Abstract
Hypermethylation of GPX3 (glutathione peroxidase 3) promoter has been identified in various cancers. However, the pattern of GPX3 promoter methylation in chronic myeloid leukemia (CML) remains unknown. Our study was aimed to investigate the methylation status of GPX3 promoter and its clinical relevance in CML. Real-time quantitative methylation-specific PCR and bisulfite sequencing PCR was performed to detect the level of GPX3 methylation in 80 CML patients and 44 controls. GPX3 promoter in CML patients was significantly methylated compared with controls (P = 0.007). GPX3 highly methylated patients showed significantly older age than GPX3 lowly methylated patients (P = 0.037). However, patients with GPX3 methylation had significantly lower white blood cells than those with low GPX3 methylation (P = 0.006). BCR-ABL transcript in GPX3 highly methylated patients was a little lower than that in GPX3 lowly methylated patients (P = 0.161). No significant differences were observed in the frequency of GPX3 methylation in the different stages of CML (P = 1.000). Significantly negative correlation was observed between GPX3 expression and GPX3 methylation (R = -0.442, P = 0.004). GPX3 mRNA level in K562 cell line was significantly increased after 5-aza-2’-deoxycytidine treatment, and GPX3 methylation level was decreased. GPX3 hypermethylation is frequent in CML and is negatively associated with its expression.
Keywords: GPX3, expression, methylation, chronic myeloid leukemia
Introduction
Chronic myeloid leukemia (CML) is a clonal disorder of the leukemic stem cells characterized by excessive proliferation of stem cells in hematopoietic tissues [1]. The main feature of CML is the Philadelphia chromosome (Ph) originating from a reciprocal translocation between chromosome 9 and 22 that gives rise to the BCR-ABL fusion gene [2-4]. Although the molecular pathogenesis of CML is well understood, the underlying mechanism that leads to the gene translocation is remains poorly understood [1]. Genomic DNA methylation plays a crucial role in affecting chromosomal stability and influences euchromatin-heterochromatin interactions [5]. Aberrant DNA methylation has been identified in various hematopoietic malignancies including CML [6,7]. Moreover, hypermethylation of quiet a few tumor suppressor genes (TSGs) have been found to be involved not only in the pathogenesis but also in the disease progression of CML [8-10].
GPX3 (glutathione peroxidase 3), located at the chromosome 5, is most studied member of GPX family [11]. GPX3 is well known to reduce redundant reactive oxygen species (ROS) against oxidative damages to host cells [11]. Down-regulation of GPX3 expression has been found in a variety of malignant tumors [12-22]. Moreover, hypermethylation of GPX3 promoter with its role in regulating GPX3 expression has been identified in several cancers [15-22]. However, the methylation pattern of GPX3 promoter is rarely investigated in CML. Our study was intended to detect GPX3 promoter methylation status and analyze its clinical relevance in CML patients.
Materials and methods
Patients
80 CML patients treated at the Affiliated People’s Hospital of Jiangsu University as well as 44 healthy donors were included in the current study. The diagnosis and clinical stages of CML were established according to WHO criteria [23]. Bone marrow (BM) was collected form all the patients and healthy donors after written informed consents were obtained. Karyotypes were analyzed on R-banded metaphases. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature. BCR-ABL mRNA level detection was quantified using RQ-PCR established previously [24]. The clinical and laboratory characteristics of the CML patients were shown in Table 1.
Table 1.
Association between GPX3 promoter methylation and clinical features in CML patients
| Patient’s parameters | Status of GPX3 promoter methylation | |||
|---|---|---|---|---|
|
| ||||
| Low (n = 68) | High (n = 12) | Total (n = 80) | P value | |
| Sex, male/female | 41/27 | 7/5 | 48/32 | 1.000 |
| Median age, years (range) | 48.5 (15-83) | 57 (35-75) | 51 (15-83) | 0.037 |
| Median WBC, ×109/L (range) | 63.9 (0.9-321.9) | 7.7 (1.5-35.7) | 58.5 (0.9-321.9) | 0.006 |
| Median hemoglobin, g/L (range) | 97.5 (47-146) | 103 (93-152) | 101 (47-152) | 0.183 |
| Median platelets, ×109/L (range) | 363.5 (30-1175) | 186.5 (22-870) | 359.5 (22-1175) | 0.376 |
| Cytogenetics | 1.000 | |||
| t(9;22) | 32 (47%) | 6 (50%) | 38 (48%) | |
| t(9;22) with additional alteration | 8 (12%) | 1 (8%) | 9 (11%) | |
| Normal karyotype | 6 (9%) | 1 (8%) | 7 (9%) | |
| No data | 22 (32%) | 4 (33%) | 26 (32%) | |
| Staging | 1.000 | |||
| CP | 54 (79%) | 10 (83%) | 64 (80%) | |
| AP | 4 (6%) | 0 (0%) | 4 (5%) | |
| BC | 10 (15%) | 2 (17%) | 12 (15%) | |
| BCR/ABL transcript | 211.0 (0.0-6375.8) | 89.2 (32.4-809.4) | 118.5 (0.0-6375.8) | 0.161 |
WBC: white blood cells; CP: chronic phase; AP: accelerated phase; BC: blast crisis.
Cell line, cell culture and 5-aza-dC treatment
Human leukemic cell line (K562) was cultured in IMDM medium containing 10% fetal calf serum and grown at 37°C in 5% CO2 humidified atmosphere. 5-aza-2’-deoxycytidine (5-aza-dC) (Sigma-Aldrich, Steinheim, USA) diluted in dimethyl sulfoxide (DMSO) was added in four flasks of K562 cells once a day at different final concentrations of 0 μM, 0.1 μM, 1 μM, 10 μM and 50 μM. All cells were cultured until harvested for extraction of RNA and DNA.
RNA isolation, reverse transcription and RQ-PCR
BM mononuclear cells (BMMNCs) were extracted by Lymphocyte Separation Medium (TBD sciences, Tianjin, China). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription (RT) was performed on iCycler Thermal Cycler (Eppendorf, Hamburg, Germany) with a 40 μL volume composed of 5× buffer 10 mM, dNTPs 10 mM, random hexamers 10 μM, RNAsin 80 U, and 200 U of MMLV reverse transcriptase (MBI Fermentas, Hanover, USA). The RT system was incubated for 10 min at 25°C, 60 min at 42°C, and then stored at -20°C.
GPX3 mRNA level was detected by real-time quantitative PCR (RQ-PCR) on a 7300 Thermo cycler (Applied Biosystems, CA, USA). RQ-PCR reaction with a volume of 20 μL consisted of cDNA 20 ng, 10 μM of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), 0.4 μM of ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA), and 0.8 μM of primers (forward 5’-GCCGGGGACAAGAGAAGT-3’ and reverse 5’-GAGGACGTATTTGCCAGCAT-3’) [16]. The RQ-PCR reaction was carried out at 95°C for 5 min, followed by 45 cycles at 95°C for 10 s, 63°C for 30 s, 72°C for 30 s, and 80°C for 30 s to collect fluorescence, finally followed by 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. Both positive and negative controls were included in each assay. Relative GPX3 transcript levels were calculated by the following equation: NGPX3 = (EGPX3)ΔCT GPX3 (control-sample) ÷ (EABL)ΔCT ABL (control-sample). The parameter efficiency (E) was counted by the formula E = 10(-1/slope) (the slope referred to CT versus cDNA concentration plot).
DNA isolation, chemical modification and RQ-MSP
Genomic DNA was isolated using genomic DNA purification kit (Gentra, Minneapolis, MN, USA) and was modified using the CpGenome DNA Modification Kit (Chemicon, Ternecula, Canada) according to the manufacturer’s instructions. The primers used for the methylated (M) [5’-TATGTTATTGTCGTTTCGGGAC-3’ (forward); 5’-GTCCGTCTAAAATATCCGACG-3’ (reverse)] and unmethylaed (U) [5’-TTTATGTTATTGTTGTTTTGGGATG-3’ (forward); 5’-ATCCATCTAAAATATCCAACACTCC-3’ (reverse)] GPX3 promoters were reported previously [20]. Real-time quantitative methyaltion-specific PCR (RQ-MSP) reaction contained 0.8 μM of primers, 10 μM of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), 0.4 μM of ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA), and 20 ng of modified DNA was also performed a 7300 Thermo cycler (Applied Biosystems, CA, USA). The reaction condition was 95°C for 5 min, 40 cycles for 10 s at 95°C, 30 s at 64°C (M) or 58°C (U), 72°C for 30 s, and 80°C (M) or 76°C (U) for 30 s, eventually a melting program of one cycle at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. The normalized ratio (NM-GPX3) was applied to assess the level of GPX3 promoter methylation in samples. NM-GPX3 was calculated using the following formula: NM-GPX3 = (EM-GPX3)ΔCT M-GPX3 (control-sample) ÷ (EALU)ΔCT ALU (control-sample).
BSP
Bisulfite sequencing PCR (BSP) reaction system was composed of 1×PCR buffer (KCl 0.25 mM), dNTP Mixture 6.25 μM, primers 0.5 μM, hot start DNA polymerase 0.75 U (Takara, Tokyo, Japan), and modified DNA 20 ng. The primers for bisulfite modified GPX3 promoter were 5’-ATTTTGGAGTTAAAAGAGGAAG-3’ (forward) and 5’-CTACCTAATCCCTAACCACC-3’ (reverse). The program was carried out at 98°C for 10 s, 40 cycles for 10 s at 98°C, 30 s at 56°C, 72°C for 30 s, and followed by a final 7 min extension step at 72°C. The PCR products were analyzed on 2% agarose gels. Each purified product was cloned into pMD19-T Vector (Takara, Tokyo, Japan), and was transfected into DH5A competent cells (ExCell Bio, Shanghai, China). Eight clones from each sample were sequenced (BGI Tech Solutions Co., Shanghai, China).
Statistical analysis
SPSS 18.0 software package (SPSS, Chicago, IL) was applied to perform statistical analyses. Mann-Whitney’s U test was used to compare the difference of continuous variables in two groups. Pearson Chi-square analysis or Fisher exact test was employed to compare the difference of categorical variables. Correlation analysis between GPX3 expression and its promoter methylation was performed by spearman rank correlation test. For all analyses, a two-tailed P value less than 0.05 was determined as statistically significant.
Results
GPX3 promoter methylation in controls and CML patients
According to the results of RQ-MSP, GPX3 presented lowly methylated promoter in 44 controls with median level of 0.005 (range 0.000 to 1.000). However, GPX3 promoter was significantly methylated in 80 CML patients (median 0.019, range 0.000-11.919) compared with controls (P = 0.007, Figure 1). The representative electrophoresis results of RQ-MSP products were shown in Figure 2.
Figure 1.
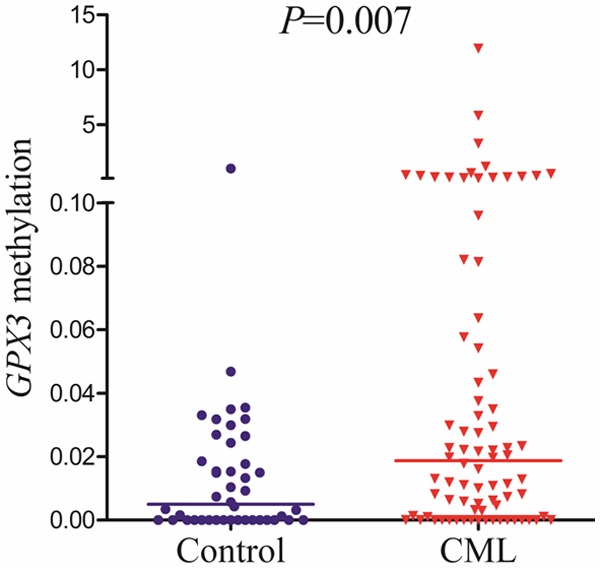
Relative methylation levels of GPX3 in controls and CML patients.
Figure 2.
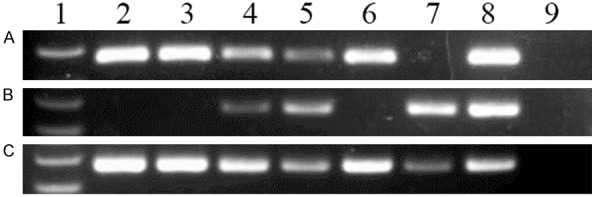
Electrophoresis results of RQ-PCR and RQ-MSP products in controls and CML patients. 1: Gene RulerTM 100bp DNA ladder; 2-3: controls; 4-7: CML patients; 8: positive control; 9: negative control. A: GPX3 expression; B: GPX3 methylation; C: GPX3 unmethylation.
To confirm the RQ-MSP results and further investigate the methylation density of GPX3 promoter, BSP was performed in two controls and four CML patients (two GPX3 methylated patients and two GPX3 unmethylated patients). Both the two controls and the two unmethylated CML patients showed extremely low density of GPX3 promoter, while the two methylated CML patients presented higher density of GPX3 promoter methylation (Figure 3).
Figure 3.
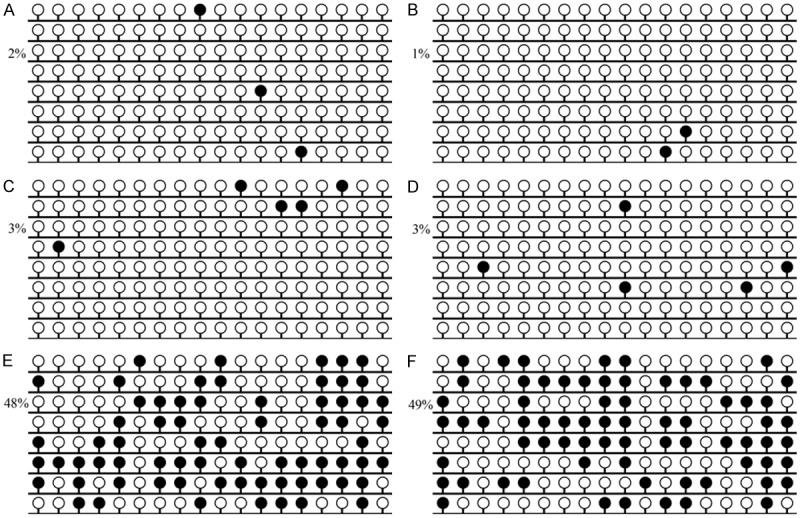
Methylation density of GPX3 promoter in controls and CML patients. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide. A, B: controls; C, D: hypomethylated CML patients; E, F: methylated CML patients.
Correlation between GPX3 methylation and clinical characteristics in CML patients
GPX3 methylation ratio more than the value of 0.184 (defined as the mean + SD in controls) was set to determine GPX3 promoter methylation in CML patients. According to the value of 0.183 (defined as mean + SD in normal controls), the whole CML patients were divided into two groups: high GPX3 methylation (GPX3 M) and low GPX3 methylation (GPX3 U). Only 2% (1/44) normal controls presented methylated GPX3 promoter. While, methylation of GPX3 promoter was identified in 15% (12/80) CML patients. There were no significant differences in sex, hemoglobin, platelets, and cytogenetics between the GPX3 M and GPX3 U patients (P > 0.05, Table 1). BCR-ABL transcript in GPX3 M patients was a little lower than that in GPX3 U patients (median 89.2 vs 211.0, P = 0.161). No significant difference was observed in the frequency of GPX3 methylation among the different stages of CML (P = 1.000). However, GPX3 M cases presented significantly older age than GPX3 U cases (P = 0.037). Moreover, GPX3 M patients had lower white blood cells than GPX3 U patients (P = 0.006).
GPX3 methylation correlated with its expression
GPX3 expression was detected in 40 CML patients as well as 44 controls with available mRNA. GPX3 transcript level in CML patients (median 0.286, range 0.000-9.114) was decreased compared to normal controls (median 0.348, range 0.069-9.258) (P = 0.169). Significantly negative correlation was observed between GPX3 expression and its promoter methylation in CML patients (R = -0.442, P = 0.004, Figure 4).
Figure 4.
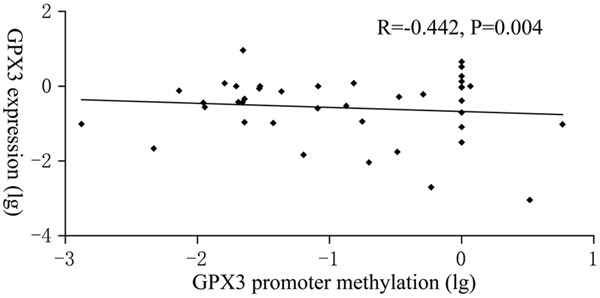
The association between GPX3 expression and its promoter methylation.
Epigenetic mechanism regulating GPX3 expression in leukemic cell line
K562 cell line treated by 5-aza-dC was selected to further verify the epigenetic mechanism regulating GPX3 expression in CML. Before 5-aza-dC treatment, K562 showed weakly GPX3 expression and fully methylated GPX3 promoter (Figure 5). As expected, GPX3 expression was significantly up-regulated after 5-aza-dC treatment, meanwhile, GPX3 promoter methylation level was decreased (Figure 5).
Figure 5.
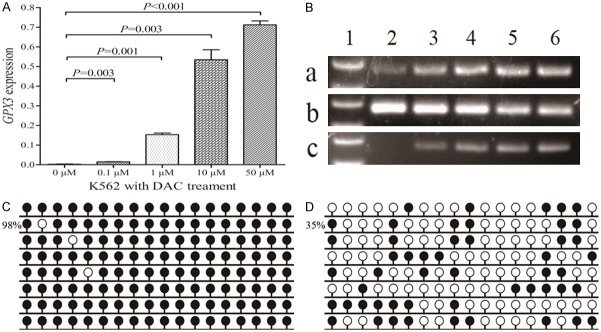
GPX3 expression and methylation in K562 cell line before and after 5-aza-dC treatment. A: GPX3 relative expression levels. B: Electrophoresis results of RQ-PCR and RQ-MSP products. 1: Gene RulerTM 100bp DNA ladder; 2: 0 μM; 3: 0.1 μM; 4: 1 μM; 5: 10 μM; 6: 50 μM. a: GPX3 expression; b: GPX3 methylation; c: GPX3 unmethylation. C: GPX3 methylation density before treatment. D: GPX3 methylation density after treatment (50 μM).
Discussion
Abnormal expression of various TSGs and/or oncogenes plays a vital role in the process of carcinogenesis. Epigenetic mechanisms including alterations in DNA methylation, histone modifications and small noncoding microRNAs refer to the mechanism that regulate gene expression without changing the primary DNA sequence. Aberrant epigenetic alterations probably occur at an early stage in cancer development, and they are widely described as essential factors in cancer progression [25]. DNA methylation, the most common and studied epigenetic alteration, is considered as a stable gene-silencing mechanism during DNA replication [25]. Silencing of TSGs by methylation of promoter-associated CpG islands has been reported in many kinds of human cancers [25]. Recent advances in epigenetics provide new insight into the discovery of putative cancer biomarkers for early detection, disease monitoring, prognosis, and risk assessment [25].
Tumor suppressor role of GPX3 has been identified in several cancers [26-28]. The epigenetic mechanism regulating GPX3 expression has been preliminarily studied. GPX3 expression regulated by its promoter methylation has been identified a host of solid tumors [15-21]. Moreover, a recent study has also revealed the mechanism in the regulation of GPX3 expression in multiple myeloma [22]. Our investigation not only observed the significant correlation between GPX3 expression and its promoter methylation in CML patients but also verified that GPX3 expression could be reactivated by 5-aza-dC treatment in the leukemic cell line K562. These results confirmed that hypermethylation of GPX3 promoter-associated CpG islands was one of the epigenetic mechanisms in silencing GPX3 expression in CML. Accordingly, the application of demethylation chemical agents could reactivate silenced tumor suppressors, which may be a novel therapeutic strategy for the clinical treatment of CML.
Clinical significance of GPX3 promoter hypermethylation has been abundantly demonstrated in various solid tumors. Li et al indicated the early diagnostic value of GPX3 promoter methylation in esophageal squamous cell carcinoma [29]. Peng et al demonstrated that GPX3 promoter methylation was associated with lymph node metastasis in gastric carcinomas [16]. Chen et al revealed that GPX3 promoter hypermethylation correlated with head and neck cancer (HNC) chemoresistance and served as a potential prognostic indicator for HNC patients treated with cisplatin-based chemotherapy [18]. Moreover, Kaiser et al also disclosed the prognostic value of GPX3 promoter methylation in multiple myeloma [22]. In the current study, we investigated the status of GPX3 promoter methylation in CML patients and demonstrated that GPX3 promoter hypermethylation is a frequent event in CML patients. However, our study failed to reveal the contribution of GPX3 promoter hypermethylation to the progression of CML, which suggests that aberrant GPX3 methylation might be an early event in CML. Recently, accumulating studies have disclosed that aberrant DNA methylation of numerous TSGs/oncogenes contributed to the progression of CML [8-10]. Li et al demonstrated SHP-1 promoter hypermethylation by dysregulating AKT, MAPK, MYC and JAK2/STAT5 signaling played a key role in the progression of CML [8]. Roman-Gomez et al indicated that the crucial role of LINE-1 promoter hypomethylation in the progression of CML by the activation of both sense and antisense transcriptions [9].
In conclusion, our study reveals that GPX3 hypermethylation is frequent in CML and is negatively associated with its expression.
Acknowledgements
This study was supported by National Natural Science foundation of China (81270630, 81172592), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), 333 Project of Jiangsu Province (BRA2013136), Science and Technology Infrastructure Program of Zhenjiang (SS2012003), Medical Key Talent Project of Zhenjiang, Social Development Foundation of Zhenjiang (SH2013042, SH2013082, SH2014044, SH2014086), and Jiangsu Government Scholarship for Overseas Studies.
Disclosure of conflict of interest
None.
References
- 1.Hehlmann R, Hochhaus A, Baccarani M, European L. Chronic myeloid leukaemia. Lancet. 2007;370:342–50. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- 2.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukemia. Nat Rev Cancer. 2005;5:172–83. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 3.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8:341–50. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 4.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–3. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 6.Fong CY, Morison J, Dawson MA. Epigenetics in the hematologic malignancies. Haematologica. 2014;99:1772–83. doi: 10.3324/haematol.2013.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deneberg S. Epigenetics in myeloid malignancies. Methods Mol Biol. 2012;863:119–37. doi: 10.1007/978-1-61779-612-8_7. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Yang L, Pan Y, Yang J, Shang Y, Luo J. Methylation and decreased expression of SHP-1 are related to disease progression in chronic myelogenous leukemia. Oncol Rep. 2014;31:2438–46. doi: 10.3892/or.2014.3098. [DOI] [PubMed] [Google Scholar]
- 9.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 10.Jelinek J, Gharibyan V, Estecio MR, Kondo K, He R, Chung W, Lu Y, Zhang N, Liang S, Kantarjian HM, Cortes JE, Issa JP. Aberrant DNA methylation is associated with disease progression, resistance to imatinib and shortened survival in chronic myelogenous leukemia. PLoS One. 2011;6:e22110. doi: 10.1371/journal.pone.0022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289–303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Zheng Z, Yingji S, Kim H, Jin R, Renshu L, Lee DY, Roh MR, Yang S. Downregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancer. Oncol Rep. 2014;31:2587–92. doi: 10.3892/or.2014.3152. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZL, Yang L, Zou Q, Yuan Y, Li J, Liang L, Zeng G, Chen S. Positive ALDH1A3 and negative GPX3 expressions are biomarkers for poor prognosis of gallbladder cancer. Dis Markers. 2013;35:163–72. doi: 10.1155/2013/187043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agnani D, Camacho-Vanegas O, Camacho C, Lele S, Odunsi K, Cohen S, Dottino P, Martignetti JA. Decreased levels of serum glutathione peroxidase 3 are associated with papillary serous ovarian cancer and disease progression. J Ovarian Res. 2011;4:18. doi: 10.1186/1757-2215-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamed MM, Sabet S, Peng DF, Nouh MA, El-Shinawi M, El-Rifai W. Promoter hypermethylation and suppression of glutathione peroxidase 3 are associated with inflammatory breast carcinogenesis. Oxid Med Cell Longev. 2014;2014:787195. doi: 10.1155/2014/787195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng DF, Hu TL, Schneider BG, Chen Z, Xu ZK, El-Rifai W. Silencing of glutathione peroxidase 3 through DNA hypermethylation is associated with lymph node metastasis in gastric carcinomas. PLoS One. 2012;7:e46214. doi: 10.1371/journal.pone.0046214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min SY, Kim HS, Jung EJ, Jung EJ, Jee CD, Kim WH. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 2012;32:3169–75. [PubMed] [Google Scholar]
- 18.Chen B, Rao X, House MG, Nephew KP, Cullen KJ, Guo Z. GPX3 promoter hypermethylation is a frequent event in human cancer and is associated with tumorigenesis and chemotherapy response. Cancer Lett. 2011;309:37–45. doi: 10.1016/j.canlet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Falck E, Karlsson S, Carlsson J, Helenius G, Karlsson M, Klinga-Levan K. Loss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinoma. Cancer Cell Int. 2010;10:46. doi: 10.1186/1475-2867-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Y, Wang Y, Li P, Zhu S, Wang J, Zhang S. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig Dis Sci. 2011;56:681–8. doi: 10.1007/s10620-010-1369-0. [DOI] [PubMed] [Google Scholar]
- 21.Lee OJ, Schneider-Stock R, McChesney PA, Kuester D, Roessner A, Vieth M, Moskaluk CA, El-Rifai W. Hypermethylation and loss of expression of glutathione peroxidase-3 in Barrett’s tumorigenesis. Neoplasia. 2005;7:854–61. doi: 10.1593/neo.05328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser MF, Johnson DC, Wu P, Walker BA, Brioli A, Mirabella F, Wardell CP, Melchor L, Davies FE, Morgan GJ. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood. 2013;122:219–26. doi: 10.1182/blood-2013-03-487884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 24.Qian J, Wang YL, Lin J, Yao DM, Xu WR, Wu CY. Aberrant methylation of the death-associated protein kinase 1 (DAPK1) CpG island in chronic myeloid leukemia. Eur J Haematol. 2009;82:119–23. doi: 10.1111/j.1600-0609.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 25.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–11. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, Luo JH. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67:8043–50. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- 27.Barrett CW, Ning W, Chen X, Smith JJ, Washington MK, Hill KE, Coburn LA, Peek RM, Chaturvedi R, Wilson KT, Burk RF, Williams CS. Tumor suppressor function of the plasma glutathione peroxidase GPX3 in colitis-associated carcinoma. Cancer Res. 2013;73:1245–55. doi: 10.1158/0008-5472.CAN-12-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi X, Ng KT, Lian QZ, Liu XB, Li CX, Geng W, Ling CC, Ma YY, Yeung WH, Tu WW, Fan ST, Lo CM, Man K. Clinical significance and therapeutic value of glutathione peroxidase 3 (GPx3) in hepatocellular carcinoma. Oncotarget. 2014;5:11103–20. doi: 10.18632/oncotarget.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang F, Wang N, Yang H, Zheng Y, Zhang J. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS One. 2014;9:e103162. doi: 10.1371/journal.pone.0103162. [DOI] [PMC free article] [PubMed] [Google Scholar]


