Abstract
Mutational studies of human DNA helicase B (HDHB) have suggested that its activity is critical for the G1/S transition of the cell cycle, but the nature of its role remains unknown. In this study, we show that during G1, ectopically expressed HDHB localizes in nuclear foci induced by DNA damaging agents and that this focal pattern requires active HDHB. During S and G2/M, HDHB localizes primarily in the cytoplasm. A carboxy-terminal domain from HDHB confers cell cycle-dependent localization, but not the focal pattern, to a reporter protein. A cluster of potential cyclin-dependent kinase phosphorylation sites in this domain was modified at the G1/S transition and maintained through G2/M of the cell cycle in vivo, coincident with nuclear export of HDHB. Serine 967 of HDHB was the major site phosphorylated in vivo and in vitro by cyclin-dependent kinases. Mutational analysis demonstrated that phosphorylation of serine 967 is crucial in regulating the subcellular localization of ectopically expressed HDHB. We propose that the helicase of HDHB operates primarily during G1 to process endogenous DNA damage before the G1/S transition, and it is largely sequestered in the cytoplasm during S/G2.
INTRODUCTION
Human genetic diseases such as cancer predisposition and aging are associated with Bloom's and Werner's syndrome helicases (reviewed by Hickson, 2003), the xeroderma pigmentosum helicases XPB/XPD (reviewed by Friedberg, 2001), and BACH1 helicase (Cantor et al., 2001). DNA helicases play a critical part in DNA replication, repair, and recombination, but assigning an unknown helicase to one or more of these pathways and elucidating its functions has been challenging in higher eukaryotes with limited genetic accessibility. Human DNA helicase B (HDHB) cDNA was recently cloned based on its homology to a mouse ortholog (Tada et al., 2001; Taneja et al., 2002). The predicted HDHB protein is 1087 amino acids in length. It seems to be composed of three functional domains: an amino terminal domain of unknown function, a central helicase domain with the seven conserved motifs of helicase superfamily 1, and a 130-amino acid carboxy-terminal domain containing seven putative cyclin-dependent kinase (CDK) phosphorylation sites. Purified recombinant HDHB displays strong ATPase activity that is dependent on single-stranded DNA, but little or no activity in the presence of RNA or double-stranded DNA. HDHB exhibits robust 5′-3′ DNA helicase activity that requires its Walker A and B motifs (Taneja et al., 2002). The properties of HDHB suggest its involvement in one or more DNA processing pathways, but its specific function(s) remains unknown.
One possible role for HDHB might lie in DNA replication, as suggested by early work on mouse DNA helicase B (Matsumoto et al., 1995; Saitoh et al., 1995; Seki et al., 1995). Like DNA helicase B purified from mouse cells, recombinant HDHB interacts physically with human DNA polymerase α-primase, stimulating primase activity on single-stranded DNA coated with human replication protein A (Taneja et al., 2002). Consistent with a possible role in initiation of DNA replication, microinjection of purified HDHB protein with the Walker A or B mutation into cells in early G1 inhibited S phase entry (Taneja et al., 2002). Injection of wild-type HDHB protein into the nucleus of cells in G1 had no detectable effect on DNA synthesis. These results raised the question of whether HDHB might be assembled with prereplication proteins on chromatin in early G1, much like minichromosome maintenance proteins MCM2-7 (reviewed by Bell and Dutta, 2002), to facilitate DNA unwinding during S phase.
Another clue to HDHB function may lie in its amino acid sequence homology with known helicases (Taneja et al., 2002). No obvious orthologs of HDHB have been found in lower eukaryotes, but the helicase motifs of HDHB are remarkably similar to those of prokaryotic helicases involved in recombination, such as Escherichia coli RecD and bacteriophage T4 Dda (Chedin and Kowalczykowski, 2002; Dillingham et al., 2003; Taylor and Smith, 2003). RecD helicase also serves a regulatory role in bacterial recombination, governing RecBC nuclease activity and polarity in response to Chi sites in the DNA. This sequence similarity raised the question of whether HDHB may function in one or more of the DNA recombination pathways in human cells (reviewed by Khanna and Jackson, 2001; West, 2003).
If HDHB is, in fact, a DNA-specific helicase involved in DNA replication or recombination pathways, a simple prediction is that active forms of HDHB should reside in the nucleus. To test this prediction, we have investigated the subcellular localization and phosphorylation of ectopically expressed HDHB. We show that HDHB is primarily nuclear in G1 and cytoplasmic in S phase, that it resides in nuclear foci induced by DNA damage, that the focal pattern requires HDHB activity, and that HDHB localization is regulated by CDK phosphorylation.
MATERIALS AND METHODS
Plasmids
pGFP-HDHB and mutant derivatives of it were created by inserting the full-length HDHB cDNA as a BglII/NotI fragment (Taneja et al., 2002) into the NotI site of the pEGFP-C1 vector (BD Biosciences Clontech, Palo Alto, CA). pFLAG-HDHB was constructed by inserting a HindIII/NotI fragment containing full-length HDHB cDNA into the NotI site of pFlag-CMV2 vector (Eastman Kodak, Rochester, NY). Tagged HDHB-ΔSLD (1-1039) was constructed by cleaving the tagged HDHB plasmid with NruI after the coding sequence for residue 1034 and with NotI in the polylinker and replacing the small fragment by a duplex adaptor oligonucleotide with a blunt end encoding residues 1035-1039, a stop codon, and an overhanging NotI-compatible 5′ end. To create pFLAG-HDHB (1-874), StuI-digested pFLAG-HDHB DNA was treated with Klenow polymerase to generate blunt ends and ligated into the pFLAG-CMV2 vector. To generate pEGFP-βGal, a DNA fragment encoding E. coli β-galactosidase (βGal) was amplified by polymerase chain reaction (PCR) from the pβGal-control vector (BD Biosciences Clontech) and inserted in frame at the 3′ end of the green fluorescent protein (GFP) coding sequence in pEGFP-C1, by using the HindIII restriction site. The HDHB coding sequence for amino acids 1040-1087(SLD) and 957-1087(PSLD) were PCR amplified and inserted in frame at the 3′ end of the βGal cDNA in pEGFP-βGal to create pGFP-βGal-SLD and pGFP-βGal-PSLD, respectively. The HDHB Walker A and Walker B mutants, MutA and MutB, were described previously (Taneja et al., 2002). The nuclear export signal (NES) mutants and phosphorylation site mutants were created in the HDHB cDNA by site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA) according to the manufacturer's protocol by using Pfu Turbo polymerase (Stratagene) and oligonucleotides containing the desired DNA sequence changes as primers in the PCR reactions. The correct DNA sequence of the substitution mutations was confirmed by DNA sequencing. Oligonucleotide sequences used for mutant construction are available upon request.
Antibodies
Anti-HDHB antibody was generated against purified recombinant HDHB (Bethyl Laboratories, Montgomery, TX) and affinity purified on immobilized HDHB (Harlow and Lane, 1988). Initial characterization of these antibodies revealed that they were not ideal for indirect immunofluorescence or immunoprecipitation, but detected purified recombinant HDHB and endogenous HDHB in human cell extracts by Western blotting (our unpublished data).
Cell Culture, Synchronization, and Microinjection
U2OS cells were cultured as exponentially growing monolayers in Dulbecco-modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA) at 37°C. Exponentially growing U2OS cells were arrested at G1/S in incubation in DMEM containing 5 mM thymidine (Sigma-Aldrich, St. Louis, MO), for 24 h. To release the cells into S phase, we aspirated the medium, washed the cells three times with warm DMEM plus 10% FBS, and incubated them in fresh DMEM plus 10% FBS. Exponentially growing U2OS cells were arrested in G2/M for 16 h in DMEM containing 30 ng/ml nocodazole (Sigma-Aldrich). To release cells into G1, mitotic cells were collected by gently shaking them off, washed three times with DMEM plus 10% FBS, and then plated on glass coverslips for microinjection, or in culture dishes for further manipulation. Cell cycle synchronization was verified by flow cytometry as described previously (Taneja et al., 2002). In experiments to block nuclear protein export, cells were cultured for 3 h in DMEM containing 10 ng/ml leptomycin B (LMB) (gift from Dr. M. Yoshida, RIKEN, Wako, Saitama, Japan) and 10 μM cycloheximide (Calbiochem, San Diego, CA) to prevent new protein synthesis. Cells plated on glass coverslips were microinjected as described previously (Herbig et al., 1999) except that plasmid DNA rather than protein was injected.
Fluorescence Microscopy
For indirect immunofluorescence staining, cells were washed three times with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde in PBS for 20 min, permeabilized for 5 min by using 0.2% Triton X-100, and incubated with 10% FBS in PBS for 45 min. FLAG-HDHB was detected by staining with mouse monoclonal anti-FLAG antibody (Sigma-Aldrich) at a dilution of 1:100 in PBS plus 10% FBS for 2 h at room temperature. After washing, the cells were incubated with Texas Red-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:100 in PBS plus 10% FBS for 1 h at room temperature. After three washes, the cells were incubated for 10 min with Hoechst 33258 at a concentration of 2 μM in PBS. Coverslips were mounted in ProLong Antifade (Molecular Probes, Eugene, OR). Images were obtained with a Hamamatsu digital camera by using the Openlab 3.0 software (Improvision, Lexington, MA) on the Axioplan 2 Imaging system (Carl Zeiss, Thornwood, NY). The number of cells that exhibited each pattern of subcellular localization was counted and expressed as a percentage of the total number of cells scored (100-150 cells in each experiment). The subcellular distribution of each protein was quantitatively evaluated in at least two independent experiments.
For GFP fluorescence, cells were washed three times with PBS and fixed with 3.7% formaldehyde in PBS containing 2 μM Hoechst 33258 for 20 min. Coverslips were mounted in ProLong Antifade (Molecular Probes), and fluorescent images were taken and evaluated as described above.
For Triton X-100 extraction, cells were washed twice with cold cytoskeleton buffer (CSK, 10 mM HEPES, pH 7.4,, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2) and extracted for 5 min on ice with 0.5% Triton X-100 in CSK buffer (supplemented with 1× protease inhibitors) and then fixed as described above.
DNA Damage Response Assay
U2OS cells (80-90% confluent) were transfected with pGFP-HDHB according to the manufacturer's protocol (LipofectAMINE 2000; Invitrogen). At 4 h after transfection, cells were treated with dimethyl sulfoxide (DMSO) (control), 20 μM etoposide, 10 μM camptothecin, or 1 μM mitomycin C. After 20 h, cells were extracted with Triton X-100 buffer and then fixed for immunofluorescence as described above. Distinctive GFP-HDHB nuclear foci were counted in more than 100 cells in each independent assay.
Electroporation
Asynchronously growing U2OS cells (5 × 106) were trypsinized, collected by centrifugation, and resuspended in 800 μl of 20 mM HEPES, pH 7.4, 0.7 mM Na2HPO4/NaH2PO4, 137 mM NaCl, 5 mM KCl, 6 mM glucose at a final pH of 7.4. Ten micrograms of DNA was added, and the mixture was transferred to a 0.4-cm electroporation cuvette (Bio-Rad, Hercules, CA). Electroporation was performed using a Gene Pulser II apparatus and Gene Pulser II RF module (Bio-Rad) at 300 V, 600 μF. Cells were then plated in tissue culture dishes, and 1 h later, washed with fresh medium and cultured for another 23 h.
Metabolic Phosphate Labeling
U2OS cells (2.5 × 106) were transiently transfected with wild-type or mutant FLAG-HDHB by electroporation. After 24 h, cells were incubated in phosphate-depleted DMEM (Invitrogen) for 15 min and then radiolabeled with [32P]H3PO4 (0.35 mCi/ml of medium; Valeant Pharmaceuticals, Costa Mesa, CA) for 4 h. Phosphate-labeled FLAG-HDHB was immunoprecipitated from extracts, separated by 7.5% SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane as described below.
Cell Extracts, Immunoprecipitation, and Western Blotting
At 24 h after transfection, FLAG-HDHB-transfected cultures to be analyzed by immunoprecipitation and immunoblotting were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 0.1% NP-40, 1 mM dithiothreitol [DTT], 25 mM NaF, 100 μg/ml phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml aprotinin, 1 μg/ml leupeptin) (0.5 ml/35-mm or 1 ml/60-mm dish or 75-cm flask). The extract was scraped off the dish, incubated for 5 min on ice, and centrifuged for 10 min at 14,000 × g. Samples of the supernatant (0.5-1 mg of protein) were incubated with 10 μl of anti-FLAG agarose (Sigma-Aldrich) on a rotator for 2 h at 4°C. The agarose beads were washed three times with lysis buffer. Immunoprecipitated proteins were transferred to a PVDF membrane and analyzed by Western blotting with anti-HDHB serum (1:5000), anti-cyclin E antibody (1:1000), and anti-cyclin A antibody (1:1000) (Santa Cruz Biotechnology, Santa Cruz, CA), and chemiluminescence (SuperSignal; Pierce Chemical, Rockford, IL).
For selective nuclear and cytoplasmic protein extraction, 80-90% confluent U2OS cells were harvested by trypsinization and washed with PBS. They were resuspended and lysed in 10 mM Tris-HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.25 M sucrose, 10% glycerol, 75 μg/ml digitonin, 1 mM DTT, 10 mM NaF, 1 mM Na3VO4, 100 μg/ml PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin for 10 min on ice, and centrifuged at 1000 × g for 5 min. The supernatant fraction was collected as the cytosolic extract. The pellet was washed, resuspended in high salt buffer (10 mM Tris-HCl, pH 7.5, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1% NP-40, 100 μg/ml PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin), and rocked for 10 min at 4°C. After sonication, the suspended material, containing both soluble and chromatin-bound protein, was analyzed as nuclear extract. Proteins in the nuclear and cytoplasmic extracts were analyzed by 8.5% SDS-PAGE, followed by Western blotting with antibodies against α-tubulin, proliferating cell nuclear antigen (PCNA) (both from Santa Cruz Biotechnology), and recombinant HDHB.
Protein Phosphatase Reactions
FLAG-HDHB bound to anti-FLAG beads was incubated with 100 U of λ-phosphatase (New England Biolabs, Beverly, MA) in phosphatase buffer (50 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 0.01% NP-40) for 1 h at 30°C. The reaction was carried out in the presence or absence of phosphatase inhibitors (5 mM Na3VO4, 50 mM NaF). The proteins were separated by 7.5% SDS-PAGE (acrylamide/bisacrylamide ratio, 30:0.36), and HDHB was detected by Western blotting with anti-HDHB serum and chemiluminescence.
Tryptic Peptide Mapping and Phosphoamino Acid Analysis
At 24 h after transfection, radiolabeled FLAG-HDHB-transfected cultures to be used for immunoprecipitation and phosphoamino acid or phosphopeptide mapping were processed as described above, except that lysis buffer was substituted by radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 1% SDS, 50 mM NaF, 1 mM EDTA, 5 mM Na3VO4, 100 μg/ml PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin). Immunoprecipitated proteins were separated by 7.5% SDS-PAGE and transferred to PVDF membranes. The membranes containing radiolabeled HDHB were rinsed well with deionized H2O twice before visualization of phosphoproteins by autoradiography. The phosphoproteins were then excised, and the membrane pieces were rewet with methanol followed by water. The membranes were blocked with 50 mM NH4HCO3 containing 0.1% Tween 20 (Sigma-Aldrich) for 30 min at room temperature and washed three times with 50 mM NH4HCO3 before enzymatic cleavage of phosphoproteins from the PVDF with l-(tosylamido-2-phenyl)ethyl chloromethyl ketone-treated bovine pancreatic trypsin (Worthington Biochemicals, Lakewood, NJ). The peptides were then subjected to two-dimensional phosphopeptide mapping or phosphoamino acid analysis as described in detail previously (Xia et al., 2003).
Cyclin-dependent Kinase Reactions In Vitro
Kinase reactions using purified cyclin/CDK (200 pmol/h) (provided by R. Ott and C. Voitenleitner, Vanderbilt University, Nashville, TN) and purified recombinant HDHB (Taneja et al., 2002) as the substrate were performed as described previously (Voitenleitner et al., 1999).
RESULTS
HDHB Resides in Nuclear Foci or in the Cytoplasm
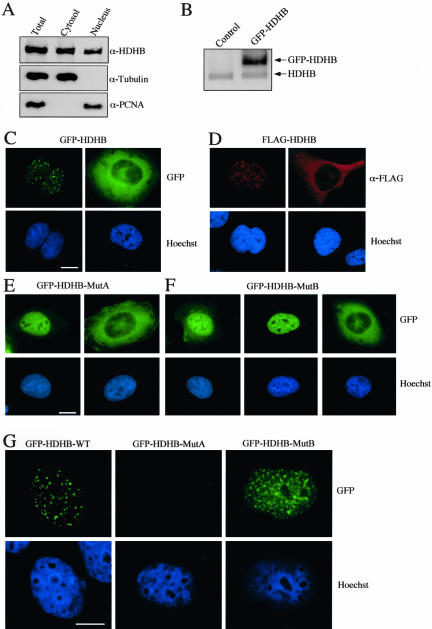
To determine the subcellular localization of endogenous HDHB, nuclear and cytoplasmic proteins were selectively extracted from human U2OS cells, separated by denaturing gel electrophoresis, and analyzed by Western blotting (Figure 1A). The presence of PCNA and α-tubulin in each extract was first monitored to assess the extraction procedure. PCNA was enriched in the nuclear extract and not in the cytoplasmic fraction, whereas α-tubulin was found primarily in the cytoplasmic fraction, validating the fractionation. HDHB was detected in both the nuclear and cytoplasmic fractions (Figure 1A). These results could indicate either that HDHB was distributed throughout the cell or that a mixed population of cells contained HDHB in either the nucleus or the cytoplasm.
Figure 1.
HDHB localizes in the nucleus or cytoplasm. (A) Cytoplasmic and nuclear extracts of U2OS cells were analyzed by denaturing gel electrophoresis and Western blotting with antibody against recombinant HDHB, α-tubulin, and PCNA. Immunoreactive proteins were detected by chemiluminescence. (B) GFP-HDHB transiently expressed in U2OS cells in contrast to endogenous HDHB. Control cells were transfected with pEGFP-C1 vector alone. (C) GFP-tagged HDHB, (D) FLAG-tagged HDHB, (E) Walker A (MutA), and (F) Walker B mutants (MutB) of GFP-HDHB transiently expressed in microinjected U2OS cells were visualized by fluorescence microscopy. Nuclei were stained with Hoechst dye. Bar, 10 μm. (G) U2OS cells transiently expressing GFP-HDHB wt, MutA, and MutB were extracted with 0.5% Triton X-100 before fixation and fluorescence microscopy. Nuclei were stained with Hoechst dye. Bar, 10 μm.
To distinguish between these alternatives, we wished to localize HDHB in situ in single cells. Because endogenous HDHB was not detectable by indirect immunofluorescence with our antisera (our unpublished data), we expressed GFP- and FLAG-tagged HDHB in human U2OS cells by transient transfection. Transiently overexpressed tagged HDHB accumulated in greater amounts than the endogenous HDHB within 24 h (Figure 1B). Because prolonged overexpression of tagged or untagged HDHB was cytotoxic, all experiments were conducted in the shortest time period possible (usually 24 h). Tagged HDHB localization was analyzed in individual cells by fluorescence microscopy. Both GFP-HDHB and FLAG-HDHB displayed two major patterns of localization, either in the nucleus in discrete foci or in the cytoplasm (Figure 1, C and D). The localization patterns of HDHB tagged with the GFP protein were not detectably different than those of HDHB tagged with the FLAG peptide. GFP-HDHB transiently expressed in primary human fibroblasts was also observed in either the nucleus or the cytoplasm (our unpublished data).
To test whether the subcellular localization of HDHB depended on its biochemical activity, the conserved lysine of the Walker A motif in GFP-HDHB was substituted by alanine (MutA) or the conserved glutamate of the Walker B motif was replaced by glutamine (MutB), crippling HDHB helicase activity (Taneja et al., 2002). Although DNA polymerase alpha-primase associated with both mutants, ATP stimulated single-stranded DNA binding of MutB, but not MutA (Taneja et al., 2002). These mutant forms of GFP-HDHB accumulated either in the nucleus, sparing the nucleoli, or in the cytoplasm of transfected cells, with few cells showing GFP-HDHB in both compartments (Figure 1, E and F). Interestingly, nuclear foci of GFP-HDHB were not obvious with the Walker A mutant (Figure 1E). The focal pattern of the Walker B mutant was more variable, with some cells resembling the Walker A pattern (Figure 1F, left) and others containing a small number of foci less prominent than in cells expressing wild-type GFP-HDHB (Figure 1F, middle). Interestingly, when the cells were extracted with detergent before fixation to remove soluble HDHB, GFP-HDHB wt maintained the distinct focal staining pattern, whereas MutB displayed a more obvious focal staining pattern, but with many smaller foci than those of the wild type (Figure 1G). MutA staining was lost, indicating that MutA was largely soluble and not bound to nuclear structures. The results suggest that nucleotide binding of HDHB is required for nuclear focus formation and that the foci are bound to detergent-insoluble nuclear structures.
GFP-HDHB Nuclear Focus Formation Increases upon DNA Damage
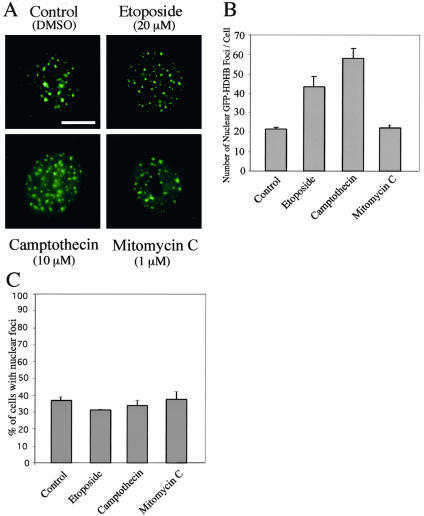
Because HDHB localization in nuclear foci depends on its biochemical activity, we reasoned that HDHB is likely to execute its function in those nuclear foci. The specificity of HDHB for DNA as a substrate and its sequence homology with bacterial RecD and T4 dda proteins (Taneja et al., 2002) led us to suspect that HDHB might be involved in DNA damage signaling, processing, or repair. Moreover, DNA damage induces nuclear foci that are thought to contain damage-sensing and -processing proteins (Nelms et al., 1998; van den Bosch et al., 2003). To test whether HDHB may reside in DNA damage foci, U2OS cells transiently expressing GFP-HDHB were treated with the DNA damaging agents etoposide, camptothecin, and mitomycin C, or with DMSO as a control (Figure 2). The number of large GFP-HDHB nuclear foci per cell more than doubled upon exposure to etoposide and camptothecin compared with the control (Figure 2B), strongly suggesting that HDHB was recruited to foci generated by DNA damage. The observed increase in foci was dose dependent (our unpublished data). The number of cells displaying nuclear foci was similar in control cultures and those treated with damaging agents (Figure 2C). Mitomycin C treatment had little effect on GFP-HDHB nuclear foci, suggesting that only specific types of DNA damage may attract HDHB.
Figure 2.
GFP-HDHB nuclear focus formation increases upon DNA damage. (A) U2OS cells transiently expressing GFP-HDHB were treated with DMSO (control), 20 μM etoposide, 10 μM camptothecin, or 1 μM mitomycin C as indicated and visualized by fluorescence microscopy. (B) Number of large distinct GFP-HDHB nuclear foci per cell in two independent assays is shown, with SD in brackets. (C) Percentage of cells in each culture containing GFP-HDHB nuclear foci is shown. The results are from two independent assays, with SD in brackets.
Identification of a Cell Cycle-dependent Subcellular Localization Domain in HDHB
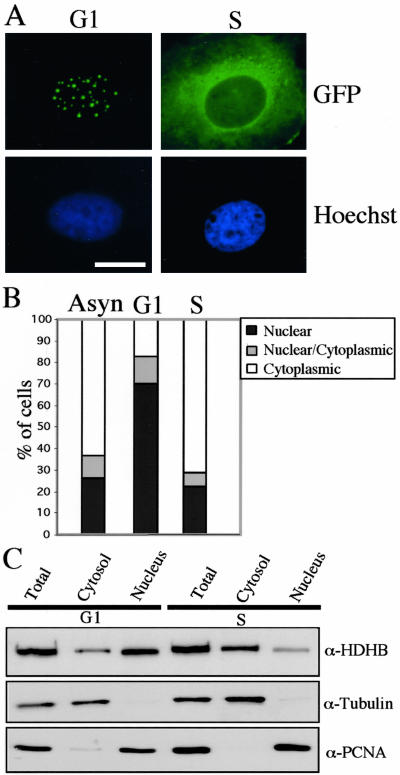
The ability of Walker A and Walker B mutants of HDHB to inhibit the onset of S phase (Taneja et al., 2002), together with the nuclear or cytoplasmic localization of tagged HDHB (Figure 1), raised the question of whether the subcellular localization of HDHB might be cell cycle dependent. To test this possibility, we arrested U2OS cells in G2/M with nocodazole, released them into G1 for 3 h, and then microinjected pGFP-HDHB DNA into their nuclei. GFP-HDHB expression was easily detectable 6 h later, when ∼70% of G1 phase cells had accumulated the fusion protein primarily in the nuclei (Figure 3, A and B). In contrast, when cells were synchronized at G1/S with thymidine, released into S phase, and then microinjected with pGFP-HDHB DNA, >70% of S phase cells had accumulated the fusion protein predominantly in the cytoplasm (Figure 3, A and B). Selective extraction of U2OS cells in G1 and S phase revealed that endogenous HDHB was mostly nuclear in G1 and mostly cytoplasmic in S phase (Figure 3C). However, endogenous HDHB was clearly detectable in both subcellular fractions in G1 and in S. These results indicate that the subcellular localization of HDHB is regulated in the cell cycle and that GFP-tagged HDHB reflects the localization of the endogenous untagged helicase.
Figure 3.
Subcellular localization of GFP-HDHB is cell cycle dependent. (A) GFP-HDHB transiently expressed in U2OS cells in G1 or S phase was visualized by fluorescence microscopy. Nuclei were stained with Hoechst dye. Bar, 10 μm. (B) Subcellular localization of GFP-tagged HDHB in asynchronous, G1, and S phase U2OS cells was quantified. The number of GFP-positive cells with a given distribution pattern was expressed as a percentage of the total number of GFP-positive cells in that experiment. (C) Cytoplasmic and nuclear extracts of synchronized U2OS cells (G1 and S phase) were analyzed by denaturing gel electrophoresis and Western blotting with antibody against recombinant HDHB, α-tubulin, and PCNA. Immunoreactive proteins were detected by chemiluminescence.
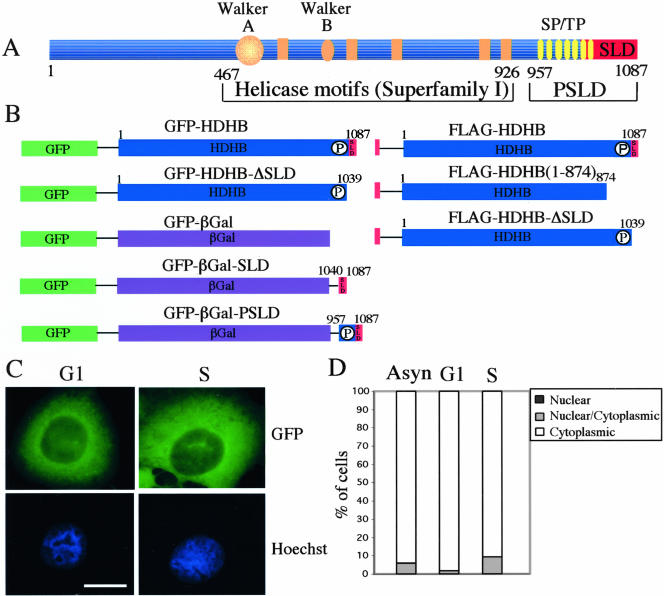
There are two primary mechanisms to target a protein to the nucleus or cytoplasm in a cell cycle-dependent manner. One is that the protein carries its own nuclear location signal (NLS) and/or NES, motifs that are recognized by nuclear import or export machinery (Gorlich and Kutay, 1999; Hood and Silver, 2000; Fabbro and Henderson, 2003; Weis, 2003). Another is that the protein lacks a targeting signal but can bind to another protein that has a NES and/or NLS. Prompted by the identification of C-terminal nuclear location signals in Bloom's syndrome helicase and other RecQ-family helicases (Hickson, 2003), we identified a possible subcellular localization domain (SLD) at the extreme C terminus of HDHB (Figure 4A). To determine whether this putative SLD was important for HDHB localization, we generated a truncation mutant of HDHB (GFP-HDHB-ΔSLD) that lacks the C-terminal 48 residues containing the SLD (Figure 4B). The expression vector was microinjected into U2OS cells in G1 or S phase, and the subcellular localization of the fusion protein was examined by fluorescence microscopy 6 h later (Figure 4C). More than 95% of the cells accumulated the fusion protein in the cytoplasm, regardless of the cell cycle timing of HDHB expression (Figure 4, C and D). This result suggests that HDHB may carry an NLS that is impaired or abolished by the C-terminal deletion in GFPHDHB-ΔSLD.
Figure 4.
Identification of a domain required for nuclear localization of HDHB. (A) Schematic representation of the HDHB protein showing seven potential phosphorylation sites for CDK (SP or TP), the putative subcellular localization domain (SLD) and phosphorylated SLD (PSLD), the Walker A and Walker B motifs. Amino acid residue numbers are indicated below the protein. (B) GFP- and FLAG-tagged HDHB and their C-terminal truncation mutants are depicted. The C terminus of HDHB SLD and PSLD was fused to a GFP-βGal reporter to create GFP-βGal-SLD and GFP-βGal-PSLD, respectively. (C) GFP-HDHB-ΔSLD was transiently expressed in U2OS cells in G1 or S phase and visualized by fluorescence microscopy. Nuclei were stained with Hoechst dye. Bar, 10 μm. (D) The subcellular localization of GFP-HDHB-ΔSLD in asynchronous, G1, and S phase U2OS cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that experiment.
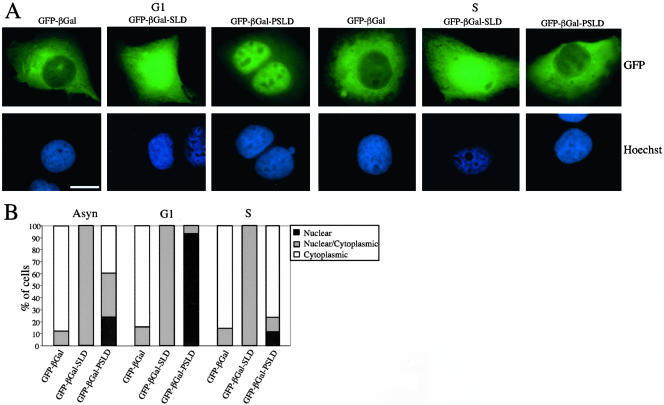
To determine whether the C-terminal domain of HDHB was sufficient for nuclear localization, we used bacterial βGal as a reporter protein because it has a molecular mass (112 kDa) close to that of HDHB and does not contain subcellular localization signals (Kalderon et al., 1984). As a control, we created a GFP-βGal expression vector (Figure 4B) and monitored the subcellular localization of the fusion protein after microinjection of the expression vector into U2OS cells. As expected, GFP-βGal protein accumulated primarily in the cytoplasm (Figure 5). In contrast, GFP-βGal-SLD was found in both the nucleus and cytoplasm in asynchronous or synchronized U2OS cells (Figure 5), suggesting that SLD contains a NLS but was not sufficient for nuclear localization of the reporter protein. Reasoning that perhaps the neighboring potential CDK phosphorylation sites might affect subcellular localization in the cell cycle (Figure 4A), we constructed GFP-βGal-PSLD, in which the C-terminal 131 residues of HDHB, containing the putative SLD and the cluster of potential CDK phosphorylation sites, were appended to the C terminus of GFP-βGal (Figure 4B). When GFP-βGal-PSLD plasmid DNA was transiently expressed in asynchronous and synchronized U2OS cells, GFP-βGal-PSLD was found in the nucleus in >90% of G1 phase cells, and in the cytoplasm in >70% of S phase cells (Figure 5). In contrast with the focal pattern observed for nuclear GFP-HDHB in G1, GFP-βGal-PSLD protein was distributed evenly throughout the nucleus in G1, sparing only the nucleoli (Figure 5). Together, these data suggested that the subcellular localization of HDHB is dependent on the cell cycle and that the C-terminal PSLD domain of HDHB plays a major role in regulating the subcellular localization of the protein.
Figure 5.
GFP-βGal-PSLD subcellular localization pattern varies with the cell cycle. (A) GFP-βGal, GFP-βGal-SLD, and GFP-βGal-PSLD were transiently expressed in U2OS cells in G1 (left) and S phase (right) of the cell cycle. Nuclei were stained with Hoechst dye. Bar, 10 μm. (B) Subcellular localization of GFP-βGal, GFP-βGal-SLD, and GFP-βGal-PSLD in asynchronous, G1, and S phase U2OS cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that experiment.
Identification of a Functional Rev-Type NES in HDHB
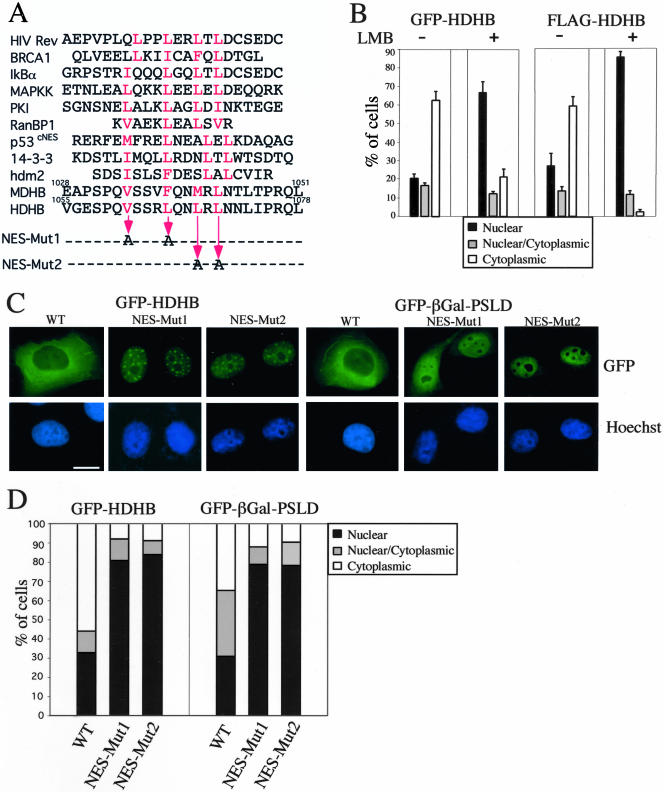
A number of proteins that shuttle between the nucleus and cytoplasm have been demonstrated to contain a NES similar to the prototype NES of HIV rev protein (Figure 6A). Proteins containing a rev-type NES require the export factor chromosome region maintenance 1 (CRM1) (also called exportin 1) to bind and transport proteins from the nucleus to the cytoplasm (reviewed by Weis, 2003). A fungal metabolite, LMB, specifically inhibits CRM1 activity in nuclear protein export (Wolff et al., 1997; Kudo et al., 1998). Inspection of the phosphorylation and subcellular localization domain (PSLD) sequence in HDHB revealed a putative rev-type NES (LxxxLxxLxL, where L also can be I, V, M, or F, and x is any amino acid) (Figure 6A). To determine whether the cytoplasmic localization of HDHB requires a functional NES, expression plasmids for GFP-HDHB or FLAG-HDHB DNA were microinjected into asynchronous, G1, and S phase cells in the presence and absence of LMB. The localization of the fusion proteins was examined by fluorescence microscopy and quantified. In the presence of LMB, both fusion proteins accumulated in the nucleus independently of the cell cycle (Figure 6B; our unpublished data), consistent with the possibility that HDHB contains a rev-type NES that functions through CRM1. However, it is also possible that HDHB may not be a direct cargo of CRM1 and that its export may be indirectly mediated through some other protein(s).
Figure 6.
Identification of a functional rev-type NES in SLD of HDHB. (A) Alignment of the putative NES in HDHB with those identified in other cell cycle-related proteins (Fabbro and Henderson, 2003). Superscripts above the amino acid sequence indicate residue numbers. Letters in red are the conserved aliphatic residues in the NES. Two pairs of residues in the putative NES in HDHB were mutated to alanine as indicated by the thin arrows to create Mut1 and Mut2. (B) GFP- and FLAG-tagged HDHB were transiently expressed in asynchronously growing U2OS cells with (+) or without (-) LMB to inhibit CRM1-mediated nuclear export. The subcellular localization of GFP-HDHB and FLAG-HDHB in asynchronous, G1, and S phase cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that sample. (C) GFP-HDHB and GFP-βGal-PSLD carrying the Mut1 or Mut2 mutations, or the corresponding proteins without the mutations, were transiently expressed in asynchronous U2OS cells and visualized by fluorescence microscopy. Cells showing the most frequently observed fluorescence pattern are shown. Nuclei were stained with Hoechst dye. Bar, 10 μm. (D) The subcellular localization of wild-type and mutant GFP-HDHB and GFP-βGal-PSLD in asynchronous U2OS cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that sample.
To assess whether the putative NES in HDHB was functional, we mutated Val/Leu and Leu/Leu of the NES motif to alanine to create NES mutants 1 and 2 (Figure 6A). GFP-HDHB and GFP-βGal-PSLD harboring these NES mutations were transiently expressed in either asynchronous or synchronized U2OS cells. Both NES mutant fusion proteins accumulated in the nucleus in >80% of cells, no matter when they were expressed in asynchronous or synchronized cells (Figure 6, C and D; our unpublished data). The focal pattern of nuclear GFP-HDHB was somewhat more diffuse with the NES mutants than with wild-type GFP-HDHB (compare Figure 6C with 1B). The results indicate that the NES mutations specifically impaired the export of both GFP-HDHB and GFP-βGal-PSLD, arguing that the PSLD region of HDHB contains a functional NES.
FLAG-HDHB Is Phosphorylated in a Cell Cycle-dependent Manner In Vivo
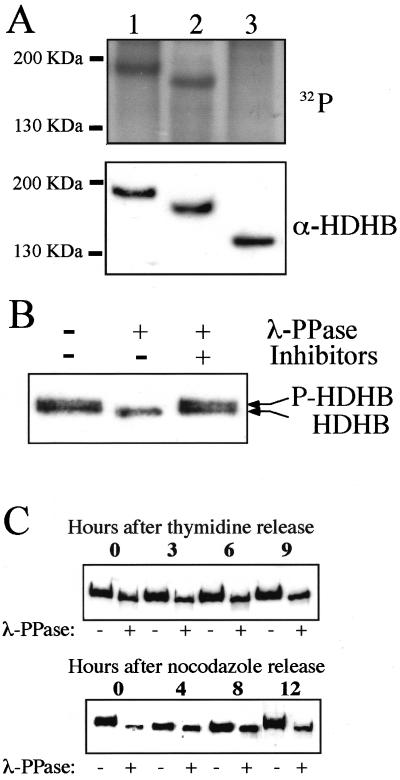
The cluster of potential CDK phosphorylation sites in the PSLD domain of HDHB (Figure 4A) suggested that phosphorylation of HDHB might regulate its subcellular localization in the cell cycle. If so, one would expect the PSLD region of HDHB to be phosphorylated in a cell cycle-dependent manner. To first test whether HDHB undergoes phosphorylation in PSLD, U2OS cells were transiently transfected with expression plasmids for wild-type and C-terminally truncated forms of FLAG-HDHB, radiolabeled with phosphate, and then FLAG-HDHB was immunoprecipitated from cell extracts. Immunoprecipitated proteins were analyzed by denaturing gel electrophoresis, immunoblotting, and autoradiography (Figure 7A). A radiolabeled band of FLAG-HDHB was detected at the same position as the immunoreactive HDHB band (Figure 7A, lanes 1). Truncated FLAG-HDHB lacking SLD also was robustly phosphorylated in vivo (lanes 2), whereas truncated FLAG-HDHB (1-874) lacking PSLD was not significantly phosphorylated (lanes 3). These results demonstrate that SLD is not required for HDHB phosphorylation, whereas PSLD is required, and suggest that the phosphorylation sites probably reside in PSLD.
Figure 7.
Cell cycle-dependent phosphorylation of FLAG-HDHB in vivo. (A) U2OS cells transiently expressing FLAG-HDHB (lane 1) and its truncation mutants 1-1039 (lane 2) and 1-874 (lane 3) were labeled with [32P]orthophosphate. Cell extracts were immunoprecipitated with anti-FLAG resin. The precipitated proteins were separated by 7.5% SDS-PAGE, transferred to a PVDF membrane, and detected by autoradiography (top) or Western blotting (bottom). The positions of marker proteins of known molecular mass are indicated at the left. (B) FLAG-HDHB expressed in U2OS cells was immunoprecipitated with anti-FLAG resin, incubated with (+) or without (-) λ-PPase in the presence (+) or absence (-) of phosphatase inhibitors, as indicated, and analyzed by SDS-PAGE and immunoblotting with anti-HDHB antibody. (C) U2OS cells expressing FLAG-HDHB were arrested at G1/S (top) or at G2/M (bottom), and then released from the block. FLAG-HDHB was harvested at the indicated time points, immunoprecipitated with anti-FLAG resin, treated with (+) or without (-) λ-PPase, and analyzed as in B.
To examine the timing of HDHB phosphorylation in the cell cycle, it would be convenient to detect phosphorylation without the use of radiolabeling. Because phosphorylation often reduces the electrophoretic mobility of a protein in denaturing gels, we immunoprecipitated transiently expressed FLAG-HDHB and examined its mobility before and after treatment with λ-phosphatase (λ-PPase) (Figure 7B). Without λ-PPase treatment, FLAG-HDHB was detected in Western blots in two very closely migrating bands (lane 1), whereas dephosphorylated FLAG-HDHB migrated as a single band at the mobility of the faster band of the doublet (lane 2). When λ-PPase inhibitors were present in the reaction, FLAG-HDHB migrated as a doublet identical to the mock-treated protein (lane 3). These data suggest that the electrophoretic mobility of FLAG-HDHB was reduced by phosphorylation and that this assay may be suitable to track HDHB phosphorylation in the cell cycle.
To determine whether HDHB is phosphorylated in a cell cycle-dependent manner, U2OS cells transiently expressing FLAG-HDHB were arrested in G1/S by adding thymidine to the medium or in G2/M by adding nocodazole to the medium. The cells were released from the blocks for different time periods, and FLAG-HDHB was immunoprecipitated from cell extracts. The immunoprecipitated material was incubated with or without λ-PPase and then analyzed by denaturing gel electrophoresis and Western blotting (Figure 7C). The mobility of FLAG-HDHB from cells arrested at G1/S was increased by λ-PPase treatment, suggesting that the protein was phosphorylated at G1/S (Figure 7C, top). The mobility shift after phosphatase treatment of FLAG-HDHB was maintained after release from the G1/S block as the cells progressed into G2 (Figure 7C, top). Cells arrested at G2/M also displayed a strong mobility shift after phosphatase treatment (Figure 7C, bottom). However, after the cells were released into G1 for 4 and 8 h, FLAG-HDHB migrated as a single band that was much less affected by phosphatase treatment (Figure 7C, bottom). By 12 h after release from the G2/M block, when most of the cells were entering S phase (our unpublished data), the mobility of FLAG-HDHB was again increased by phosphatase treatment, restoring the pattern observed in nocodazole-arrested cells (Figure 7C, bottom). These results strongly suggest that phosphorylation of FLAG-HDHB is cell cycle dependent, with maximal phosphorylation from G1/S through G2/M and minimal phosphorylation during G1.
Serine 967 Is the Major Phosphorylation Site of Ectopically Expressed HDHB
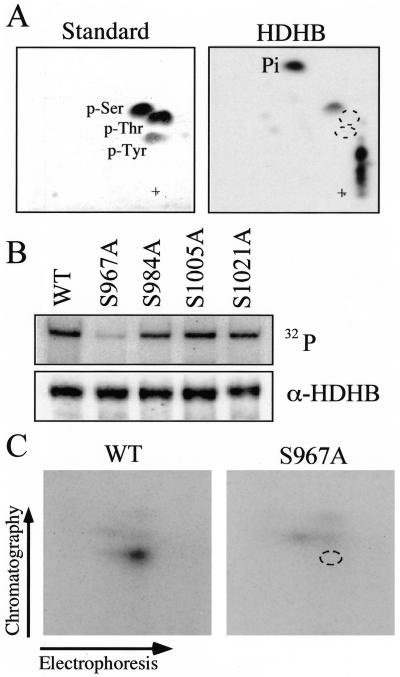
To map the phosphorylation sites in FLAG-HDHB, we first wished to determine what amino acid residues were modified. Phosphoamino acid analysis of in vivo-radiolabeled FLAG-HDHB revealed that phosphoserine(s) was the major phosphoamino acid of FLAG-HDHB in vivo (Figure 8A). Assuming that the cell cycle-dependent phosphorylation sites of HDHB are located in PSLD between residues 874 and 1039 (Figure 7A), that these sites are modified by CDKs, and that phosphoserine is the major amino acid modified (Figure 8A), only four of the seven potential CDK sites would remain as candidate sites. To test each of these sites individually, FLAG-HDHB expression plasmids with the corresponding serine-to-alanine mutations were constructed. Cells transiently transfected with these plasmids were radiolabeled with orthophosphate in vivo and FLAG-HDHB was immunoprecipitated and analyzed by autoradiography and Western blotting (Figure 8B). The results showed that FLAG-HDHB and three of the mutant proteins were phosphorylated approximately equally, whereas the S967A mutant protein was only weakly phosphorylated (Figure 8B). This result suggested that S967 might be the primary site of HDHB phosphorylation in vivo. Consistent with this interpretation, an electrophoretic mobility shift after phosphatase treatment of immunoprecipitated FLAG-HDHB was detected with three of the mutant proteins, but not with S967A protein (our unpublished data).
Figure 8.
Identification of a major in vivo phosphorylation site in HDHB. (A) Phosphoamino acid markers (left) and phosphoamino acids from in vivo 32P-labeled FLAG-HDHB (right) were separated in two dimensions and visualized by autoradiography. Some incompletely hydrolyzed phosphopeptides remained near the origin (+). (B) Wild-type and mutant FLAG-HDHB proteins were radiolabeled with orthophosphate in vivo, immunoprecipitated, separated by SDS-PAGE, and analyzed by autoradiography (top) and immunoblotting with anti-HDHB (bottom). (C) Tryptic phosphopeptides of 32P-labeled wild-type and S967A mutant FLAG-HDHB were separated in two dimensions and visualized by autoradiography.
To confirm that S967 was the major phosphorylation site in HDHB in vivo, tryptic phosphopeptide mapping was carried out with wild-type and S967A mutant FLAG-HDHB that had been metabolically radiolabeled with orthophosphate (Figure 8C). One predominant radiolabeled peptide and a weakly labeled peptide were observed with the wild-type protein (Figure 8C, left). The predominant phosphopeptide was absent in the S967A protein, but the weakly labeled peptide remained detectable (Figure 8C, right). The results provide additional evidence that serine 967 is a prominent phosphorylation site in HDHB in vivo.
Identification of Cyclin E/CDK2 as a Kinase That Potentially Modifies HDHB in G1/S
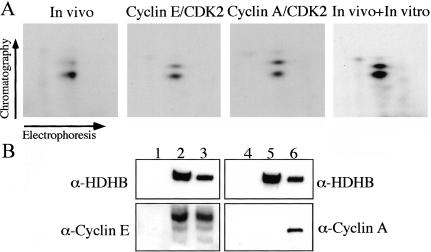
To test whether CDKs can actually modify HDHB, as suggested by the timing of HDHB phosphorylation in the cell cycle and the identification of S967 as a primary site of modification, we incubated purified cyclin E/CDK2 or cyclin A/CDK2 with purified recombinant HDHB and radiolabeled ATP in vitro. After the kinase reactions, the proteins were separated by denaturing gel electrophoresis, transferred to a PVDF membrane, and detected by autoradiography. The results revealed that recombinant HDHB could be phosphorylated strongly by both cyclin E/CDK2 and cyclin A/CDK2 (our unpublished data). The radiolabeled HDHB bands were then further processed for tryptic phosphopeptide mapping. Peptides from each digestion were separated in two dimensions, either individually or after mixing with tryptic peptides from in vivo phosphorylated FLAG-HDHB, and visualized by autoradiography (Figure 9A). HDHB peptides phosphorylated by cyclin E/CDK2 and cyclin A/CDK2 yielded patterns essentially identical to those observed in the in vivo labeled peptide map, with one major spot and one minor spot (Figure 9A). When the in vitro and in vivo labeled peptides were mixed and separated on one chromatogram, they comigrated (Figure 9A, right). These data argue that the major phosphopeptides modified in vitro by cyclin E/CDK2 and cyclin A/CDK2 in purified recombinant HDHB were the same ones modified in vivo in FLAG-HDHB.
Figure 9.
Identification of cyclin E/CDK2 as the potential G1/S kinase of HDHB S967. (A) Tryptic phosphopeptides from FLAG-HDHB phosphorylated in vivo as in Figure 7C, or recombinant HDHB phosphorylated in vitro by purified cyclin E/CDK2 or cyclin A/CDK2, were separated in two dimensions, either individually or as a mixture, and visualized by autoradiography. (B) Proteins that coimmunoprecipitated with FLAG vector (lanes 1 and 4) or FLAG-HDHB (lanes 2 and 5) expressed in U2OS cells were analyzed by immunoblotting with antibodies against HDHB (lanes 1-6), cyclin E (lanes 1-3), or cyclin A (lanes 4-6). One-tenth of the cell lysate used for immunoprecipitation was analyzed in parallel as a positive control (lanes 3 and 6).
Because cyclin E activity in human cells rises in late G1, whereas cyclin A activity rises later coincident with the onset of S phase (Pines, 1999; Erlandsson et al., 2000), it was important to try to distinguish whether one of these kinases might preferentially modify HDHB. Cyclin subunits frequently form a complex with the substrate proteins that they target for phosphorylation (Endicott et al., 1999; Takeda et al., 2001). To test whether cyclin E or cyclin A could associate with HDHB, FLAG-HDHB and associated proteins were immunoprecipitated from extracts of cells transfected with either FLAG-HDHB expression vector or empty FLAG-vector as a control. The cell extracts and the immunoprecipitated material were analyzed by Western blotting (Figure 9B). Cyclin E clearly coprecipitated with FLAG-HDHB, but cyclin A did not (Figure 9B, lanes 2 and 5), suggesting that FLAG-HDHB may interact preferentially with cyclin E in vivo. It is conceivable that this interaction may be required for phosphorylation of HDHB by cyclin E/CDK2 in vivo, and if so, mutations in HDHB that prevent its association with cyclin E would abrogate phosphorylation by cyclin E/CDK2. To test the possibility that the FLAG-HDHB mutant S967A was not phosphorylated in vivo (Figure 8, B and C) due to an inability to bind to cyclin E, FLAG-HDHB-S967A and associated proteins were immunoprecipitated from extracts of transfected cells and analyzed by Western blotting. Coprecipitation of cyclin E with the mutant protein was as robust as with wild-type FLAG-HDHB (our unpublished data).
Phosphorylation of Serine 967 Is Critical for Regulation of HDHB Localization
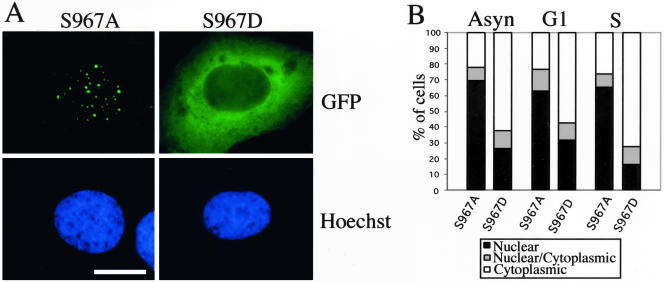
The data mentioned above indicate that subcellular localization and phosphorylation of ectopically expressed HDHB were regulated in a cell cycle-dependent manner with maximal phosphorylation of HDHB from G1/S to G2/M, coinciding with the period when HDHB accumulated in the cytoplasm. These results, together with the identification of S967 as the major in vivo phosphorylation site in HDHB, suggest that phosphorylation of S967 may regulate the subcellular localization of HDHB. To test this idea, expression plasmids for wild-type GFP-HDHB and the mutants S967A, S984A, S1005A, and S1021A were microinjected into synchronized U2OS cells. Wild-type GFP-HDHB accumulated in nuclear foci of cells in G1, but in the cytoplasm of cells in S phase as expected (our unpublished data). However, regardless of cell cycle timing, GFP-HDHB-S967A localized in nuclear foci in ∼70% of the fluorescent cells (Figure 10, A and B). The other three substitution mutants localized in either the nucleus or the cytoplasm like wild-type GFP-HDHB (our unpublished data). In an attempt to mimic the phosphorylation of S967, we mutated serine 967 to aspartic acid, expressed GFP-HDHB-S967D in asynchronous and synchronized U2OS cells, and examined the subcellular distribution of the mutant fusion protein. About 60% of the cells expressing GFP-HDHB-S967D displayed cytoplasmic fluorescence in asynchronous, G1 phase, and S phase cells (Figure 10, A and B), demonstrating that the S967D mutation mimicked phosphorylated S967. The data strongly suggest that phosphorylation of serine 967 is critical in regulating the subcellular localization of HDHB.
Figure 10.
Subcellular localization of HDHB is regulated by phosphorylation of S967. (A) GFP fluorescence in U2OS cells transiently expressing GFP-HDHB with the S967A or S967D mutation was examined by fluorescence microscopy. Nuclei were stained with Hoechst dye. Bar, 10 μm. (B) Subcellular localization of GFP-HDHB S967A and S967D expressed in asynchronous, G1, and S phase U2OS cells was quantified.
DISCUSSION
HDHB Resides in Nuclear Foci Inducible by DNA Damage
During G1 of the cell cycle, wild-type GFP-HDHB resides in prominent nuclear foci that are associated with detergent-insoluble nuclear structures (Figures 1, 2, 3). The topoisomerase inhibitors etoposide and camptothecin induced many more HDHB nuclear foci, suggesting that the helicase activity may be recruited to process sites of DNA damage (Figure 2). Many proteins involved in DNA double strand break repair and recombination show a similar focal relocalization in response to DNA damage at different times in the cell cycle, including Rad52, RPA, Mre11, Ddc1 Ddc2, Rad9, Rad24, Rad51, Rad53, Rad54, and Rad55 (Lisby et al., 2003). In support of this notion, one of the helicase-defective point mutants (MutA) did not associate with detergent-insoluble foci (Figure 1), indicating that focus formation depended on the biochemical activity of HDHB. Also consistent with this interpretation, the PSLD peptide of HDHB, which lacks the helicase domain (Figure 4A), directed a βGal reporter to the nucleus during G1, but it did not produce the focal localization pattern (Figure 5). Intriguingly, MutB also localized in a focal pattern in a detergent-resistant manner, indicating that the ability of HDHB to bind to single-stranded DNA in the presence of ATP may be sufficient for the focal localization. These results suggest that the main function of HDHB may be to process endogenous DNA damage, possibly caused by incomplete topoisomerase reactions, during G1 phase of the cell cycle. This interpretation provides a simple explanation for the ability of helicase-defective HDHB mutants to block G1/S progression when the mutant protein is injected into cells in early G1 (Taneja et al., 2002). The MutB form of HDHB would associate with the damage, but it could not process it properly, interfering with the activity of the endogenous helicase and leading to cell cycle arrest in late G1. Further work will be required to explore the proposed role of HDHB in DNA damage processing.
Two major DNA damage repair pathways in eukaryotes are error-free homologous recombination and error-prone nonhomologous end-joining (NHEJ) (Khanna and Jackson, 2001; West, 2003). Homologous recombination events are somehow suppressed during G1, whereas NHEJ pathways are favored during G0, G1, and early S phase (Takata et al., 1998; Adachi et al., 2001; Karanjawala et al., 2003). Srs2, a member of helicase superfamily 1 like HDHB, has been shown to inhibit excessive homologous recombination in yeast and also may play a role in NHEJ (Hegde and Klein, 2000, and references therein). Srs2 can disrupt Rad51 nucleoprotein filaments in vitro (Krejci et al., 2003; Veaute et al., 2003), suggesting a mechanism by which Srs2 could conceivably suppress homologous recombination. If HDHB resembled Srs2 in interacting with human Rad51 nucleoprotein filaments to disrupt them, one might expect to observe Rad51 colocalization with HDHB in nuclear foci. However, Rad51 nuclear foci were observed only after the G1/S transition (Yuan et al., 2003; Gu, Podust, and Fanning, unpublished data), arguing that the nuclear foci of GFP-HDHB in G1 cells have some other function. However, the small fraction of endogenous HDHB present in the nucleus during S phase could conceivably interfere with homologous recombination.
Another possible function considered for HDHB was DNA replication (Taneja et al., 2002). However, the predominantly cytoplasmic localization of HDHB during S and G2/M argues that HDHB is probably not directly involved in genomic DNA replication (Figure 3). Consistent with this idea, the helicase-defective HDHB did not affect DNA synthesis when injected into the nucleus of cells in late G1 or S phase (Taneja et al., 2002), probably because the injected protein was phosphorylated by CDK2 associated with cyclins E or A and rapidly targeted to the cytoplasm. Indeed, export to the cytoplasm may serve as a means to sequester HDHB when its function is not needed or might be detrimental in the nucleus. HDHB activity is not affected by CDK phosphorylation in vitro (Taneja and Fanning, unpublished data), and prolonged overexpression of helicase-proficient HDHB mutants that cannot be exported prevents the G1/S transition and eventually results in apoptosis (Gu and Fanning, unpublished data).
A C-Terminal Domain of HDHB Confers Cell Cycle-dependent Localization
A 131-residue domain, PSLD, is sufficient to target HDHB or a βGal reporter to either the nucleus or the cytoplasm in a cell cycle-dependent manner (Figure 5). A rev-type NES resides in this domain (Figure 6), but its activity or accessibility to the nuclear export machinery depends on phosphorylation of PSLD, primarily on serine 967, at the G1/S transition (Figures 7, 8, 9, 10). S967 is a perfect match to the consensus CDK substrate recognition motif (S/T)PX(K/R). Both cyclin E/CDK2 and cyclin A/CDK2 can modify HDHB in vitro, but the ability of cyclin E/CDK2 to complex with HDHB in cell extracts suggests that it may be the initial kinase that modifies HDHB at the G1/S transition (Figure 9). Phosphorylation of HDHB in PSLD seems to persist through the latter part of the cell cycle, correlating well with the predominantly cytoplasmic localization of HDHB. However, we cannot yet distinguish whether HDHB undergoes dephosphorylation in early G1 (Figure 7C) or is perhaps targeted for proteolysis and rapidly resynthesized in early G1, when it would enter the nucleus in a hypophosphorylated form.
Another open question is the mechanism by which phosphorylation of PSLD regulates nuclear export of HDHB. Our data provide strong evidence that PSLD contains active targeting signals that are independent of protein context (Figure 4-5). Because mutant HDHB with an inactivated NES is nuclear even when it is expressed during S phase and thus presumably phosphorylated (Figure 6), we suggest that the NLS is probably not inactivated or masked by phosphorylation and that the primary target of CDK regulation is the NES. Extending this reasoning, we speculate that the NES may be masked during G1 when the CDK motifs in PSLD are unmodified and that the NES is liberated when S967 becomes phosphorylated, leading to NES recognition by nuclear export factors (Figures 5 and 6). How might phosphorylation of PSLD liberate the NES? Structural studies of a rev-type NES have shown that it forms an amphipathic α-helix, with the leucines aligned on one side of the helix and charged residues on the other side (Rittinger et al., 1999). Because the SLD of HDHB contains both the rev-type NES and an NLS, and the basic residues likely to serve as the NLS are interspersed through the NES, the NES and NLS may reside on opposite faces of an amphipathic helix. Additional sequences in PSLD would mask the NES intramolecularly, allowing only the NLS to be recognized. Phosphorylation of S967 would alter the conformation of the mask in PSLD to expose the NES, without affecting exposure of the NLS. Detailed studies of PSLD in the future will be needed to test this speculative model.
Acknowledgments
We thank Drs. R. Ott, C. Voitenleitner, G. Grosveld, B. Henderson, and M. Yoshida for reagents, and members of the Fanning laboratory for constructive discussions. We gratefully acknowledge support from the National Institutes of Health grant GM-52948 to E.F., grant CA-55724 to A.B.R., and the Vanderbilt-Ingram Cancer Center through the Cancer Center Support grant CA-69485.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0227. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0227.
Abbreviations used: βGal, β-galactosidase; λ-PPase, λ-phosphatase; CDK, cyclin dependent kinase; CRM1, chromosome region maintenance 1; GFP, green fluorescent protein; HDHB, human DNA helicase B; LMB, leptomycin B; Mut, mutant; NES, nuclear export signal; NHEJ, non-homologous end-joining; NLS, nuclear localization signal; PVDF, polyvinylidene difluoride; PSLD, phosphorylation and subcellular localization domain; SLD, subcellular localization domain.
References
- Adachi, N., Ishino, T., Ishii, Y., Takeda, S., and Koyama, H. (2001). DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku 70, Implications for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 98, 12109-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Cantor, S.B., et al. (2001). BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 10, 149-160. [DOI] [PubMed] [Google Scholar]
- Chedin, F., and Kowalczykowski, S.C. (2002). A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43, 823-834. [DOI] [PubMed] [Google Scholar]
- Dillingham, M.S., Spies, M., and Kowalczykowski, S.C. (2003). RecBCD enzyme is a bipolar DNA helicase. Nature 423, 893-897. [DOI] [PubMed] [Google Scholar]
- Endicott, J.A., Noble, M.E.M., and Tucker, J.A. (1999). Cyclin-dependent kinases: inhibition and substrate recognition. Curr. Opin. Struct. Biol. 9, 738-744. [DOI] [PubMed] [Google Scholar]
- Erlandsson, F., Linnman, C., Ekholm, S., Bengtsson, E., and Zetterberg, A. (2000). A detailed analysis of cyclin A accumulation at the G1/S border in normal and transformed cells. Exp. Cell. Res. 259, 86-95. [DOI] [PubMed] [Google Scholar]
- Fabbro, M., and Henderson, B.R. (2003). Regulation of tumor suppressors by nucleo-cytoplasmic shuttling. Exp. Cell Res. 282, 59-69. [DOI] [PubMed] [Google Scholar]
- Friedberg, E.C. (2001). How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1, 22-33. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607-660. [DOI] [PubMed] [Google Scholar]
- Harlow, E. and Lane, D. P. (1988). Antibodies: A Laboratory Manual, Cold Spring, NY: Cold Spring Harbor Laboratory Press.
- Hegde, V., and Klein, H. (2000). Requirement for the SRS2 DNA helicase gene in non-homologous end-joining in yeast. Nucleic Acids Res. 14, 2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig, U., Marlar, C.A., and Fanning, E. (1999). The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell 10, 2631-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson, I.D. (2003). RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3, 169-178. [DOI] [PubMed] [Google Scholar]
- Hood, J.K., and Silver, P.A. (2000). Diverse nuclear transport pathways regulate cell proliferation and oncogenesis. Biochim. Biophys. Acta 1471, M31-M41. [DOI] [PubMed] [Google Scholar]
- Kalderon, D., Roberts, B.L., Richardson, W.D., and Smith, A.E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39, 499-509. [DOI] [PubMed] [Google Scholar]
- Karanjawala, Z. E., Adachi, N., Irvine, R.A., Oh, E.K., Shibata, D., Schwarz, K., Hsieh, C.L., and Lieber, M.R. (2003). The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implication for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair 1, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Khanna, K.K., and Jackson, S.P. (2001). DNA double-strand breaks: signaling, repair, and the cancer connection. Nat. Genet. 27, 247-254. [DOI] [PubMed] [Google Scholar]
- Krejci, L., Van Komen, S., Li, Y., Villemain, J., Reddy, M.S., Klein, H., Ellenberger, T., and Sung, P. (2003). DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305-309. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E.P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540-547. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Mortensen, U.H., and Rothstein, R. (2003). Colocalization of mutiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5, 572-577. [DOI] [PubMed] [Google Scholar]
- Matsumoto, K., Seki, M., Masutani, C., Tada, S., Enomoto, T., and Ishimi, Y. (1995). Stimulation of DNA synthesis by mouse DNA helicase B in a DNA replication system containing eukaryotic replication origins. Biochemistry 34, 7913-7922. [DOI] [PubMed] [Google Scholar]
- Nelms, B.E., Maser, R.S., MacKay, J.F., Lagally, M.G., and Petrini, J.H.J. (1998). In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280, 590-592. [DOI] [PubMed] [Google Scholar]
- Pines, J. (1999). Four-dimensional control of the cell cycle. Nat. Cell Biol. 1, E73-E79. [DOI] [PubMed] [Google Scholar]
- Rittinger, K., Budman, J., Xu, J., Volinia, S., Cantley, L.C., Smerdon, S.J., Gamblin, S.J., and Yaffe, M.B. (1999). Structural analysis of 14–3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14–3-3 in ligand binding. Mol. Cell 4, 153-166. [DOI] [PubMed] [Google Scholar]
- Saitoh, A., Tada, S., Katada, T., and Enomoto, T. (1995). Stimulation of mouse DNA primase-catalyzed oligoribonucleotide synthesis by mouse DNA helicase B. Nucleic Acids Res. 23, 2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Kohda, T., Yano, T., Tada, S., Yanagisawa, J., Eki, T., Ui, M., and Enomoto, T. (1995). Characterization of DNA synthesis and DNA-dependent ATPase activity at a restrictive temperature in temperature-sensitive tsFT848 cells with thermolabile DNA helicase B. Mol. Cell. Biol. 15, 165-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, S., Kobayashi, T., Omori, A., Kusa, Y., Okumura, N., Kodaira, H., Ishimi, Y., Seki, M., and Enomoto, T. (2001). Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells. Nucleic Acids Res. 29, 3835-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata, M., Sasaki, M.S., Sonoda, E., Morrison, C., Hashimoto, M., Utsumi, H., Yamaguchi-Iwai, Y., Shinohara, A., and Takeda, S. (1998). Homologous recombination and non-homologous end-joining pathways of D.N.A. double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, D.Y., Wohlschlegel, J.A., and Dutta, A. (2001). A bipartite substrate recognition motif for cyclin-dependent kinases. J. Biol. Chem. 276, 1993-1997. [DOI] [PubMed] [Google Scholar]
- Taneja, P., Gu, J., Peng, R., Carrick, R., Uchiumi, F., Ott, R.D., Gustafson, E., Podust, V.N., and Fanning, E. (2002). A dominant-negative mutant of human DNA helicase B blocks the onset of chromosomal DNA replication. J. Biol. Chem. 277, 40853-40861. [DOI] [PubMed] [Google Scholar]
- Taylor, A.F., and Smith, G.R. (2003). RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423, 889-893. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., Bree, R.T., and Lowndes, N.F. (2003). The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4, 844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute, X., Jeusset, J., Soustelle, C., Kowalczykowski, S.C., Le Cam, E., and Fabre, F. (2003). The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309-312. [DOI] [PubMed] [Google Scholar]
- Voitenleitner, C., Rehfuess, C., Hilmes, M., O'Rear, L., Liao, P.C., Gage, D.A., Ott, R., Nasheuer, H.P., and Fanning, E. (1999). Cell cycle-dependent regulation of human DNA polymerase alpha-primase activity by phosphorylation. Mol. Cell. Biol. 19, 646-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K. (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441-451. [DOI] [PubMed] [Google Scholar]
- West, S.C. (2003). Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell. Biol. 4, 1-11. [DOI] [PubMed] [Google Scholar]
- Wolff, B., Sangier, J.J., and Wang, Y. (1997). Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of human immunodeficiency virus type-1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4, 139-147. [DOI] [PubMed] [Google Scholar]
- Xia, X., Mariner, D.J., and Reynolds, A.B. (2003). Adhesion-associated and PKC-modulated changes in serine/threonine phosphorylation of p120-catenin. Biochem. 42, 9195-9204. [DOI] [PubMed] [Google Scholar]
- Yuan, S.F., Chang, H., and Lee, E.Y. (2003). Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM-/- and cAbl-/- cells. Mutat. Res. 525, 85-92. [DOI] [PubMed] [Google Scholar]