Abstract
Accumulating evidence has shown that microRNAs (miRNAs) deregulation is commonly observed in human malignancies and crucial to cancer metastasis. Herein, we demonstrated that miR-126 play a suppressor role in human breast cancer cells invasion through the direct repression of a disintegrin and metalloprotease 9 (ADAM9). MiR-126 expression was investigated in forty cases of breast cancer specimens by real-time PCR. Transwell assay was conducted to explore the effects of miR-126 on the invasion of human breast cancer cell lines. The impact of miR-126 overexpression on putative target ADAM9 was subsequently confirmed by Western blot analysis. Our results indicated that miR-126 expression was frequently down-regulated in breast cancer specimens compared with adjacent normal tissues (P<0.05). Overexpression of miR-126 significantly reduced (P<0.05) the protein levels of ADAM9, further suppressed (P<0.05) breast cancer cell invasion in vitro. Meanwhile, knockdown of ADAM9 by small interfering RNA (siRNA) also inhibited (P<0.05) breast cancer cell invasion. Thus, our study revealed that miR-126 may act as a tumor suppressor via inhibition of cell invasion by downregulating ADAM9 in breast cancer development.
Keywords: MiR-126, breast cancer, invasion, ADAM9
Introduction
Breast cancer is a major cause of cancer-related death in women worldwide [1,2]. Over the past years, more than one million cases are reported annually, with almost half of cases in developed countries [3]. The breast cancer development is characterized by aggressive local invasion, early metastasis, and multidrug resistance to chemotherapy [4-6]. Therefore, a better understanding of the molecular mechanism underlying its growth and early metastasis are particularly important.
MicroRNAs (miRNAs) are small noncoding RNAs of approximately 21-25 nucleotides that regulate the translation of mRNAs to proteins [7]. Mature miRNAs play important regulatory roles in cell growth, proliferation, differentiation and cell death. As regulators of gene expression, deregulation and upregulation of miRNAs has been linked to tumor progression, and could be function as tumor suppressors and oncogenes during the tumor development [8,9]. In breast cancer, distinct expression profiles of miRNAs have been widely studied and demonstrated association with specific molecular subtypes and clinicopathological characteristics [10,11], including miR-126 [12]. However, the biological functions of the deregulated miRNAs in tumor progression, especially for tumor metastasis have not yet been completely defined.
Cancer metastasis is a complex process involving the interplay between cancer cells and surrounding components of extracellular matrix [13]. A disintegrin and metalloproteases (ADAM) are modular type I transmembrane proteins with a number of characteristic domains, including a metalloprotease, disintegrin-like and transmembrane domain [14]. Recent studies have shown a close correlation between increased ADAMs and cancer progression [15]. Till now, twenty-one members of ADAM family have been identified in humans. Recently ADAM9 has been reported on the upregulation in various types of cancers, including colon [16], pancreas [17], and breast [18], tightly associated with cancer progression and metastasis [16-18].
In the present study, we discovered that miR-126 was significantly downregulated in breast cancer tissues compared with adjacent normal tissues. Meanwhile, overexpression of miR-126 and knockdown of ADAM9 both inhibited the invasion of breast cancer cell lines. Based on these results, we postulate the role of the miR-126/ADAM9 axis in the invasion abilities of breast cancers.
Material and methods
Clinical breast cancer specimens
Twenty pairs of primary breast cancer and corresponding adjacent normal tissues were obtained from patients who underwent primary surgical treatment at the Affiliated Tumor Hospital of Zhengzhou University (Zhengzhou, China) from 2012 to 2014. All samples were histologically confirmed by staining with hematoxylin-eosin. The study protocol was approved by the Scientific and Ethical Committee of Zhengzhou University. The specimens were snap frozen in liquid nitrogen and stored at -80°C.
Cell lines and cell culture
Two human breast cancer cell lines (MCF-7 and MDB-MA-231) used in the present study were purchased from Cell Center of Shanghai Institute of Life Science, Chinese Academy of Science (Shanghai, China). All Cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (Gibco) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transfection
Transfection of miR-126 mimics or non-specific miR control (NC) (GenePharma, Shanghai, China) was performed using X-treme GENE Transfection Reagent (Roche, Indianapolis, IN) according to the manufacturer’s instructions. Briefly, cells were seeded into 6-well plates until 60% confluent and transiently transfected with 20 and 40 nM miR-126 mimics or NC. Then cells were incubated at 37°C for 48 hours prior to further analysis.
Transfection of ADAM9 specific siRNA and scrambled siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) was carried using Lipofectamine TM 2000 (Invitrogen Co., Carlsbad, CA) according to the manufacturer’s instructions. Cells with 60% confluence were cultured in RPMI-1640 medium without any antibiotics overnight at 37°C in 5% CO2 and then transfected with both ADAM9 and scrambled siRNAs. After 4-6 hours, RPMI-1640 containing 10% FBS was added to each well and the cells were cultured for an additional 48 hours.
RNA isolation and real-time RT-PCR
Total RNA was isolated using TRIzol® reagent (Invitrogen) according to the manufacturer’s protocol. Expression of miR-126 was assessed by quantitative real-time RT-PCR, and the small nuclear RNA U6 was used as an internal control. Total RNA was reverse transcribed to cDNA using a PrimeScript RT reagent kit (Takara, Dalian, China). Quantitative real-time PCR (qPCR) was performed using a SYBR Premix Ex Taq™ II kit (Takara) on an ABI7900HT System.
Cell proliferation assay
For the cell proliferation assay, approximately 3000 cells after transfection with miR-126 mimics or non-specific miR control were seeded in 96-well plates. After 48 hours incubation, cell proliferation rates were detected by MTS assay according to the manufacturer’s protocol. Wells containing known cell numbers (0, 1000, 2000, 5000, 10,000, 20,000 or 40,000 cells/well) were treated in the similar fashion to establish standard curves.
Cell invasion assay
Cell invasion was evaluated by using a 24-Multiwell BD Falcon FluoroBlok Insert System (8.0 μm pores; BD Biosciences, San Jose, CA). Approximately 5×104 cells after transfection were plated onto the upper compartment of chamber coated with Matrigel. The lower compartment was filled with complete RPMI-1640 medium. The chamber was incubated at 37°C for 16 hours and the filters were removed. Cells invaded on the bottom of inserts were fixed, stained, and counted under a microscope. The numbers of invaded cells were counted using the MetaMorph image analysis software (Molecular Devices, Sunnyvale, CA).
Western blot analysis
Cells after treatment were washed in ice-cold PBS and added to RIPA lysis and extraction buffer (Thermo Fisher Scientific, Franklin, MA). The protein samples (10-20 μg) were separated on 10% SDS-PAGE gels and electrically transferred to polyvinylidene difluoride membrane (Immobilon PVDF, Millipore Corp., Bedford, MA). The membranes were probed with the ADAM9 antibody (1:1000; Abcam, Cambridge, MA) and followed by reprobing with ACTIN (1:3000; Santa Cruz Biotechnology, San Diego, CA) as an endogenous loading control. Western blot studies were run in at least three independent experiments.
Statistical analysis
Statistical analyses were carried out using SPSS software (version 16, SPSS, Inc, Chicago, IL); All data were expressed as means ± SD. Between-group differences were analyzed using Student’s t-test. Western blot assays were tested using one-way ANOVA. P<0.05 was considered to be statistically significant.
Results
MiR-126 was frequently down-regulated in clinical specimens
We firstly examined miR-126 expression levels in 40 frozen samples from breast cancer patients (20 cancers and 20 adjacent normal controls) by TaqMan real time RT-PCR. Our results showed that miR-126 expression is significantly (P<0.05) reduced in breast cancer tissues in comparison with adjacent normal tissues (Figure 1). Meanwhile, we did not find any correlation of miR-126 expression with clinical characteristics of breast cancer (data not shown).
Figure 1.
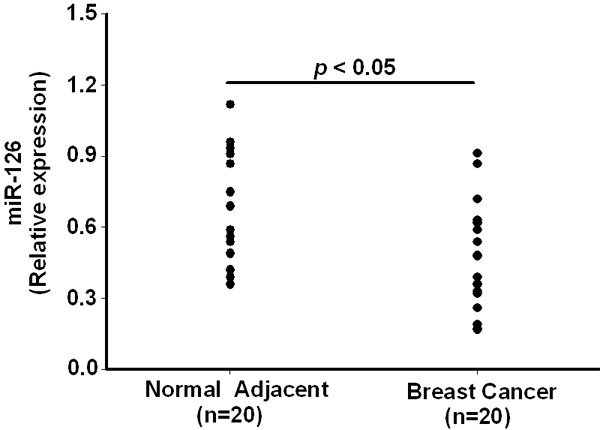
Expression of miR-126 in breast cancer and adjacent normal tissues. Decreased miR-126 expression in breast cancer tissues compared to the adjacent normal tissues from a panel of 20 breast cancer patients. Data are presented as means ± SEM.
Effect of miR-126 on breast cancer cell invasion and proliferation
To validate whether miR-126 regulated cancer cell invasion, we performed a trans-well assay by transfecting miR-126 mimics or NC into MCF-7 and MDA-MB-231 cells. As shown in Figure 2, the miR-126 levels were highly increased in these two breast cancer cell lines after transient transfection. As expected, the increased expression of miR-126 induced significant inhibition (P<0.05) of cell invasion in both MCF-7 and MDA-MB-231 cells (Figure 3A and 3B). However, we did not observe significant alteration in the cell proliferation after 48 hours incubation by transfecting miR-126 mimics or NC into these cells (Figure 3C).
Figure 2.
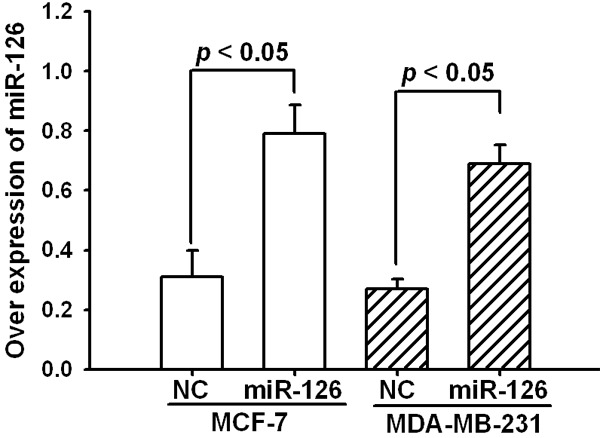
Overexpression of miR-126 by transfecting miR-126 mimics. The expression level of miR-126 was detected by real-time PCR after 48 hours transfection and normalized to that of U6. Data are presented as means ± SEM.
Figure 3.
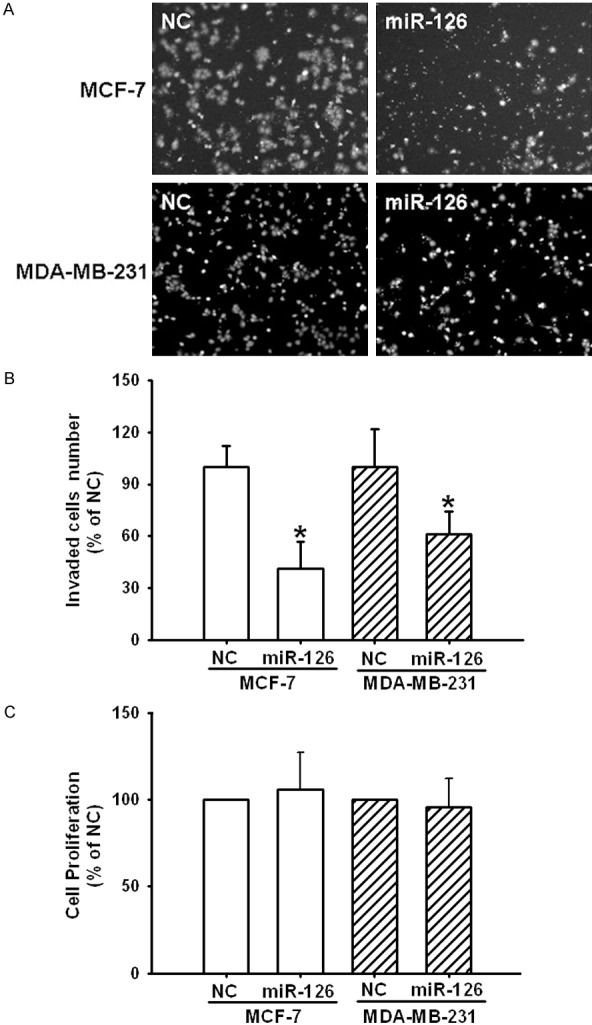
Overexpression of miR-126 decreases invasion but not proliferation in MCF-7 and MDB-MA-231 cell lines. A. Photographs depict the invasion of MCF-7 and MDB-MA-231. B. Quantified data are expressed as means ± SEM from 3 independent experiments. C. Cell proliferation was detected with MTS assay, and the graphs show the results of three independent experiments. *Different (P<0.05) from their controls, respectively.
Overexpression of miR-126 decreased ADAM9 protein level
To explore the mechanism of invasion inhibition induced by miR-126, we investigated whether miR-126 could regulate ADAM9 expression in MCF-7 and MDA-MB-231 cells. After transfected with miR-126 mimics at different concentrations (0, 20, 40 nM) and then examined ADAMP9 expression levels. As shown in Figure 4, overexpression of miR-126 directly induced a dose-dependent (P<0.05) decrease in ADAM9 protein level.
Figure 4.
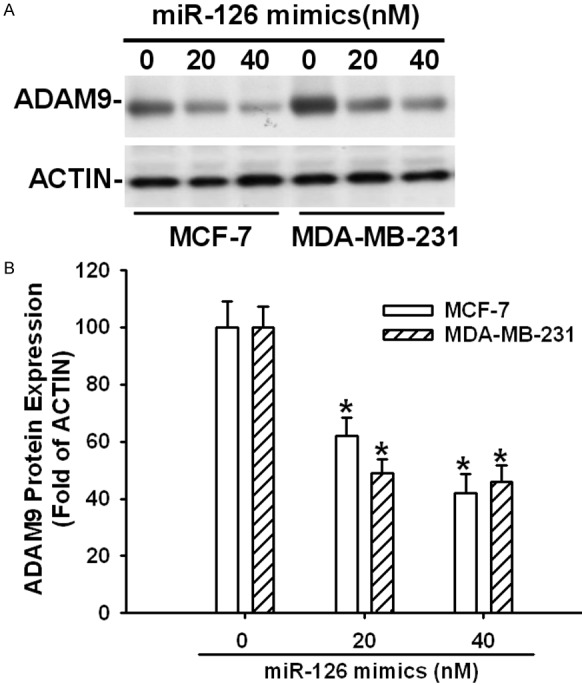
Overexpression of miR-126 decreased the protein levels of ADAM9 in MCF-7 and MDB-MA-231 cell lines. A. Representative Western blot analysis of ADAM9. B. Relative expression of ADAM9 was normalized to the endogenous control ACTIN. Data are presented as means ± SEM. *Different (P<0.05) from the control group.
Inhibition of ADAM9 protein levels by ADAM9 siRNA suppressed cell invasion
To determine whether miR-126 mimics reduced cell invasion through downregulation of ADAM9; we used ADAM9 siRNA to knockdown endogenous ADAM9 expression and investigated cell invasion ability in vitro. Western blot experiment demonstrated that the expression of ADAM9 in both MCF-7 and MDA-MB-231 were significantly reduced after siRNA transfection for 48 hours (Figure 5A). For invasion assay, the numbers of invaded cells were markedly decreased in ADAM9 siRNA treated group compared to Scrambled siRNA control group (P<0.05) (Figure 5B).
Figure 5.
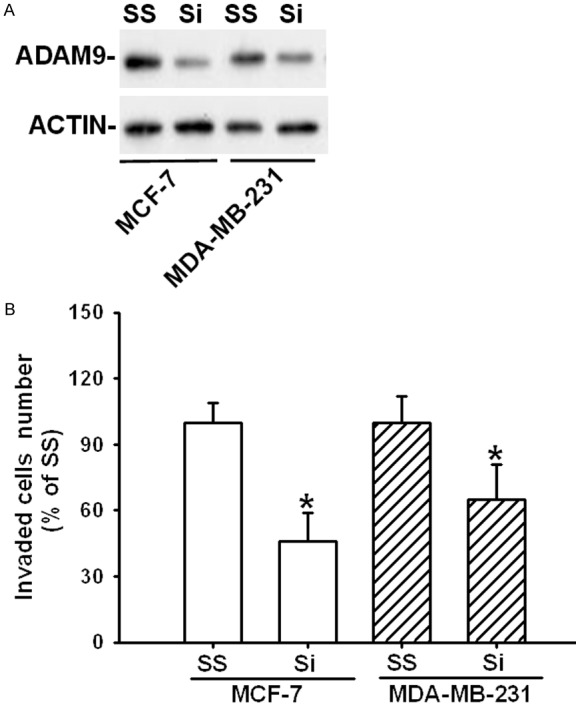
Effects of ADAM9 siRNA on cell invasion in MCF-7 and MDB-MA-231 cell lines. A. The protein levels of ADAM9 were detected by Western blot after transfected with ADAM9 specific siRNA in cells. B. Quantified data of cell invasion are expressed as means ± SEM. *Different (P<0.05) from scrambled control. Si: ADAM9 siRNA; SS: scrambled siRNA.
Discussion
In the present study, we observed a significant downregulation of miR-126 expression in human breast cancer tissues as compared with adjacent normal tissues. Overexpression of miR-126 inhibited the invasion of breast cancer cell lines in vitro. Moreover, we found that miR-126 directly targeted and down-regulated a key molecule involved in metastasis, ADAM9.
Aberrant expression of miRNAs has been deeply involved in couple of human diseases, including cancer [19,20]. It was reported that the miR-126 expression was significantly downregulated in different types of cancer [21-23]. In agreement with these reports, our current study found that miR-126 was frequently downregulated in breast cancer clinical specimens by real-time PCR detection. Unfortunately, we did not find significant correlation between miR-126 low expression and clinicopathological features (data not shown). These results also confirmed by another study that miR-126 expression was significantly downregulated in breast cancer tissues; however, in this study they found this low expression of miR-126 was associated with cancer metastasis [24].
To further reveal the functional roles of miR-126 in breast cancer metastasis, we ectopically raised the miR-126 level in two breast cancer cell lines to investigate its effect on cell invasion. Consistent with our expectation, increasing the expression levels of miR-126 directly inhibited cell invasion in both MCF-7 and MDA-MB-231 without alteration of cell proliferation, revealing its potential role as a suppressor of metastasis in breast cancer development. These findings were similar to the investigation by Tavazoie et al. that overexpression of miR-126 reduced both breast cancer cell growth in vitro and metastases in vivo. Furthermore, they included that loss of miR-126 was closely correlated with reduced metastasis-free survival [25]. Overall, these findings indicated that miR-126 could behave as a tumor suppressor in breast cancer growth and metastasis.
It is well known that miR-126 tightly involved in the different type of cancers development through regulation of various target mRNAs. Couple of studies has demonstrated a close relationship regarding the overexpression of miR-126 and ADAM9 silence in osteosarcoma, bladder cancer, and pancreatic cancer cells [26-28]. Meanwhile, we also observed that ADAM9 could be a potential target of miR-126 from miR databases (TargetScan and miRanda). Due to the strong involvement of ADAM9 in the metastatic process, more and more studies focused on the regulatory mechanism of ADAM9 in cancer metastasis [29,30]. Our current study demonstrated that the miR-126 suppression of ADAM9 is closely related to invasion of breast cancer cells. These findings were in agreement with several of recent reports that ADAM9 is involved in the negative regulation of breast cancer cell invasion [31,32]. Combining these findings, expression level of miR-126 may be inversely correlated with the metastatic potential of breast cancer, which may contribute to inhibition of breast cancer metastasis.
In summary, we proved that miR-126 was markedly downregulated in human breast cancer tissues. Increasing the expression of miR-126 may lead to breast cancer cell invasion suppression in vitro through targeting ADAM9. Though there is still much to learn about the role of miR-126 in breast cancer tumorigenesis, miR-126 provides us with a new metastatic suppressor and has potential as a new therapeutic target in metastatic breast cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Matsen CB, Neumayer LA. Breast cancer: a review for the general surgeon. JAMA Surg. 2013;148:971–980. doi: 10.1001/jamasurg.2013.3393. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Cyr AE, Margenthaler JA. Molecular profiling of breast cancer. Surg Oncol Clin N Am. 2014;23:451–62. doi: 10.1016/j.soc.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Gradishar WJ. Treatment of metastatic breast cancer. J Natl Compr Canc Netw. 2014;12:759–61. doi: 10.6004/jnccn.2014.0184. [DOI] [PubMed] [Google Scholar]
- 6.Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: current perspectives. Breast Cancer (Dove Med Press) 2014;6:1–13. doi: 10.2147/BCTT.S37638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 8.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–7. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 10.Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477–87. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 11.Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C, Junkermann H, Schneeweiss A, Burwinkel B. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS One. 2013;8:e76729. doi: 10.1371/journal.pone.0076729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y. Endothelial specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–64. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 13.Goubran HA, Kotb RR, Stakiw J, Emara ME, Burnouf T. Regulation of tumor growth and metastasis: the role of tumor microenvironment. Cancer Growth Metastasis. 2014;7:9–18. doi: 10.4137/CGM.S11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- 15.Wagstaff L, Kelwick R, Decock J, Edwards DR. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front Biosci (Landmark Ed) 2011;16:1861–72. doi: 10.2741/3827. [DOI] [PubMed] [Google Scholar]
- 16.Hatkevich T, Ramos J, Santos-Sanchez I, Patel YM. Overexpression of ADAM9 enhances growth factor-mediated recycling of E-cadherin in human colon cancer cell line HT29 cells. Exp Cell Res. 2006;312:331–9. doi: 10.1016/j.yexcr.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Yamada D, Ohuchida K, Mizumoto K, Ohhashi S, Yu J, Egami T, Fujita H, Nagai E, Tanaka M. Increased expression of ADAM 9 and ADAM 15 mRNA in pancreatic cancer. Anticancer Res. 2007;27:793–9. [PubMed] [Google Scholar]
- 18.Lendeckel U, Kohl J, Arndt M, Carl McGrath S, Donat H, R€ocken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol. 2005;131:41–8. doi: 10.1007/s00432-004-0619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plank M, Maltby S, Mattes J, Foster PS. Targeting translational control as a novel way to treat inflammatory disease: the emerging role of microRNAs. Clin Exp Allergy. 2013;43:981–99. doi: 10.1111/cea.12170. [DOI] [PubMed] [Google Scholar]
- 21.Khella HW, Scorilas A, Mozes R, Mirham L, Lianidou E, Krylov SN, Lee JY, Ordon M, Stewart R, Jewett MA, Yousef GM. Low expression of miR-126 Is a prognostic marker for metastatic clear cell renal cell carcinoma. Am J Pathol. 2015;185:693–703. doi: 10.1016/j.ajpath.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Song KL, Chang H, Chen L. Decreased expression of microRNA-126 is associated with poor prognosis in patients with cervical cancer. Diagn Pathol. 2014;9:220. doi: 10.1186/s13000-014-0220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Zhou Y, Feng X, Yang P, Yang J, An P, Wang H, Ye S, Yu C, He Y, Luo H. Low expression of microRNA-126 is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2014;53:358–65. doi: 10.1002/gcc.22146. [DOI] [PubMed] [Google Scholar]
- 24.Hafez MM, Hassan ZK, Zekri AR, Gaber AA, Al Rejaie SS, Sayed-Ahmed MM, Al Shabanah O. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac J Cancer Prev. 2012;13:591–598. doi: 10.7314/apjcp.2012.13.2.591. [DOI] [PubMed] [Google Scholar]
- 25.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, He A, Zhang Q, Tao C. miR-126 inhibits cell growth, invasion, and migration of osteosarcoma cells by downregulating ADAM-9. Tumour Biol. 2014;35:12645–54. doi: 10.1007/s13277-014-2588-3. [DOI] [PubMed] [Google Scholar]
- 27.Jia AY, Castillo-Martin M, Bonal DM, Sánchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer. 2014;110:2945–54. doi: 10.1038/bjc.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 29.Lin CY, Chen HJ, Huang CC, Lai LC, Lu TP, Tseng GC, Kuo TT, Kuok QY, Hsu JL, Sung SY, Hung MC, Sher YP. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014;74:5229–43. doi: 10.1158/0008-5472.CAN-13-2995. [DOI] [PubMed] [Google Scholar]
- 30.Shintani Y, Higashiyama S, Ohta M, Hirabayashi H, Yamamoto S, Yoshimasu T, Matsuda H, Matsuura N. Overexpression of ADAM9 in non-small cell lung cancer correlates with brain metastasis. Cancer Res. 2004;64:4190–6. doi: 10.1158/0008-5472.CAN-03-3235. [DOI] [PubMed] [Google Scholar]
- 31.Micocci KC, Martin AC, Montenegro Cde F, Durante AC, Pouliot N, Cominetti MR, Selistre-de-Araujo HS. ADAM9 silencing inhibits breast tumor cell invasion in vitro. Biochimie. 2013;95:1371–8. doi: 10.1016/j.biochi.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Fry JL, Toker A. Secreted and membrane-bound isoforms of protease ADAM9 have opposing effects on breast cancer cell migration. Cancer Res. 2010;70:8187–98. doi: 10.1158/0008-5472.CAN-09-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]


