumors on the lung surface were counted. The lung tissues were fixed in 4% paraformaldehyde and stored in 70% ethanol. Then, the specimens were embedded in paraffin, sectioned, stained with hematoxylin and eosin (H&E), and subjected to histopathological examination. The protocol for the animal experiments was inspected and approved by the Institutional Animal Care and Use Committee of Tongji University.
Statistical analysis
All data were presented as mean ± SEM, and analyzed by Student’s t-test using Graphpad Prism 5 software. Only a p value of less than 0.05 was considered significant. “*” indicates P<0.05; “**” indicates P<0.01.
Results
LncRNA-AP001631.9 is positively correlated with the FOXM1 levels in gastric cancer cells
To verify the regulatory relationship between FOXM1 and LncRNA-AP001631.9, several gastric cancer cell lines upregulating or downregulating FOXM1 were prepared, and q-RT PCR analysis was performed to measure the RNA levels of LncRNA-AP001631.9 in these cells. The data showed that LncRNA-AP001631.9 RNA rates were decreased in MGC803 and AGS cells in which FOXM1 expression was stably downregulated (Figure 1A). In contrast, the LncRNA-AP001631.9 RNA levels were increased in AGS and SGC7901 in which FOXM1 levels were stably upregulated (Figure 1B). These results suggested that the LncRNA-AP001631.9 level is positively correlated with the FOXM1 expression, indicating that FOXM1 may be a transcription activator of LncRNA-AP001631.9.
Figure 1.
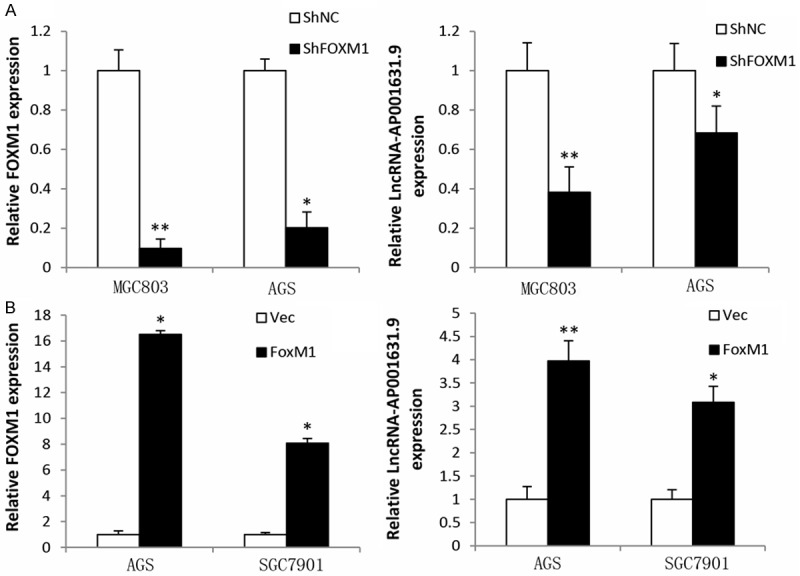
FOXM1 expression levels. A. FOXM1 expression levels declined in MGC803 and AGS cells infected with LV-shFOXM1, while LncRNA-AP001631.9 expression was decreased after FOXM1 knockdown. B. FOXM1 expression levels were elevated in AGS and SGC7901 cells infected with LV-FOXM1, whereas those of LncRNA-AP001631.9 were increased after FOXM1 overexpression.
LncRNA-AP001631.9 has a significant effect on gastric cancer cell migration
To determine whether LncRNA-AP001631.9 had an effect on gastric cancer migration, the small interference RNA targeting LncRNA-AP001631.9 (siLncRNA-AP001631.9) and the negative control (siNC) were transfected into MGC803 and AGS cells, respectively; the transfection efficiency is depicted in Figure 2A. Then, a transwell migration assay was performed, and the final data showed that LncRNA-AP001631.9 knockdown significantly inhibited the migratory ability of MGC803 and AGS cells (Figure 2B). In addition, we transfected a pCDH-LncRNA-AP001631.9 plasmid and an empty vector into BGC823 and SGC7901 cells, respectively; the transfection efficiency is presented in Figure 3A. Next, a transwell migration assay was conducted, which revealed that LncRNA-AP001631.9 overexpression significantly promoted the migration capacity of BGC823 and SGC7901 cells (Figure 3B). These findings suggested that overexpression of FRLnc1 enhanced the migration of gastric cancer cells.
Figure 2.
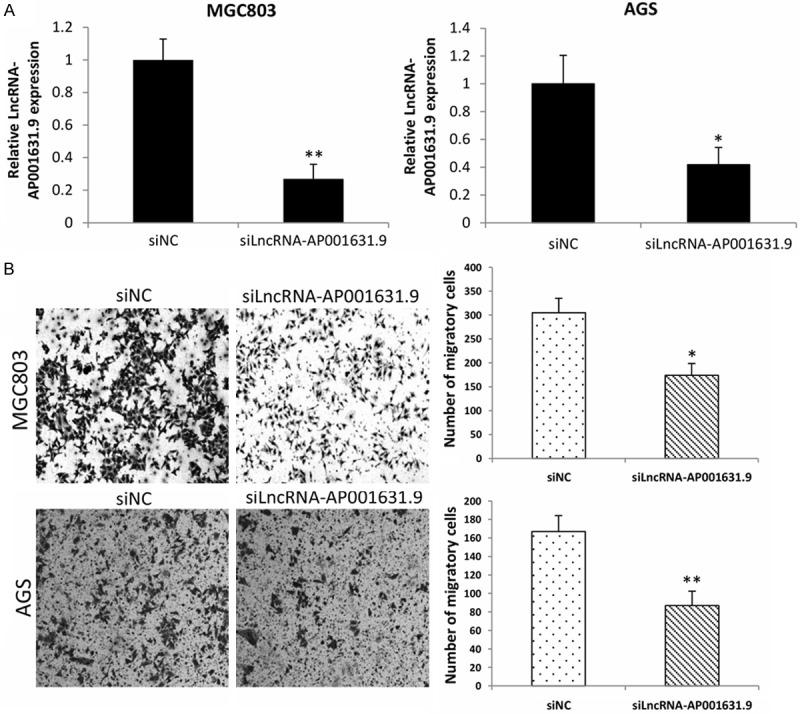
Relative expression levels of LncRNA-AP001631.9 in MGC803 and AGS cells, and numbers of the migrated cells. A. Relative expression levels of LncRNA-AP001631.9 were declined in MGC803 and AGS cells after transfection with small interference RNA target LncRNA-AP001631.9. B. The migrated cell number of MGC803 and AGS was significantly decreased when LncRNA-AP001631.9 knockdown.
Figure 3.
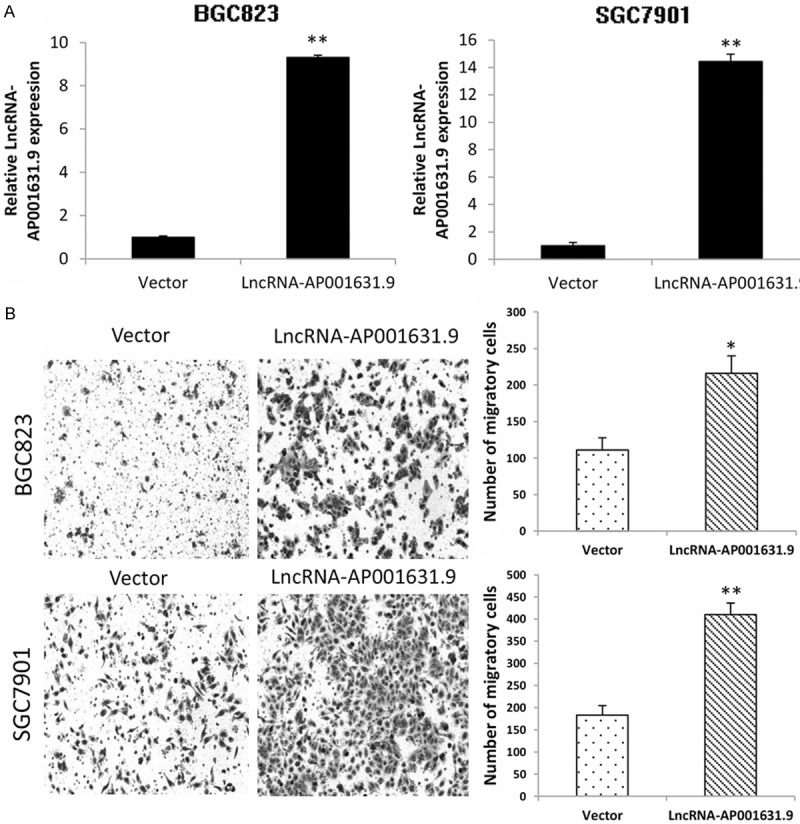
Relative overexpression of LncRNA-AP001631.9 in BGC823 and SGC7901 cells and the increase in cell migration. A. The relative expression of LncRNA-AP001631.9 was upregulated in BGC823 and SGC7901 cells after the transfection with LncRNA-AP001631.9 overexpression plasmids. B. The number of migrated BGC823 and SGC7901 cells was significantly increased when LncRNA-AP001631.9 was overexpressed.
LncRNA-AP001631.9 promotes the mobility of gastric cancer cells
To further verify the functional role of LncRNA-AP001631.9 in gastric cancer cell transferability, a wound healing assay was performed subsequently. Consistently with the transwell assay results, the SGC7901 cells transfected with pCDH-LncRNA-AP001631.9 exhibited a faster movement rate compared with that in the negative control (Figure 4). However, the AGS cells transfected with siLncRNA-AP001631.9 did not manifest significant variations in the movement speed, compared with those of the negative control.
Figure 4.
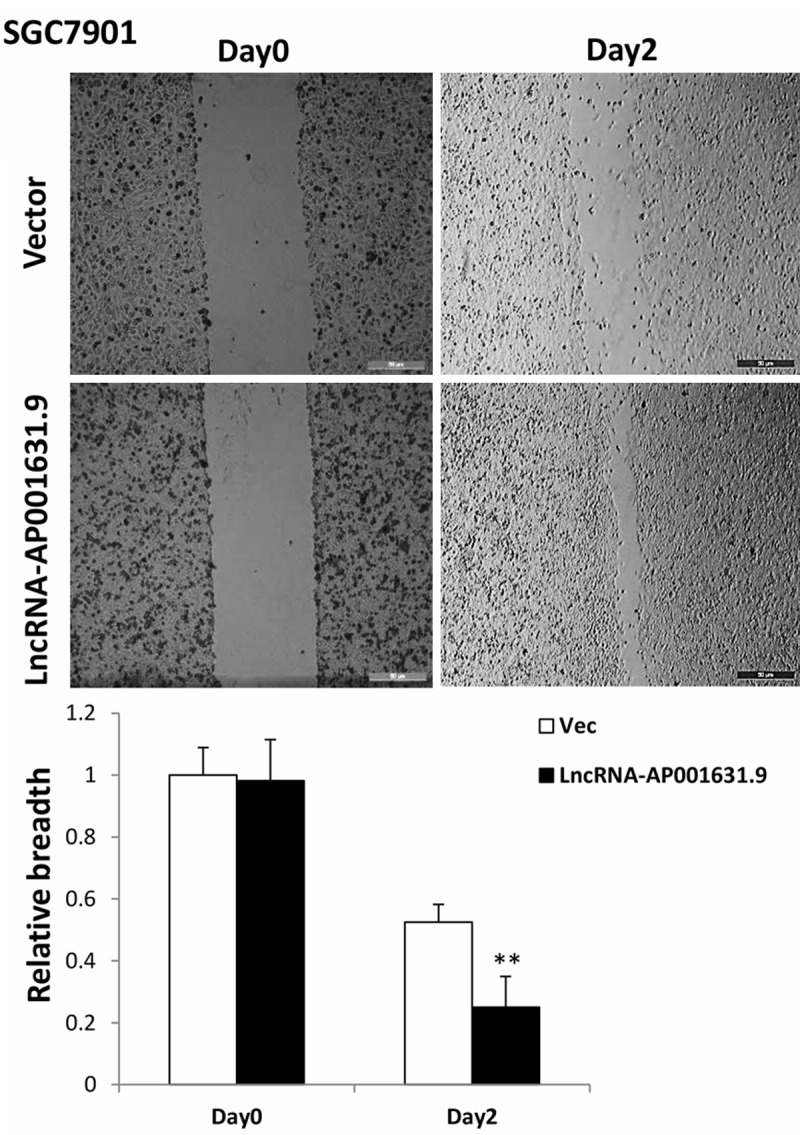
Results from the wound healing assay in SGC7901 cells. A wound healing assay was performed in SGC7901 cells transfected with LncRNA-AP001631.9; the mobility capacity was enhanced after the overexpression of LncRNA-AP001631.9.
LncRNA-AP001631.9 influences gastric cancer cell metastasis in vivo
To investigate in vivo the role of LncRNA-AP001631.9 in metastasis of gastric cancer cells, stable cells (1.5×106) of SGC7901-LncRNA-AP001631.9 were injected into the tail veins of nude mice. Six weeks after the injection, the mice were euthanized, and their lungs were removed and photographed. The metastatic nodules on the surface of the lungs were counted. The number of tumors on the lung surface from the mice receiving SGC7901-LncRNA-AP001631.9 stable cells was significantly higher than that of the control group (Figure 5A and 5B). The results from the histological analysis confirmed the presence of metastatic tumors in the lungs of these mice, as shown in Figure 5C. On the other hand, we did not find any visible tumors on the lung surfaces of the mice that were injected with MGC803 stably infected with shRNA-LncRNA-AP001631.9, regardless of whether the mice received MGC803-shLncRNA-AP001631.9 or control cells (data not shown). The overexpression experiment indicated that LncRNA-AP001631.9 could promote pulmonary metastasis in vivo.
Figure 5.
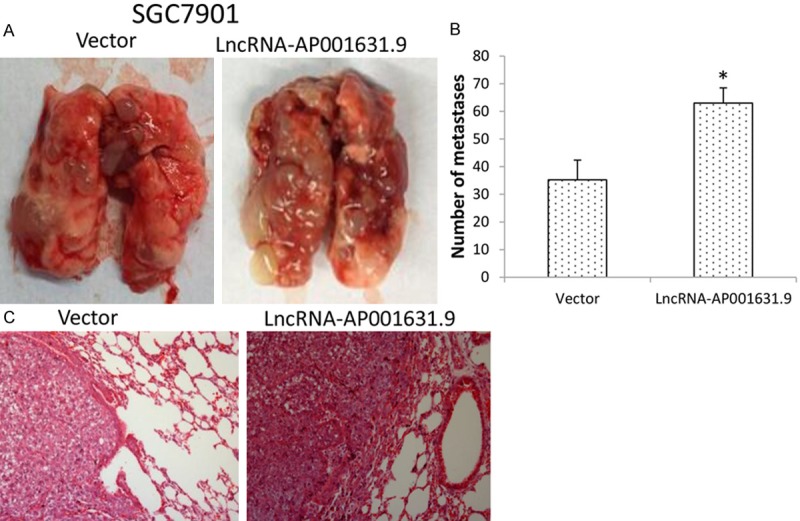
Lung metastatic tumor nodules, their mean values, and the stained lung sections. A. Lung metastatic tumor nodules, observed on the lung surface of nude mice that received tail vein injection with SGC7901-Vec or SGC7901-LFRLnc1 cells. B. The mean value of tumor nodules from each experimental group (n=5). C. The hematoxylin and eosin (HE) stained lung sections.
LncRNA-AP001631.9 is frequently increased in gastric cancer samples
By using qRT-PCR, we further investigated the expression of LncRNA-AP001631.9 in 36 paired gastric cancer tissue samples and their adjacent non-cancerous tissues. In 15 of 36 samples LncRNA-AP001631.9 was evidenced to be at least 1.5-fold upregulated in comparison with the adjacent non-cancerous tissues. Approximately 8 of 36 cancer specimens were downregulated, whereas the other 13 paired samples displayed no significant difference in LncRNA-AP001631.9 expression levels. The expression pattern is illustrated in Figure 6. Altogether, we did not find any correlation between FRLnc1 expression and gastric cancer metastasis, tumor size, tumor grade, or other clinicopathologic characteristics. This can be possibly attributed to the small amount of clinical samples.
Figure 6.
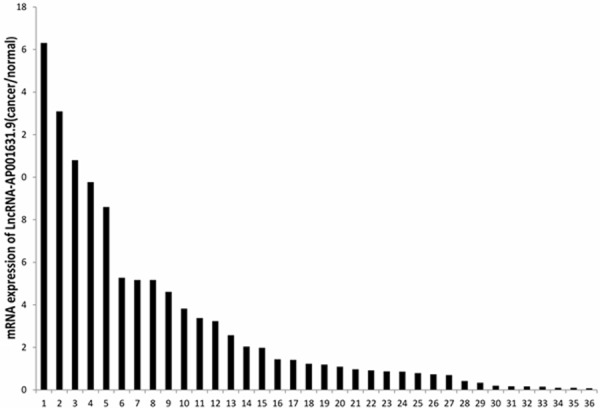
The FRLnc1 expression pattern in 36 paired gastric cancer samples. qRT-PCR was performed to detect the expression level of LncRNA-AP001631.9 in cancer tissues and their adjacent non-cancerous tissues. β-actin was used as an internal loading control. The data are shown as the upregulated fold change (C/N) of LncRNA-AP001631.9 expression levels. The values “≥1.5” indicate overexpression, “≤0.66” represents underexpression, and “>0.66 but <1.5” denotes no significant variation.
Discussion
Currently, accumulating studies have indeed demonstrated that LncRNAs, initially considered transcriptional noise, play pivotal roles in carcinogenesis. A part of them are linked with the diagnostic and prognostic prediction for cancer patients and are gradually identified as novel biomarkers in a diverse range of malignancies [22]. Despite the substantial body of existing studies have uncovered the mysteries of many LncRNAs, research that has been concentrated on examination of the relationship between LncRNAs and FOXM1, as well as investigations on their functional role in gastric cancer have never been reported before. The deregulation of FOXM1 has been confirmed as a substantial factor in the development of carcinomas, which is likely achieved through its effects on tumor initiation, progression, epithelial-to-mesenchymal transition (EMT), metastasis, angiogenesis, and drug resistance [16].
In the present study, we used a microarray assay to screen out a FOXM1-related LncRNA (LncRNA-AP001631.9) in gastric cancer cell lines. We demonstrated that LncRNA-AP001631.9 and FOXM1 have a positive correlation. The LncRNA-AP001631.9 expression levels in GC cells were in keeping with those of FOXM1. We further examined the potential role of LncRNA-AP001631.9 in gastric cancer development by transwell and wound healing assays. The results suggested that LncRNA-AP001631.9 was a motivator in GC cell migration; its overexpression obviously enhanced the migratory ability of GC cells in vitro, whereas silencing of LncRNA-AP001631.9 inhibited this phenotype change. In addition, after the tail vein injection with GC cells was administered in nude mice, we found that LncRNA-AP001631.9 can indeed facilitate lung metastasis in vivo. Finally, we evaluated the expression levels of LncRNA-AP001631.9 in 36 paired GC tissue samples by qRT-PCR and established that about 42% of the samples had a significant increase in LncRNA-AP001631.9.
In conclusion, LncRNA-AP001631.9 is a novel FOXM1-related LncRNA, which is frequently upregulated in gastric cancer tissues. It exhibits a significant effect on gastric cancer cell migration, facilitates the mobility of gastric cancer cells, and promotes metastasis in vivo. The data obtained support the concept that LncRNA-AP001631.9 plays a critical role in gastric cancer development and progression, and it might have the potential to be a novel biomarker for gastric cancer diagnosis or prognosis. However, further, more comprehensive research is required to uncover the underlying mechanism by which LncRNA-AP001631.9 promotes gastric cancer metastasis.
Acknowledgements
This work was supported in part by grants from National Natural Science Foundation of China (81272917 and 81272241), Shanghai medical key specialty (ZK2012A26), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq20142014-04) and Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (PWRl2013-02).
Disclosure of conflict of interest
None.
References
- 1.Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K, Yamashita M, Taniguchi H, Nosho K, Suzuki H, Yasuda H, Shinomura Y, Itoh F. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol. 2014;20:3927–3937. doi: 10.3748/wjg.v20.i14.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464:367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 4.Wang XN, Liang H. Some problems in the surgical treatment of gastric cancer. Chin J Cancer. 2010;29:369–373. doi: 10.5732/cjc.009.10629. [DOI] [PubMed] [Google Scholar]
- 5.Karim-Kos HE, de Vries E, Soerjomataram I, Lemmens V, Siesling S, Coebergh JW. Recent trends of cancer in Europe: a combined approach of incidence, survival and mortality for 17 cancer sites since the 1990s. Eur J Cancer. 2008;44:1345–1389. doi: 10.1016/j.ejca.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano A. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. J Cell Physiol. 2007;211:287–295. doi: 10.1002/jcp.20982. [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro H, Bordeira-Carrico R, Seixas S, Carvalho J, Senz J, Oliveira P, Inacio P, Gusmao L, Rocha J, Huntsman D, Seruca R, Oliveira C. Allele-specific CDH1 downregulation and hereditary diffuse gastric cancer. Hum Mol Genet. 2010;19:943–952. doi: 10.1093/hmg/ddp537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther. 2013;23:15–20. doi: 10.1089/nat.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- 15.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- 16.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 17.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 19.Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, Ngan HY. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 20.Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, AbdulJabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Jehan Z, Bavi P, Siraj AK, Al-Kuraya KS. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol. 2011;178:537–547. doi: 10.1016/j.ajpath.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]


