Abstract
Malignant melanoma is the deadliest form of all skin cancers. Recently, microRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression by targeted repression of transcription and translation and play essential roles during cancer development. Our study showed that miR-135a is upregulated in malignant melanoma tissues and cell lines by using Real-time PCR assay. Enforced expression of miR-135a in malignant melanoma cells promotes cell proliferation, tumorigenicity, and cell cycle progression, whereas inhibition of miR-135a reverses the function. Additionally, we demonstrated FOXO1 is a direct target of miR-135a and transcriptionally down-regulated by miR-135a. Ectopic expression of miR-135a led to downregulation of the FOXO1 protein, resulting in upregulation of Cyclin D1, and downregulation of P21Cip1 and P27Kip1 through AKT pathway. Our findings suggested that miR-135a represents a potential onco-miRNA and plays an important role in malignant melanoma progression by suppressing FOXO1 expression.
Keywords: miR-135a, FOXO1, malignant melanoma
Introduction
Malignant melanoma is an aggressive malignancy associated with a high mortality rate and often occurs in the skin, with a clear familial aggregation [1]. Although surgical excision is an effective therapy against localized disease, the median survival of patients with metastatic malignant melanoma, following treatment with radiation and chemotherapy drugs, is only 6-9 months [2]. Therefore, investigation of the biology of malignant melanoma initiation and progression remains urgently needed to develop effective therapeutic strategies.
Recent studies have shown that microRNAs (miRNAs) play important roles in the development, invasion and metastasis of malignant melanoma [3]. Since it exists stably in the plasma/serum, miRNA appears promising as a new biomarker for the diagnosis and treatment of cancer [3]. Cumulative evidence has been reported that miRNAs, function as tumor promoters or suppressors, regulated a wide range of biologic processes such as proliferation, apoptosis, invasion and so on [4]. In particular, miR-135a has been extensively studied in several cancers regulating cell proliferation, invasion and apoptosis [5,6]. Recently, miRNAs have been implicated as key regulators influencing the development of malignant melanoma [7,8]. However, although there are a large number of mechanistic reports on the molecular mechanism of malignant melanoma in the literature, the functions of miR-135a and its target gene in regulating malignant melanoma progression still have a poor knowledge.
The forkhead box O (FoxO) subfamily of forkhead transcription factors includes FOXO1, FOXO3, FOXO4 and FOXO6 in humans [9]. Importantly, FOXO proteins play important roles in tumor suppressors by their capacity to induce cell-cycle arrest, apoptosis and DNA repair as well as modulate the expression of genes responsive to oxidative stress and cell differentiation through complex pathways [10,11]. FOXO proteins are functions as post-translational modifications by phosphorylation, acetylation and ubiquitinylation. Additionally, recent studies have indicated that abnormal expression of FOXO genes is associated with the development of human tumors [12], as well as malignant melanoma [13].
In the current study, we found that miR-135a was upregulated in malignant melanoma tissue and cell lines. Ectopic overexpression of miR-135a in malignant melanoma cell lines led to the promotion of cell growth rate, tumorigenicity and cell cycle progression. Furthermore, we demonstrated that the tumor suppressor gene FOXO1 is a direct target of miR-135a. Therefore, the findings of this study revealed that overexpression of miR-135a could promote cell proliferation, tumorigenicity and cell cycle progression in malignant melanoma by directly suppressing FOXO1.
Materials and methods
Specimen collection and ethics statement
A total of 20 patients were recruited from the first affiliated hospital of medical college of Xi’an Jiaotong University, Xi’an in China. All patients provided their consent and agreement. All patients were confirmed to have primary cancer by pathology and were not subjected to preoperative radiotherapy or chemotherapy. Malignant melanoma tissues and adjacent non-tumor tissues were collected during the operation, treated with RNAlatter reagent (TaKaRa, Japan), and stored at -80°C until use. The present study was approved by the hospital institutional review board (the first affiliated hospital of medical college of Xi’an Jiaotong University) and written informed consent was obtained from all patients.
Cell culture and transfection
Human epidermal melanocytes (HEM) were purchased from Sciencell (USA) and cultured in melanocyte medium (Sciencell, USA) according to the manufacturer’s instructions. Human malignant melanoma cell lines sk-mel-1 and A375 were obtained from American Type Culture Collection (Manassas, VA, USA). A375 and sk-mel-1 cells were maintained in RPMI-1640 medium, containing 10% fetal bovine serum (HyClone, USA) and antibiotics (100 μg/ml penicillin and 100 U/ml streptomycin, Sigma-Aldrich, USA) at 37°C with 5% CO2. The miR-135a mimic and anti-miR-135a were synthesized by RiboBio Corporation (China), and transfection was performed using Lipofectamine RNAiMAX (Life Technologies, USA) according to the instructions. Cells (2×105 per well), miR-135a mimic or anti-miR-135a with a final concentration of 30 nM, and Lipofectamine RNAiMAX were seeded in a 6-well plate. Trans-fection efficiency was detected by RT-qPCR after 48 h of incubation.
Generation of stably engineered cell lines
pMSCV-miR-135a, the miR-135a expression plasmid, was generated by cloning the genomic pre-miR-135a gene into the retroviral transfer plasmid pMSCV-puro (Clontech Laboratories Inc., USA). pMSCV-miR-135a was then cotransfected with the pIK packaging plasmid into 293FT cells, using the standard calcium phosphate transfection method [14]. 36 h after cotransfection, supernatants were collected and incubated with malignant melanoma cells to be infected for 24 h in the presence of polybrene (2.5 μg/ml USA). After infection, puromycin (1.5 μg/ml, Sigma) was used to select stably transduced cells over 10 days. Stable cell lines expressing miR-135a were established according to a previously reported protocol [14].
MTT assay
Cell proliferation was assessed by the MTT assay. A375 and sk-mel-1 (1.0×104/well) cells were seeded in 96-well plates and transfected with suitable miRNAs for 24 h, 48 h, 72 h and 96 h. Then, 20 μl of 5 mg/ml MTT (in PBS) was added to each well and continually incubated for 4 h at 37°C in 5% CO2. The supernatants were removed, and formazan granules obtained from cells were dissolved in 150 μl dimethyl sulfoxide (DMSO) for 10 min. Finally, cell viability was then measured in terms of optical density (OD) at a wavelength of 495 nm. Each cell viability assay was performed in quadruplicate and repeated three times.
Preparation of cell extracts and western blotting analysis
Protein lysates were prepared on ice in RIPA buffer (Sigma, USA) with freshly added 0.1 mg/ml phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium orthovanadate and 1 mg/ml aprotinin. Protein concentrations were determined using the Bio-Rad protein assay system (Bio-Rad Laboratories, CA, USA). Cell extracts containing 30-40 μg of total protein were resolved in 10% separation gel and 5% spacer gel concentrations and transferred to 0.22 μm nitrocellulose membranes (Millipore, MA, USA). Filters were blocked for 1 h at room temperature in Buffer A (5% BSA powder in TBS-T: 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.05% Tween-20), and then incubated for 1 h at room temperature (RT) in Buffer A containing suitable dilution of rabbit anti-p-AKT, anti-AKT, anti-P21Cip1, anti-P27Kip1, anti-Cyclin D1 and anti-GAPDH antibodies. After washing in TBS-T buffer (5 min, RT), filters were incubated for 1 h at RT in Buffer A containing a suitable dilution of peroxidase conjugated anti-rabbit or anti-mouse secondary antibody (Santa Cruz, USA). After washing in TBS-T, signals were visualized with chemiluminescent substrate (Pierce, USA) according to the manufacturer’s recommendation. All results were normalized to GAPDH protein expression.
Colony formation assay
Cells (0.5×103 cells per well) were seeded into 6-well plates and cultured. After 10 days’ culture in DMEM containing fetal calf serum, cells were subsequently fixed and stained with crystal violet (0.5% w/v in ethanol, Sigma) for 5 min. The number of colonies formed was counted in 10 different fields of vision and the mean value was evaluated. Three reduplicate dishes was performed independently for each cell line.
Analysis of cell cycle analysis
1×106 Cells were harvested and washed in PBS, then fixed in 75% ice-cold ethanol. Before cell cycle analysis, cells were washed in cold PBS three times, followed by incubation in propidium iodide (20 μg/ml; Sigma) and RNase (2 μg/ml; Sigma) for 20 min at 37°C. Cell cycle analysis was performed using the BD FACSCalibur System (BD Bioscience, USA). The data were analyzed with the FlowJo 7.6 software and the cell cycle distribution was shown as the percentage of cells in the G1, S, and G2 populations.
Luciferase reporter assay
The wild type and mutant 3’UTR of FOXO1 were cloned into the pGL3 luciferase assays vector (Promega, USA) to confirm direct target association. The wild type contained binding sites of FOXO1 3’UTR with miR-135a. The sequence that was complementary to the binding sites (AUAAUUGUAUAAAGCCAU) was replaced by CACUUUGAUCCUUACGUA for mutagenesis (Figure 5A). For luciferase assays, cells (5×104) were plated in a 24-well plate and incubated for 24 h prior to transfection. Firefly luciferase constructs containing the 3’UTR (or 3’UTRmutant) of the potential miR-135a target, pRL-TK Renilla luciferase normalization control, miR-135a, anti-miR-135a or miR-CON control were cotransfected using Lipofectamine 2000 (Invitrogen). Lysates were collected 48 h after transfection and measured using a Dual-Luciferase Reporter System (Promega) according to the manufacturer protocol. Three independent experiments were performed and the data were presented as the mean ± SD.
Figure 5.
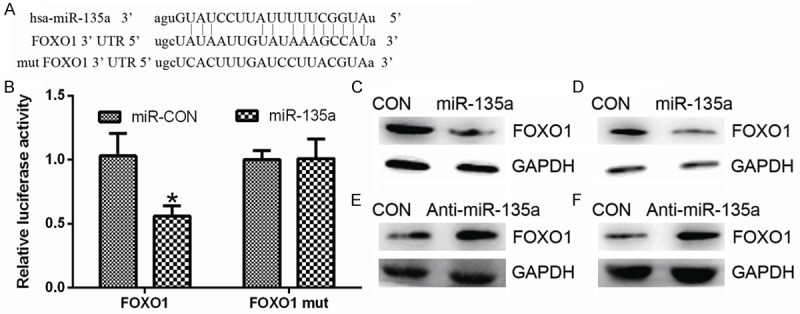
miR-135a negatively regulates FOXO1 expression in malignant melanoma cells. (A) Bioinformatic prediction of miR-135 binding sites in the 3’UTRs of FOXO1. (B) Luciferase reporter assays in HEK293 cells. Cells were transfected with 100 ng of wild-type 3’-UTR-reporter or mutant constructs together with miR-135a or anti-miR-135a as indicated. pRL-TK Renilla luciferase was used as the normalization control. (C and D) Western blotting analysis of FOXO1 in A375 (C) or sk-mel-1 (D) cells transfected with miR-135a or miR-CON. GAPDH was used as a loading control. (E and F) Western blotting analysis of FOXO1 in A375 (E) or sk-mel-1 (F) cells transfected with anti-miR-135a or miR-CON. GAPDH was used as a loading control. Differences between two groups were analyzed using Student’s t-test. *P < 0.05.
Statistical analysis of data
The prediction of miRNA targets was performed by using Target Scan (release 6.2, http://www.targetscan.org/), Pictar and RNhybrid. All results were expressed as means ± SD. One-way ANOVA was used for luciferase reporter assay analysis. All experiments were performed in triplicate. All statistical analyses were performed on SPSS 18.0. P < 0.05 was considered statistically significant.
Results
MiR-135a is upregulated in malignant melanoma tissues and cells
To investigate the relationship of miR-135a and human malignant melanoma development, we first examined the expression of miR-135a in 20 paired human malignant melanoma tissues and adjacent non-tumor skin tissues by qRT-PCR in this study. MiR-135a was significantly upregulated in malignant melanoma tissues, as compared with that detected in matched non-tumor tissues (Figure 1A), suggesting that upregulation of miR-135a might be involved in human malignant melanoma development. We next determined whether miR-135a expression was similarly correlated with the malignant melanoma cells lines. We compared miR-135a expression in three selected cell lines: HEM (human epidermal melanocyte), sk-mel-1 and A375. Indeed, miR-135a was highly upregulated in malignant melanoma cell lines, as compared with human normal epidermal melanocyte HEM cells (Figure 1B). Taken together, these data indicated that miR-135a overexpression was significantly associated with the proliferation of malignant melanoma.
Figure 1.
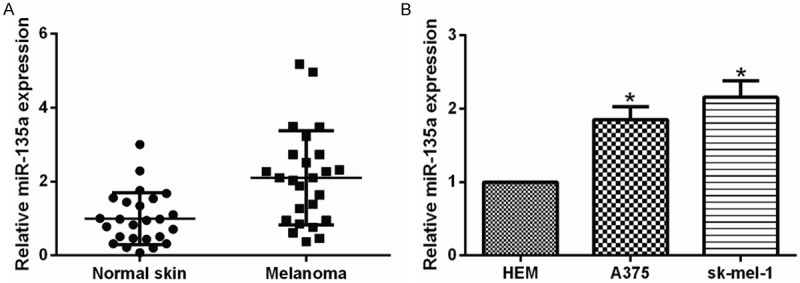
miR-135a was up-regulated in malignant melanoma tissues and cells. A. The expression of miR-135a in normal skin tissues and malignant melanoma tissues was measured using qRT-PCR. U6 snRNA was used as a loading control. B. qRT-PCR detecting the expression of miR-135a in HEM, A375 and sk-mel-1. Bars represent the mean ± SD of experiments. *P < 0.05.
Upregulation of miR-135a promotes malignant melanoma cell proliferation and cell cycle progression in vitro
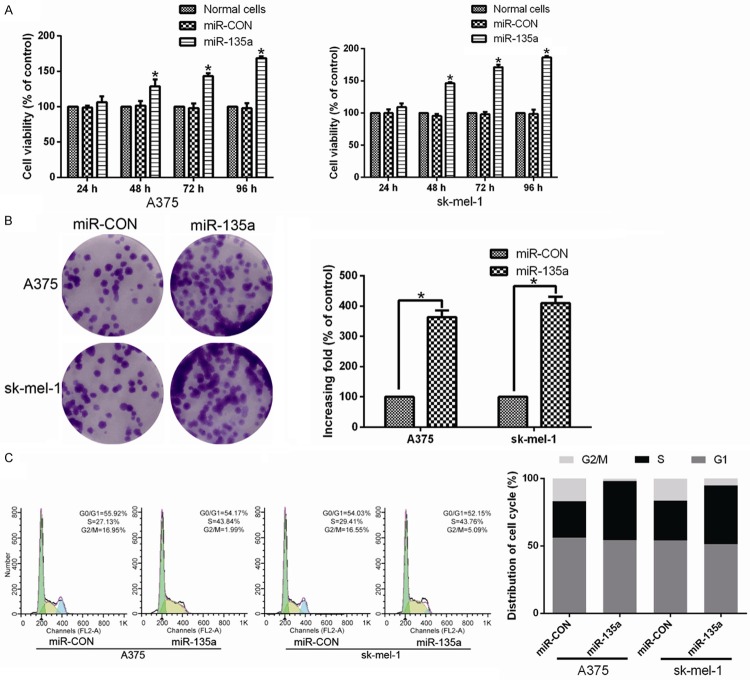
To explore the role of miR-135a in malignant melanoma proliferation, A375 and sk-mel-1 cells stably overexpressing miR-135a were established for further investigations. An MTT assay showed that ectopic expression of miR-135a significantly increased the growth rate of malignant melanoma cells (Figure 2A). Additionally, colony formation assays showed that overexpression of miR-135a enhanced the proliferation of malignant melanoma cancer cells than the control cells (Figure 2B). The cell cycle analysis of A375 and sk-mel-1 cells by flow cytometry showed a statistically significant decrease in the percentage of cells in G1/G0 phase and an increase in the percentage of cells in the S phase of the cell cycle (Figure 2C). These observations suggest that upregulation of miR-135a promoted the proliferation, tumorigenicity and cell cycle progression of malignant melanoma cells in vitro.
Figure 2.
MiR-135a induces proliferation of malignant melanoma cells. A. Effects of miR-135a on proliferation of the indicated cells, as analyzed by MTT assays. B. Representative micrographs (left) and quantifications (right) of crystal violet stained colonies formed by the indicated cells. C. Effects of ectopic miR-135a expression on cell cycle progression of the indicated cells, as analyzed by flow cytometry. All these experiments were done with A375 and sk-mel-1 cells stably overexpressing miR-135a or miR-control (miR-CON)-vector. Bars represent the mean ± SD of three independent experiments. *P < 0.05.
Inhibition of miR-135a suppresses proliferation of malignant melanoma cells
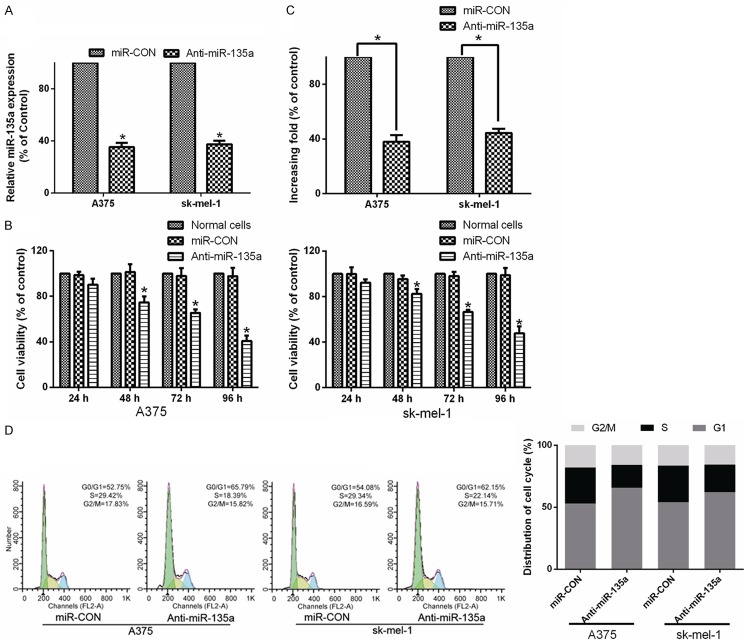
To further explore the role of miR-135a in promoting malignant melanoma cell proliferation, loss-of-function approach using a miR-135a inhibitor were performed (Figure 3A). Analysis by MTT and colony formation assays showed that downregulation of miR-135a markedly decreased the growth capacity of A375 and sk-mel-1 cells transfected with the miR-135a inhibitor, compared with that of control cells (Figure 3B and 3C). Analysis by flow cytometry showed a markedly increase in the percentage of cells in G1/G0 phase and a decrease in the percentage of cells in S phase in cells transfected with the miR-135a inhibitor, compared with control cells (Figure 3D). Taken together, these results suggested that downregulation of miR-135a suppresses the proliferation and cell cycle progression of malignant melanoma cells.
Figure 3.
Inhibition of miR-135a suppresses the proliferation and tumorigenicity of malignant melanoma cells. A. Real-time PCR analysis miR-135a in A375 and sk-mel-1 cells transfected with a miR-135a inhibitor (anti-miR-135a). Transcript levels were normalized to U6 expression. B. The effects of miR-135a inhibition (shown as Anti-miR-135a) or control (shown as miR-CON) on cell viability of malignant melanoma cells, as analyzed by MTT assays. C. Quantifications of crystal violet stained colonies formed by the indicated cells. D. The effects of miR-135a on cell cycle progression of the indicated cells analyzed by flow cytometry. Bars represent the mean ± SD of three independent experiments. *P < 0.05.
MiR-135a represses expression of the cell cycle inhibitors p21Cip1 and p27Kip1 while increases expression of cell-cycle regulator Cyclin D1
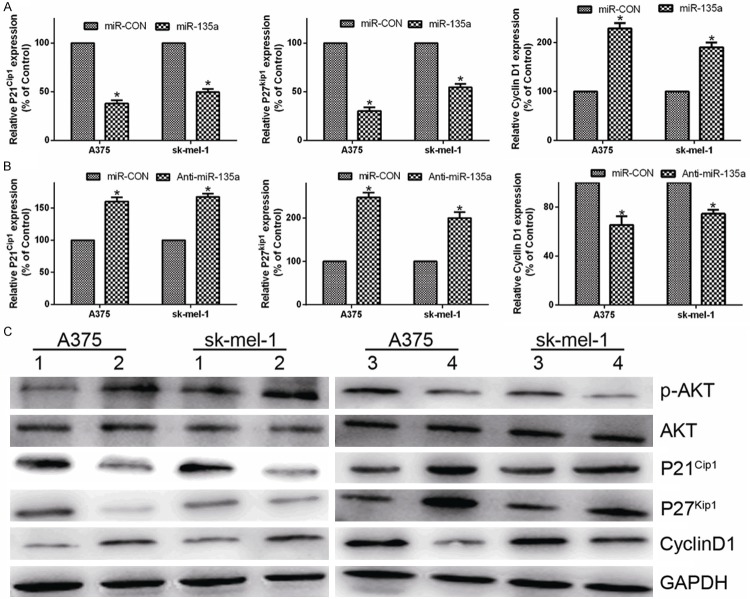
As miR-135a promoted cell proliferation, we further examined its functions on expression of the genes which regulate cell cycle and proliferation, including the CDK inhibitors p21Cip1, p27Kip1 and the CDK regulator Cyclin D1. Compared to the control transfected cells, the expression of p21Cip1 and p27Kip1 were downregulated and Cyclin D1 level was upregulated in miR-135a-transfected cells, while p21Cip1 and p27Kip1 were upregulated and Cyclin D1 level was downregulated in anti-miR-135a -transfected cells in mRNA and protein levels (Figure 4A-C). Similar results were also observed in sk-mel-1 cell, further confirming that miR-135a can promote the proliferation and cell cycle progression of malignant melanoma cells (Figure 4A-C).
Figure 4.
MiR-135a regulates expression of p21Cip1, p27Kip1 and Cyclin D1. A. Real-time PCR analysis of the mRNA expression of cell cycle regulator, p21Cip1, p27Kip1 and Cyclin D1 in malignant melanoma cells transfected with miR-135a or miR-CON. B. Real-time PCR analysis of p21Cip1, p27Kip1 and Cyclin D1 mRNA in malignant melanoma cancer cells transfected with the anti-miR-135a or miR-CON. GAPDH was used as a loading control. C. Western blotting analysis of AKT, p-AKT, p21Cip1, p27Kip1, and Cyclin D1 in A375 and sk-mel-1 cells. GAPDH was used as a loading control. Bars represent the mean ± SD values of three independent experiments; *P < 0.05.
FOXO1 activity is directly regulated by phosphorylation by Akt upon external growth signals, resulting in translocalization from the nucleus [15]. To check if miR-135a overexpression significantly alters upstream Akt signaling, we assessed Akt and phosphorylated Akt expression in malignant melanoma cells. Phosphorylated Akt levels were significantly altered upon transfection with miR-135a or anti-miR-135a (Figure 4A), demonstrating that the activity of FOXO1 was affected by changes in Akt expression or activity. These data suggest critical roles for the AKT pathways in miR-135a mediated cell biology.
MiR-135a directly targets the transcription factor FOXO1 in malignant melanoma cells
As previously reported, FOXO1 can transcriptionally regulate genes related to the cell cycle, including p21Cip1, p27Kip1 and Cyclin D1 [16,17]. In parallel, the putative target of miR-135a was predicted using the bioinformatics search (Targetscan, Pictar, RNhybrid). Among the candidate targets of miR-135a, FOXO1 were identified and the potential binding sequence for miR-135a was matched within the 3’UTR of FOXO1 (nucleotides 2889-2909; NM_002015) (Figure 5A). Indeed, miR-135a overexpression led to a remarkable decrease of luciferase activity when the pGL3-FOXO1-3’UTR luciferase reporter plasmid was co-transfected, whereas inhibition of miR-135a led to increased luciferase activity (Figure 5B). Moreover, miR-135a overexpression led to reduced FOXO1 protein expression in A375 and sk-mel-1 cells (Figure 5C and 5D). However, mutation of the miR-107 binding site abrogated the reduced luciferase expression (Figure 5B). While, downregulation of miR-135a led to increased FOXO1 protein expression in A375 and sk-mel-1 cells (Figure 5E and 5F). Collectively, these results indicating that FOXO1 is a target of miR-135a in malignant melanoma cells.
Discussion
The current study revealed that miR-135a is significantly upregulated in malignant melanoma cells and tissues, compared with normal cells and tissues. Moreover, our results showed that overexpression of miR-135a promoted the proliferation, tumorigenicity and cell cycle progression of malignant melanoma cells. In agreement with these observations, overexpression of miR-135a decreased the expression of p21Cip1, a cyclin-dependent kinase (CDK) inhibitor, and increased the expression of the cell cycle regulator cyclin D1. To explore the mechanism of miR-135a-induced cell proliferation, we investegated that miR-135a suppressed FOXO1 expression via directly targeting its 3’-UTR. Taken together, our results suggest that upregulation of miR-135a might play an essential role in promoting carcinogenesis and progression of malignant melanoma through AKT-mediated signaling.
FOXO1, a member of the forkhead box O (FOXO) subfamily of transcription factors, regulated the expression of a program of genes involved in the apoptotic response, cell cycle checkpoints and hypoxia responsiveness [18,19]. FOXO1 is also known as a tumor suppressor gene and its downregulation has been identified in various tumors, including malignant melanoma [13]. Typically, inhibition of FOXO1 in cancer is associated with the activation of higher level kinases such as AKT [19,20].
FOXO1 has been shown to be a target of regulation by many cancer-related miRNAs. These include miR-27a, miR-96, miR-182 in breast cancer [21], miR-96 in colorectal cancer [22], and miR-183 in gliomas [23]. Wu et al. found that miR-223 regulates cell proliferation through targeting and downregulating FOXO1, and upregulation of miR-370 promotes proliferation of prostate cancer cells by suppressing the endogenous FOXO1 expression [24,25]. It is also known that miR-27a, miR-96 and miR-182 have all been found to target FOXO1 directly and regulate endogenous FOXO1 protein expression in breast cancer cells, while suppression of the related microRNAs resulted in an increase in FOXO1 protein and a decrease in cell proliferation [21]. In the present study, we found that miR-135a directly targeted FOXO1, which was downregulated by the miR-135a expression. These results supported the viewpoint that miRNA can modulate the FOXO1 epigenetic expression. Moreover, consistent with previous studies, the regulation of FOXO1 by specific miRNAs is essential for tumor progression.
It has also been reported that FOXO1 is closely correlated with the phosphotidylino sitide-3-kinase (PI3K)/AKT signaling pathway [26,27]. Activation of the PI3K/AKT signaling pathway leads to FOXO1 phosphorylation, which results in dysregulation of Cyclin D1, Cyclin D2, and p21 levels and Cyclin-Dependent Kinase-4 activity [27]. According to the previous studies, our results suggested that FOXO1 is downregulated by miR-135a in malignant melanoma cells, and mediated miR-135a-induced cell growth. In addition, ectopic miR-135a inhibits the expression of p21Cip1, and induces the expression of cyclin D1, indicating a putative correlation between miR-135a and the PI3K/Akt signaling pathway. Further research is needed to elucidate the detailed molecular mechanism underlying the role of miR-135a in tumor development.
In conclusion, the current study provides novel evidence that miR-135a might have potential function to determine human epithelial cell fate. However, the function and regulation of miR-135a during malignant melanoma progression has not been unfolded. Herein, for the first time we have revealed an important link between miR-135a and malignant melanoma progression. We have shown that miR-135a functions in regulating cell growth, tumorigenicity, and cell cycle progression. Our findings suggested the essential role that miRNAs play in the pathogenesis of malignant melanoma, and further suggest that miR-135a is an onco-miRNAs and might represent a potential therapeutic target for malignant melanoma.
Disclosure of conflict of interest
None.
References
- 1.Miller AJ, Mihm MJ. Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Gogas HJ, Kirkwood JM, Sondak VK. Chemotherapy for metastatic melanoma: time for a change? Cancer. 2007;109:455–464. doi: 10.1002/cncr.22427. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg E, Besser MJ, Ben-Ami E, Shapira-Frommer R, Itzhaki O, Zikich D, Levy D, Kubi A, Eyal E, Onn A, Cohen Y, Barshack I, Schachter J, Markel G. A comparative analysis of total serum miRNA profiles identifies novel signature that is highly indicative of metastatic melanoma: a pilot study. Biomarkers. 2013;18:502–508. doi: 10.3109/1354750X.2013.816777. [DOI] [PubMed] [Google Scholar]
- 4.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang W, Jiang Y, Mu X, Xu L, Cheng W, Wang X. MiR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. 2014;26:1420–1426. doi: 10.1016/j.cellsig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Mao XP, Zhang LS, Huang B, Zhou SY, Liao J, Chen LW, Qiu SP, Chen JX. Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med. 2015;13:438. doi: 10.1186/s12967-015-0438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice DA, Higgs BW, Richman L, Jallal B, Ranade K, Yao Y. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558–1570. doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 9.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 10.Alikhani M, Alikhani Z, Graves DT, Aoki M, Jiang H, Vogt PK. FOXO1 functions as a master switch that regulates gene expression necessary for tumor necrosis factor-induced fibroblast apoptosis Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. J Biol Chem. 2005;280:12096–12102. doi: 10.1074/jbc.M412171200. [DOI] [PubMed] [Google Scholar]
- 11.Maiese K, Chong ZZ, Shang YC, Hou J. A “FOXO” in sight: targeting Foxo proteins from conception to cancer. Med Res Rev. 2009;29:395–418. doi: 10.1002/med.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanella F, Renner O, Garcia B, Callejas S, Dopazo A, Peregrina S, Carnero A, Link W. Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene. 2010;29:2973–2982. doi: 10.1038/onc.2010.58. [DOI] [PubMed] [Google Scholar]
- 14.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 16.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 17.Kops GJ, Medema RH, Glassford J, Essers MA, Dijkers PF, Coffer PJ, Lam EW, Burgering BM. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol Cell Biol. 2002;22:2025–2036. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 20.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Calvano FC, Calvano-Mendes DC, Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR, Baracat EC. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p and miR-20a-5p. Tumour Biol. 2014;35:7733–7741. doi: 10.1007/s13277-014-2025-7. [DOI] [PubMed] [Google Scholar]
- 22.Gao F, Wang W. MicroRNA-96 promotes the proliferation of colorectal cancer cells and targets tumor protein p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1) and FOXO3a. Mol Med Rep. 2015;11:1200–1206. doi: 10.3892/mmr.2014.2854. [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Current Cancer Drug Targets. 2013;13:221–231. doi: 10.2174/1568009611313020010. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M, Zhu Y, Zhao Q, Dong YW, Shao K, Wu A, Wu XZ. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586:1038–1043. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Sun H, Zeng W, He J, Mao X. Upregulation of MircoRNA-370 induces proliferation in human prostate cancer cells by downregulating the transcription factor FOXO1. PLoS One. 2012;7:e45825. doi: 10.1371/journal.pone.0045825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung LW, Zhou W, Dong JT. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 27.Aoki M, Jiang H, Vogt PK. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci U S A. 2004;101:13613–13617. doi: 10.1073/pnas.0405454101. [DOI] [PMC free article] [PubMed] [Google Scholar]