Abstract
In eukaryotes, bidirectional transport of macromolecules between the cytoplasm and the nucleus occurs through elaborate supramolecular structures embedded in the nuclear envelope, the nuclear pore complexes (NPCs). NPCs are composed of multiple copies of ∼30 different proteins termed nucleoporins, of which several can be biochemically isolated as subcomplexes. One such building block of the NPC, termed the Nup107-160 complex in vertebrates, was so far demonstrated to be composed of six different nucleoporins. Here, we identify three WD (Trp-Asp)-repeat nucleoporins as new members of this complex, two of which, Nup37 and Nup43, are specific to higher eukaryotes. The third new member Seh1 is more loosely associated with the Nup107-160 complex biochemically, but its depletion by RNA interference leads to phenotypes similar to knock down of other constituents of this complex. By combining green fluorescent protein-tagged nucleoporins and specific antibodies, we show that all the constituents of this complex, including Nup37, Nup43, Seh1, and Sec13, are targeted to kinetochores from prophase to anaphase of mitosis. Together, our results indicate that the entire Nup107-160 complex, which comprises nearly one-third of the so-far identified nucleoporins, specifically localizes to kinetochores in mitosis.
INTRODUCTION
All trafficking of macromolecules between the cytoplasm and the nucleus occurs through nuclear pore complexes (NPCs), which are supramolecular assemblies at points of fusion between the inner and outer nuclear membranes. NPCs are composed of an eightfold symmetric spoke-ring complex, cytoplasmic fibers, and a nuclear basket. Structural comparisons and mass measurements indicate that vertebrate NPCs are significantly larger than their yeast counterparts: they have a calculated mass of 60 MDa, compared with 44 MDa in Saccharomyces cerevisiae (Rout et al., 2000; Cronshaw et al., 2002). Despite this difference, identification of the full protein composition of yeast and vertebrate NPCs have revealed that they are composed in both organisms of roughly 30 different proteins, termed nucleoporins, of which about two-thirds have been conserved during evolution (Rout et al., 2000; Cronshaw et al., 2002; reviewed in Suntharalingam and Wente, 2003).
Emerging evidence indicates unexpected relationships between NPCs and kinetochores. In particular, two metaphase checkpoint proteins that are transient components of kinetochores, Mad1 and Mad2, reportedly localize at nuclear pores in interphase both in vertebrates and yeasts (Campbell et al., 2001; Ikui et al., 2002; Iouk et al., 2002). In S. cerevisiae, Mad1 and Mad2 were shown to be associated with NPCs through the Nup53p subcomplex (Iouk et al., 2002). In vertebrates, the WD (tryptophane-aspartic acid)-repeat-containing protein Rae1 (also called Gle2 or mrnp41) that interacts with the nucleoporin Nup98 (Pritchard et al., 1999) was shown to interact with the mitotic checkpoint protein mBUB1 and to be localized at kinetochores of prometaphase chromosomes (Wang et al., 2001). It was further demonstrated that loss of a single Rae1 allele causes a mitotic checkpoint defect and chromosome missegregation (Babu et al., 2003). It was also recently shown that SUMO-1 modified RanGAP1 and RanBP2/Nup358, which are associated with the cytoplasmic face of NPCs in interphase, are targeted as a complex to kinetochores and mitotic spindles in mitosis (Matunis et al., 1998; Joseph et al., 2002, 2004). RNA interference approaches in Caenorhabditis elegans early embryo and HeLa cells further revealed the involvement of RanBP2/Nup358 in chromosome congression and segregation, stable kinetochore-microtubule association, and kinetochore assembly (Askjaer et al., 2002; Salina et al., 2003; Joseph et al., 2004). Finally, a fraction of human Nup133, Nup107, and Nup85/75 was shown to associate with kinetochores in mitosis (Belgareh et al., 2001; Harel et al., 2003).
The later three nucleoporins are part of an evolutionarily conserved NPC subcomplex, termed the Nup107-160 complex in vertebrates and Nup84 complex in the yeast S. cerevisiae. Constituents of this complex are localized on both sides of the NPCs in yeast (Rout et al., 2000) and vertebrates (Belgareh et al., 2001; Enninga et al., 2003). This complex was shown to be involved in NPC distribution in S. cerevisiae, and in mRNA export both in yeast (reviewed in Doye and Hurt, 1997) and vertebrates (Vasu et al., 2001; Boehmer et al., 2003; Walther et al., 2003a). More recently, this complex was demonstrated to also be critical for NPC assembly in vertebrates (Harel et al., 2003; Walther et al., 2003a).
In S. cerevisiae, this complex is formed by seven constituents: Nup84; Nup85; Nup120; the in vivo-cleaved carboxy-terminal domain of Nup145 (Nup145-C); Seh1; a fraction of Sec13; and Nup133 (Siniossoglou et al., 1996; Teixeira et al., 1997; Allen et al., 2002; Lutzmann et al., 2002). Isolation under native conditions and in vitro reconstitution have further revealed the Y-shaped structure of this complex (Siniossoglou et al., 2000; Lutzmann et al., 2002). The vertebrate Nup107-160 complex was so far demonstrated to consist of Nup107 (homologous to ScNup84), Nup133 (homologous to ScNup133), Nup160 (homologous to ScNup120 and thus also referred in human cells as Nup120), Nup96 (homologous to ScNup145-C), Sec13 and Nup85/75 (Fontoura et al., 1999; Belgareh et al., 2001; Vasu et al., 2001; Harel et al., 2003). In addition, previous studies reported the cofractionation on sucrose gradients of Nup107, Nup96, and Sec13 together with a 37-kDa WD-repeat containing protein (Fontoura et al., 1999), subsequently identified as Nup37 (Cronshaw et al., 2002). Conversely, the S. cerevisiae Nup84 complex contains another WD-repeat nucleoporin, ScSeh1 (Siniossoglou et al., 1996), for which a potential higher eukaryote homologue has been identified (Cronshaw et al., 2002).
These data prompted us to determine whether Nup37 or Seh1 were also part of the Nup107-160 NPC subcomplex. Interestingly, this study revealed that the Nup107-160 complex contains in addition to Sec13, at least two additional WD-repeat-containing nucleoporins: Nup37 and Nup43. Although biochemical approaches did not enable us to demonstrate a stable interaction between GFP- or myc-tagged Seh1 and the Nup107-160 complex in cell lysates, small interfering RNA (siRNA) experiments revealed that depletion of Seh1 induces phenotypes reminiscent to those induced by depletion of Nup133 or Nup107. We further demonstrate that the constituents of this complex are all, including Seh1 and Sec13, targeted to kinetochores in mitosis.
MATERIALS AND METHODS
Plasmids
A human cDNA corresponding to most of the Nup160 open reading frame (ORF) (KIAA0197, GI:1228044 lacking amino acids 1295-1386 of Nup160) cloned in a pBluescrispt KS+ vector was provided by Kazusa DNA Research Institute Japan. The SalI/NotI insert was subcloned in a pGEX-4T2 vector (Amersham Biosciences, Piscataway, NJ) to produce the glutathione S-transferase (GST)-Nup160ΔC92 plasmid, and into a pEGFP3-C1 vector that contains a triple GFP concatamer constructed in pEGFP-C1 to produce the pEGFP3-Nup160ΔC92 plasmid (Daigle et al., 2001). Plasmids pGEX-4T2-Nup160-N (encoding aa 1-598 of Nup160) and pGEX-4T2-Nup160-C (encoding aa 598-1294 of Nup160) were generated using an internal BspE1 restriction site. To obtain a full-length cDNA, a NotI/PmlI fragment encoding the C-terminal domain of Nup160 was purified from the IMAGE Consortium (LLNL) cDNA clone (IMAGE: 33299, GI: 772459) (Lennon et al., 1996) obtained from the Resource Center of the German Human Genome Project at the Max-Planck-Institut for Molecular Genetics (http://www.rzpd.de) and used to replace the corresponding fragment of the pEGFP3-Nup160ΔC92 vector.
For human Seh1, part of the 3′ noncoding sequence of the IMAGE Consortium (LLNL) cDNA clone (IMAGE: 3865994, GI: 15214607) was deleted by a SpeI digestion. The cDNA from the resulting pCMV-SPORT6-Seh1-ΔSpeI vector was amplified by polymerase chain reaction (PCR) to introduce a BamHI site right after the ATG by using a 5′-specific primer (cgcggatcctttgtggctcgcagcatc) and the T7 primer. The PCR product was digested by BamHI/XhoI and ligated in the pEGFP-C1 and pEGFP3-C1 vectors. The SmaI/SpeI blunt-ended fragment of Seh1 purified from the pCMV-SPORT6-Seh1-ΔSpeI vector (including 40 amino acids upstream of the Seh1 ATG) was also inserted downstream of a 6-myc epitope tag in the pCS3+MT vector (Turner and Weintraub, 1994).
For human Nup85, an EcoRI-NotI fragment, encoding most of Nup85 ORF (starting at amino acid 17) was purified from the IMAGE Consortium (LLNL) cDNA clone (IMAGE: 753140, GI: 2054587) and subcloned in the pGEX-4T3 and pEGFP-C1 vectors.
Plasmids pEGFP-Nup43, pEGFP-Nup35, pEGFP-Aladin, and pEGFP-Nup37 were generously provided by J. Cronshaw and M. Matunis (Cronshaw et al., 2002). The Nup37 cDNA was subcloned as an EcoRI/XbaI fragment in the pGST-parallel3 vector (Sheffield et al., 1999) to produce the recombinant fusion protein GST-Nup37, and in pCS2+MT vector (Turner and Weintraub, 1994) to express 6-myc tagged fusion. Plasmid pEGFP3-Sec13 was constructed by inserting the Sec13 ORF (provided by B. Glick, University of Chicago, Chicago, IL) in frame in the pEGFP3-C vector.
Transfections, Immunoprecipitations, Sucrose Gradient Sedimentation, and Gel Filtration Chromatography
HeLa cells were grown at 37°C in Dulbecco's modified Eagles's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum, 1% l-glutamine, 100 μg/ml streptomycin, and 100 U/ml penicillin. For immunoprecipitations (IPs) and sucrose gradients, 5 × 105 cells were seeded in 10-cm dishes 24 h before transfection. Simple and cotransfections were performed by using calcium phosphate-mediated procedure as described previously (Jordan et al., 1996). Plates were harvested 72 h after transfection. Cells pellets were frozen in liquid nitrogen and lysed in 175 μl (for sucrose gradient) or 600 μl (for IP) of lysis buffer (100 mM NaCl, 1 mM EDTA, 25 mM Tris, pH 7.5, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2% Triton X-100). Lysates were centrifuged 30 min at 4°C at 14,000 × g. For preparation of mitotic extracts, HeLa S3 cells were treated for 14 h with demecolcin (60 ng/ml), leading to a mitotic index of 90-95%. Cell pellets were immediately frozen in liquid nitrogen. To confirm that the extracts used were maintained in a mitotic state upon the immunoprecipitation procedure, samples were analyzed by Western blot by using an cyclin B1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoprecipitation experiments were performed essentially as previously described (Grandi et al., 1997) by using either 25 μl of affinity-purified anti-Nup133 antibody bound to 30 μl of Affi-prep protein A matrix (Bio-Rad, Hercules, CA), or 10 μg of anti-GFP, or 30 μg of anti-myc monoclonal antibodies bound to 30 μl of protein G-Sepharose 4 Fast Flow matrix (Amersham Biosciences).
For sucrose gradients, supernatants were applied on the top of two 20-35% linear sucrose gradients (500 μl in lysis buffer) in Ultra Clear 5 × 41-mm centrifugal tubes (Beckman Coulter, Fullerton, CA). Centrifugations were performed using a SW55Ti swinging rotor (Beckman Coulter) at 100,000 × g for 16 h at 4°C. Twenty-six fractions (20 μl each) were collected starting from the top of the gradient. Fractions 11-14 were pooled and 50 μl of the resulting sample were analyzed by gel filtration chromatography by using a Superose 6 (PC 3.2/30) column (Amersham Biosciences) equilibrated with lysis buffer (100 mM NaCl, 1 mM EDTA, 25 mM Tris, pH 7.5, 1% Triton X-100 reduced form). The flow rate was 40 μl/min, and 50-μl fractions were collected. Molecular size markers used to calibrate the sucrose gradient and the Superose 6 column were thyroglobulin, 670 kDa; ferritin, 440 kDa; catalase, 230 kDa; aldolase, 160 kDa; bovine serum albumin, 67 kDa; and chymotrypsinogen A, 25 kDa.
siRNA Experiments and Quantitative Reverse Transcription (RT)-PCR
The siRNA duplex used for silencing Seh1 (aagacacatagtggatctgtatg) and a random siRNA duplex (aagagacaactgcaaatgg) were purchased from QIAGEN (Valencia, CA) or Dharmacon (Lafayette, CO). siRNA duplexes for silencing Nup133, Nup153, and Giantin were described previously (Harborth et al., 2001; Nizak et al., 2003; Walther et al., 2003a). The siRNA duplex used for silencing Tpr was provided by Ulf Nherbass (Institut Pasteur, Paris, France). Cells were transfected with Oligofectamine (Invitrogen) as described using 480 nM of siRNAs (Elbashir et al., 2001; Walther et al., 2003a). For siRNA cotransfection experiments, the Giantin siRNA was used at a 10-fold lower concentration (48 nM) than the cotransfected siRNAs. Preliminary experiments using Nup133 siRNAs and either anti-Nup133 antibodies (our unpublished data) or anti-Nup107 and mAb414 labeling (Figure 4B, a-c, and Supplemental Figure S1 A, c) demonstrated that under these experimental conditions, depletion of Giantin (assessed by the decreased Golgi labeling detected with a specific anti-Giantin scFv) could be used to identify the siRNA-treated cells. Additional controls confirmed that the recorded phenotypes were not due to the cotransfection with Giantin siRNA duplexes and that siRNA duplexes directed against Nup133 or Seh1 did not affect Giantin labeling (Supplemental Figure S1 A).
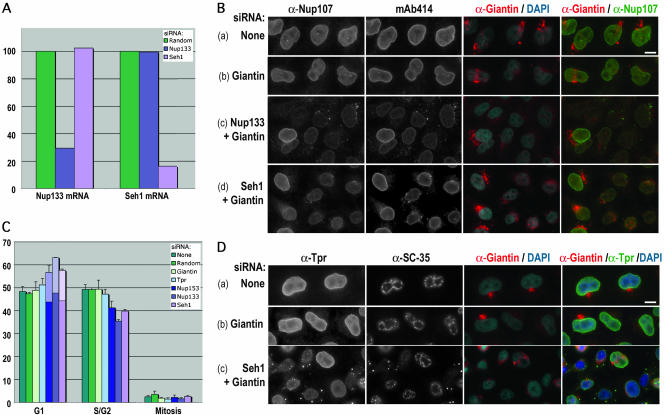
Figure 4.
Effects of Seh1 depletion in HeLa cells are reminiscent to those observed upon Nup133 depletion. (A) Quantitative RT-PCR analysis of Seh1 and Nup133 mRNAs from HeLa cells treated for 3 d with siRNA duplexes specific for Seh1 or Nup133 or a random siRNA. The amounts of Seh1 and Nup133 mRNAs were arbitrarily set at 100% for cells treated with the random siRNA. (B) Widefield microscopy images of immunofluorescence of HeLa cells treated for 3 d with no siRNA (a), Giantin siRNA duplexes (b), or Giantin siRNA duplexes together with siRNA duplexes specific for Nup133 (c) or Seh1 (d) and labeled with anti-Nup107, mAb414, and anti-Giantin antibodies. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Note in c and d the wild-type labeling of Nup107 and mAb414 in cotransfected cells that are not depleted for Giantin. As previously observed in Nup107-depleted cells (Walther et al., 2003a), cytoplasmic foci are slightly less abundant in Seh1-depleted cells compared with Nup133-depleted cells. (C) Cell cycle analyses of HeLa cells treated for 3 d with no siRNA, a random siRNA, or with siRNA duplexes specific for Tpr, Nup153, Nup133, or Seh1. S/G2 cells were detected by BrdU and cyclin B1 staining, and mitotic cells by phase contrast. The clear part of the histograms corresponds to the percentage of cells with SC-35 cytoplasmic foci. Standard deviations are indicated. (D) Widefield microscopy images of immunofluorescence of HeLa cells treated for 3 d with no siRNA (a), Giantin siRNA duplexes (b), or Giantin siRNA duplexes (c) together with siRNA duplexes specific for Seh1. Cells were labeled with anti-Tpr, anti-SC-35, anti-Giantin, and DAPI. Bars, 10 μm.
Total RNAs were isolated 72 h after siRNA transfection, by using the RNeasy mini kit (QIAGEN). mRNA were reverse-transcribed using random hexameres and the AMV reverse transcriptase (Finnzymes, Espoo, Finland). Real-time PCR was performed with the ABI PRISM 7900HT sequence detection system by using Syber Green incorporation (SYBR Green PCR-Master Mix; Applied Biosystems, Foster City, CA) and the following primer pairs: Seh1 (ttggagcgaatgcaatatca and caatggccaaggttcagatt), Nup133 (tgatggcaaagatgatccaa and tcactgagctgaatgccatc), and TBP (agtgaagaacagtccagactg and ccaggaaataactctggctcat). The relative amounts of Seh1 and Nup133 cDNAs in the samples were calculated by reference to standard curves performed for each cDNA on a calibrator sample (cells treated with a random siRNA) and normalized by reference to the TBP cDNA by using the Sequence Detection System 2.0 software (Applied Biosystems).
Antibodies and Immunofluorescence
Polyclonal antibodies against Nup85, Nup37, and Nup160 were obtained by injecting recombinant GST-Nup85 or GST-Nup37 into rabbits, and a mixture of GST-Nup160-N and GST-Nup160-C to chickens (Agro-Bio, La Ferté St. Aubin, France). Serums were depleted of GST antibodies and affinity purified using GST-Nup85, GST-Nup37 or a mixture of GST-Nup160-N and GST-Nup160-C immobilized on a NHS-activated columns. The others antibodies used were affinity-purified rabbit polyclonal antibodies directed against human Nup133 and Nup107 (Belgareh et al., 2001), Nup96/p87 serum affinity-purified by using recombinant Nup96/p87 (residues 1291-1482 of Nup196) immobilized on nitrocellulose (Fontoura et al., 1999), affinity-purified anti-Nup96 (directed against a synthetic peptide corresponding to aa 1743-1763 of Nup196) (Hase and Cordes, 2003), affinity-purified anti-Sec13 (Tang et al., 1997) and Sec31 (Tang et al., 2000), anti-Tpr (Kuznetsov et al., 2002), or anti-myc epitope (Euromedex, Souffelweyersheim, France); monoclonal antibodies anti-myc (clone 9E10; Sigma-Aldrich, St. Louis, MO), anti-GFP (clone 7.1 and 13.1; Roche Diagnostics, Indianapolis, IN), anti-SC-35 (Sigma-Aldrich), anti-cyclin B1 (Santa Cruz Biotechnology), anti-mitosin (BD Biosciences, San Jose, CA), and mAb414 (Babco, Richmond, CA); the autoimmune CREST serum was obtained from J.C. Courvalin (Institut J. Monod, Paris, France). To detect Giantin, the TA10 scFv (Nizak et al., 2003), obtained from F. Perez (Institut Curie, Paris, France), was used at 1/100 and mixed with a goat anti-myc antibody (Santa Cruz Biotechnology). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) or Molecular Probes (Eugene, OR).
Immunofluorescence on paraformaldehyde (PFA) fixed cells was performed essentially as described previously (Belgareh et al., 2001). In the course of this study, we noticed that the use of a slightly basic PFA was critical for a clear detection of Nup96 at both the NPCs and kinetochores.
For cell cycle analyses, cells were incubated for 10 min with 40 μM bromodeoxy-uridine (BrdU) and washed twice with phosphate-buffered saline before being processed for immunofluorescence as described previously (Belgareh et al., 2001), by using anti-Tpr and anti-cyclin B1 antibodies. Cells were then postfixed for 15 min with 2% PFA, treated for 10 min with 4 N HCl, and immunostained with a rat anti-BrdU antibody (Harlan Sera-Lab, Leicestershire, United Kingdom). S/G2 cells were detected by BrdU and cyclin B1 staining and mitotic cells by phase contrast. The other cells were scored as G1. For each siRNA, quantifications were based on two or three independent experiments with at least 2000 cells scored in total. To eliminate bias, no attempt was made to discriminate between transfected versus nontransfected cells.
Widefield and Deconvolution Microscopy
Images were acquired using an upright motorized widefield microscope (DMRA2; Leica, Wetzlar, Germany). Acquisitions were performed using an oil immersion objective (100× PL APO HCX, 1.4 numerical aperture) and a high-sensitive cooled interlined charge-coupled device camera (CoolSnap HQ; Roper Scientific, Trenton, NJ) operating at 20 MHz in z-streaming mode. Rapid and precise Z-positioning was accomplished by a piezo-electric motor (LVDT; Physik Instrument, Auburn, MA) mounted underneath the objective lens. The whole system was steered by MetaMorph 5 Software (Universal Imaging, Downingtown, PA). Image stacks were acquired without camera binning, with a plane spacing of 0.2 μm, leading a voxel size of 64.5 × 64.5 × 200 nm3. Deconvolution was performed by the three-dimensional deconvolution module from MetaMorph (Universal Imaging), by using the fast Iterative Constrained PSF-based algorithm.
RESULTS
The Higher Eukaryote-specific WD-Repeat-containing Nucleoporins Nup37 and Nup43 Are Stable Constituents of the Nup107-160 Complex
To determine whether human Seh1 and/or Nup37 were constituents of the Nup107-160 NPC subcomplex, HeLa cells were transiently transfected with these two nucleoporins tagged with GFP. As controls, we used either GFP alone, GFP-Nup107, GFP-Nup35, or two other WD-repeat nucleoporins, GFP-Nup43 and GFP-Aladin. As reported previously, these various nucleoporins were properly targeted at the nuclear envelope after 2 or 3 d of transfection (Cronshaw et al., 2002; see also Figure 5), with the exception of Sec13 that gave an additional signal typical for endoplasmic reticulum (ER) and ER exit sites (Hammond and Glick, 2000; Tang et al., 2000; see also Figure 8).
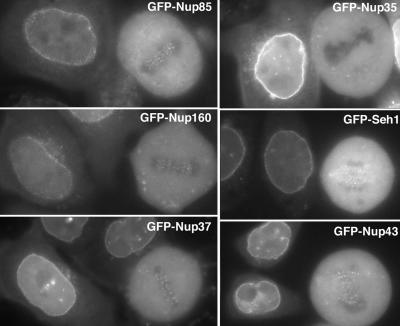
Figure 5.
Constituents of the Nup107-160 complex, but not GFP-Nup35, are targeted to kinetochores. Widefield microscopy images of live interphase and mitotic HeLa cells expressing, as indicated, GFP-Nup85, GFP-Nup35, GFP-Nup160, GFP-Seh1, GFP-Nup37, or GFP-Nup43. Whereas all these GFP-tagged nucleoporins are targeted to the nuclear envelope of interphase cells, fluorescent foci can be visualized at the metaphase plate of mitotic cells expressing these various fusions, with the exception of GFP-Nup35-expressing cells.
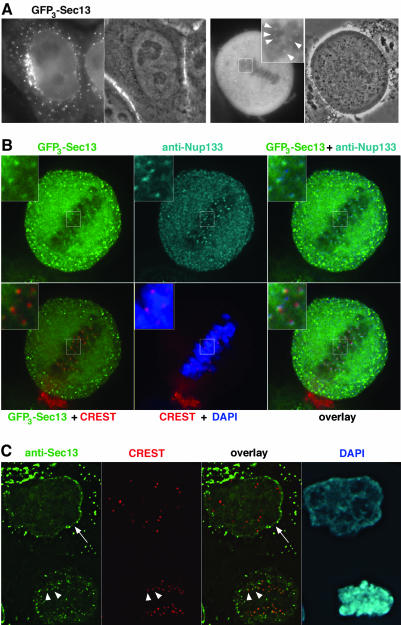
Figure 8.
A fraction of Sec13 localizes to both the nuclear envelope and kinetochores. (A) Widefield microscopy images of live interphase and mitotic HeLa cells expressing GFP3-Sec13. The fluorescent signal and the corresponding phase contrast images are shown. The inset shows a threefold enlargement of the marked area within the metaphase plate, where bright GFP3-Sec13 foci can be detected (arrowheads). (B) Deconvolved images of GFP3-Sec13-expressing cells double labeled with affinity-purified anti-Nup133 antibody and the CREST serum. DNA was stained with DAPI. Insets show a threefold enlargement of the marked area. Note the overlap between the GFP3-Sec13 and the anti-Nup133 stainings that are more peripheral compared with the CREST labeling. (C) Deconvolved images of HeLa cells double labeled with affinity-purified anti-Sec13 antibody and the CREST serum. DNA was stained with DAPI. The arrow on the interphase cell (top) points to the nuclear envelope, labeled with the Sec13 antibody, and arrowheads indicate the position of kinetochores, labeled with both the anti-Sec13 antibody and CREST serum in an anaphase cell (bottom).
Immunoprecipitations were performed using anti-GFP antibodies on total cell extracts collected 3 d after transfection. As anticipated, GFP-Nup107 immunoprecipitated the other constituents of the Nup107-160 complex, i.e., Nup85, Nup133, Nup160 (Figure 1C), and Nup96 (our unpublished data). In addition, GFP-Nup37 was able to efficiently coprecipitate all the previously characterized constituents of the Nup107-160 complex (Figure 1A; our unpublished data). Unexpectedly, a similar result also was observed with the WD-repeat-containing nucleoporin GFP-Nup43 (Figures 1B and 3). In contrast, none of the constituents of the Nup107-160 complex were detectable in any of the control precipitations (GFP-Aladin, GFP-Nup35, or GFP) (Figures 1 and 3). For Seh1 tagged with either one (Figure 1C) or three (Figure 1A) copies of GFP the amount of coprecipitated nucleoporins was low when detectable and less reproducible compared with the other nucleoporins analyzed under similar conditions. Use of a NRK cell line stably expressing GFP-Seh1 did not increase the amount of coprecipitated nucleoporins (our unpublished data), suggesting that, unlike in S. cerevisiae, Seh1 might be less stably associated with the Nup107-160 complex in extracts. However, we could not exclude that the GFP-tag may affect Seh1 assembly within the Nup107-160 complex or not be accessible to antibodies once GFP-Seh1 is assembled within the NPCs.
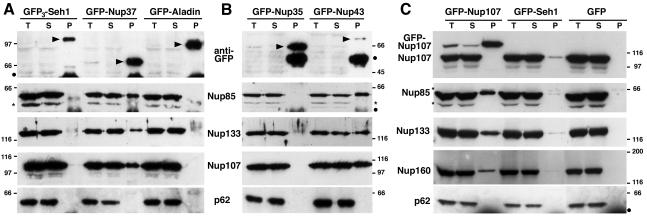
Figure 1.
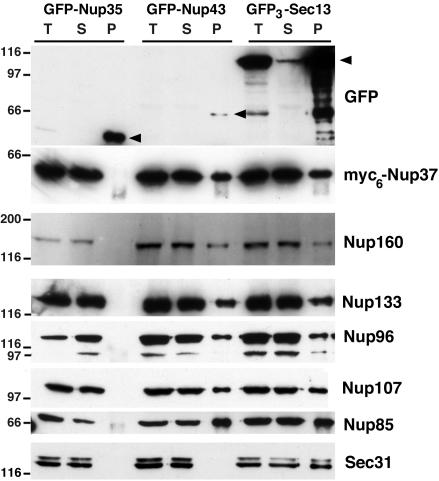
Immunoprecipitation of soluble extracts from HeLa cells expressing GFP3-Seh1, GFP-Nup37, GFP-Aladin (A); GFP-Nup35, GFP-Nup43 (B); and GFP-Nup107, GFP-Seh1, or GFP (C) by using anti-GFP antibodies. Equivalent amounts of total extracts (T) and immune supernatants (S) and tenfold equivalents of the immune pellets (P) were analyzed by immunoblot by using, as indicated, anti-GFP, Nup85, Nup133, Nup107, or Nup160 antibodies or the mAb414 antibody that mainly recognizes p62. Arrowheads in A and B point to the position of the GFP fusions. Dots indicate IgG heavy chains and stars a protein that cross-reacts with the anti-Nup85 antibody. Molecular mass markers are in kilodaltons.
Figure 3.
GFP-Nup43 and GFP3-Sec13 coimmunoprecipitate with myc6-Nup37 and the other constituents of the Nup107-160 complex. Immunoprecipitation using anti-GFP antibodies of soluble extracts from HeLa cells coexpressing GFP-Nup35, GFP-Nup43, or GFP3-Sec13, and myc6-Nup37. Equivalent amounts of total extracts (T) and immune supernatants (S) and tenfold equivalents of the immune pellets (P) were analyzed by immunoblot by using anti-GFP, anti-myc (that recognizes the myc6-Nup37 fusion), Nup85, Nup160, Nup133, Nup96, Nup107, Nup85, or Sec31 antibodies. Arrowheads indicate the position of the GFP fusions. Molecular mass markers on the left are in kilodlatons.
Because attempts to obtain anti-Seh1 antibodies were not successful, we also tagged both Nup37 and Seh1 at their amino-terminal domain with a 6-myc epitope. When transfected in HeLa cells, both constructs were properly targeted to the NPCs. Whole cell extracts from myc6-Nup37- and myc6-Seh1-expressing cells were then analyzed on sucrose gradients. As shown in Figure 2A (a, b, c, and f), endogenous Nup133, Nup85, and Nup37 all peaked at fraction 11-14 on sucrose gradients, whereas p62 was found in lighter fractions of the gradient. Unlike endogenous Nup37, the major pool of transiently transfected myc6-Nup37 was found in the lightest fractions of the sucrose gradients, whereas a minor pool (most likely corresponding to aggregated and/or mis-folded protein) was recovered at the bottom of the gradient. However, part of the myc6-Nup37 fusion protein was also found to comigrate together with Nup133 within fractions 11-14 (Figure 2A, d). In the case of myc6-Seh1, a minor pool of the fusion protein could also be found in those fractions (Figure 2A, e). Yet, immunoprecipitation experiments performed on similar samples by using either anti-myc or anti-Nup107 antibodies did not reveal any significant interaction between myc6-Seh1 and constituents of the Nup107-160 complex (our unpublished data).
Figure 2.
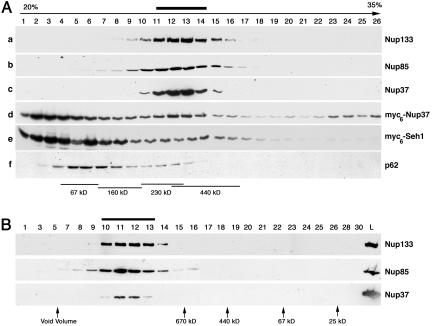
Analysis of myc6-Nup37 and myc6-Seh1 behavior by sucrose gradients. (A) Soluble extracts from HeLa cells (b and f) or from HeLa cells expressing myc6-Nup37 (a and d) or myc6-Seh1 (c and e) were loaded on continuous 20-35% sucrose gradients and centrifuged at 100,000 × g for 16 h. Fractions from the top (fraction 1) to the bottom (fraction 26) of the gradient were analyzed by Western blot by using anti-Nup133 (a), Nup85 (b), Nup37 (c), or myc (d and e) antibodies, or the mAb414 antibody (f). (B) Fractions 11-14 from the above-described sucrose gradient (b) were pooled, and 50 μl of the resulting sample was loaded on a Superose 6 (PC 3.2/30) gel filtration chromatography column. The initial sample (L) and the Superose 6 fractions were analyzed by Western blot by using anti-Nup133, Nup85, and Nup37 antibodies. Molecular size markers used to calibrate the sucrose gradient and the Superose 6 column are indicated. Thick bars denote the position of the Nup107-160 complex.
Noteworthy, whereas addition of the theoretical mass of each constituents of the Nup107-160 complex predicts a theoretical mass of ∼700 kDa, this complex migrated with an apparent specific mass lower than Ferritin (440 kDa) on sucrose gradient at equilibrium (Figure 2A). To further characterize the behavior of these nucleoporins, positive fractions from the sucrose gradient were analyzed by gel filtration chromatography. As shown in Figure 2B, Nup133, Nup85, and Nup37 were all eluted much before Thyroglobulin (670 kDa) on a Superose 6 column (Figure 2B), a result consistent with the behavior of the Xenopus Nup107-160 complex upon fractionation on Sephacryl S400-HR column (Vasu et al., 2001). The unexpected behavior of the Nup107-160 complex on sucrose gradient may reflect its elongated shape, similar to the previously described Y-shaped structure of the S. cerevisiae Nup84 complex (Lutzmann et al., 2002). In addition, the similar behavior of Nup37 and constituents of the Nup107-160 complex on both sucrose gradient and upon gel filtration is consistent with the fact that most, if not all, Nup37 associates with the Nup107-160 complex in HeLa cells.
The presence of several distinct WD-repeat nucleoporins in the Nup107-160 complex raised the possibility of the existence of distinct complexes that may contain different combinations of these WD-repeat proteins. To address this issue, HeLa cells were cotransfected with myc6-Nup37 and several GFP-tagged nucleoporins. As the other constituents of the Nup107-160 complex, myc6-Nup37 coprecipitated with GFP-Nup43 and GFP3-Sec13 (Figure 3) but not with GFP-Nup35 that does not belong to the Nup107-160 complex. Together, these results thus indicate that all Nup107-160 complexes contain several distinct WD-repeat nucleoporins, Sec13, Nup37, Nup43, and possibly Seh1.
RNA Interference of Seh1 Induces Phenotypes Similar to Nup133 RNAi
Because we could not observe a stable interaction between Seh1 and constituents of the Nup107-160 complex in vitro, we decided to address its function at NPCs by using siRNA duplexes (Elbashir et al., 2001) specifically targeting the Seh1 mRNA. Such a study previously revealed that partial depletion of Nup133, Nup107, or Nup85 leads to the decreased localization of several nucleoporins at the nuclear envelope (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003a). We could further correlate this decreased labeling to an overall decrease of NPC number in the Nup133 or Nup107-depleted cells (Walther et al., 2003a).
Quantitative RT-PCR showed that the selected Seh1 siRNA led to a strong decrease of its mRNA, whereas the level of Nup133 mRNA was not significantly affected (Figure 4A). Conversely, the level of Seh1 mRNA was not significantly altered upon treatment of HeLa cells with siRNAs targeting Nup133 mRNAs (Figure 4A). Because antibodies directed against Seh1 were not available, siRNA duplexes directed against Giantin were cotransfected with the Seh1 or Nup133 siRNA duplexes and depletion of Giantin (assessed by its decreased labeling at the Golgi) was used to identify the siRNA-treated cells (see MATERIALS AND METHODS). Using this approach, a reduction of Nup107 and mAb414 staining at the nuclear envelope could be observed in Seh1 siRNA-treated HeLa cells (Figure 4B, d). This decreased labeling was accompanied by the appearance of cytoplasmic foci, containing both Nup107 and mAb414 antigens, and reminiscent of annulate lamellae (Figure 4B). Similar results were also observed with anti-Nup133 antibodies (our unpublished data). However, the delocalization of these nucleoporins from the nuclear envelope (NE) was usually less pronounced upon Seh1 siRNA treatment compared with Nup133-depleted cells (Figure 4B, c and d, and Supplemental Figure S1, b and c). In contrast, the labeling of Nup153 at the nuclear envelope was less affected, and this nucleoporin was never found into Nup107-labeled cytoplasmic foci (our unpublished data). Finally, as also previously reported in Nup133- or Nup107-depleted cells (Walther et al., 2003a), a decrease of Tpr labeling at the NE was observed in most Seh1 siRNA-treated cells. In addition, both Tpr and the speckle protein SC-35 were found to accumulate as distinct cytoplasmic foci in some, but not all, Seh1- or Nup133-depleted cells (Figure 4D, c and Supplemental Figure S1B, b). In contrast, the nuclear targeting of Coilin was not affected in Seh1 siRNA-treated cells displaying an extreme Tpr delocalization (our unpublished data). Similarly, previous studies did not reveal any alteration of the kinetics of import of the RGG reported in Nup133-depleted cells (Walther et al., 2003). Because cytoplasmic SC-35 foci have been previously observed in cells at the end of mitosis (Spector et al., 1991), we reasoned that this labeling could reflect a cell cycle defect occurring in some Nup133- or Seh1-depleted cells. In agreement with this hypothesis, immunofluorescence analyses revealed that both Seh1 and Nup133 siRNA-treated cells moderately accumulate in a G1-like stage and that no cell displaying cytoplasmic foci of SC-35 were in S, G2, or M phase of the cell cycle (Figure 4C). Similar experiments performed using random, Giantin, or Tpr siRNA duplexes did not reveal such a G1 accumulation, nor the appearance of SC-35 in cytoplasmic foci (Figure 4C). Similarly, depletion of Nup358 did not lead to the mislocalization of SC-35 (our unpublished data). However, analysis of Nup153 siRNA-treated cells revealed a similar modest increase of G1 cells, associated with the appearance of cells with SC-35 foci in the cytoplasm (Figure 4C). Yet, the early phenotypes associated with Nup153 depletion clearly differ from those induced by Nup133, or Seh1 depletion (see DISCUSSION).
GFP-tagging Reveals the Association of Nup160, Nup37, Nup43, and Seh1 with Kinetochores in Mitosis
So far, three constituents of the Nup107-160 complex, Nup133, Nup107, and Nup85, have been localized to kinetochores in mitosis (Belgareh et al., 2001; Harel et al., 2003). Because those results suggested that the entire Nup107-160 complex might be targeted to kinetochores in mitosis, we analyzed the localization of GFP-tagged constituents of this complex in mitotic HeLa cells. In agreement with previous data demonstrating the localization of endogenous Nup85 at kinetochores, analysis of live cells revealed the targeting of GFP-tagged Nup85 to kinetochores (Figure 5, top left). In the case of Nup160, initial attempts to localize this protein by using the KIAA0197-derived pEGFP3-Nup160ΔC92 vectors, which carries an internal 92-amino acid deletion within the C-terminal domain of Nup160, did not revealed any specific localization of this fusion to either the NPCs or kinetochores. In contrast, full-length GFP3-Nup160 was targeted to both NPCs and kinetochores (Figure 5). Finally, the WD-repeat nucleoporins GFP-Nup37, GFP-Nup43, and GFP-Seh1 all gave rise to a typical dot-like labeling on mitotic chromosomes. Such a labeling appeared to be specific for the constituents of the Nup107-160 complex, because it was not observed for several other nucleoporins tested, including GFP-tagged Nup35 (Figure 5, top right), Nup98, Nup50, or the WD-repeat containing nucleoporin Aladin (our unpublished data). As previously observed for Nup133 and Nup107, this labeling reflected the localization of a minor fraction of these nucleoporins that are otherwise dispersed within the mitotic cytoplasm.
Those signals remained upon paraformaldehyde fixation of the cells, which enabled us to perform triple labeling by using the human autoimmune CREST serum and the monoclonal anti-CENP-F antibody, thereby confirming the identity of these fluorescent foci as kinetochores. As shown in Figure 6 and Supplemental Figure S2, deconvolved images of GFP-Nup37 and GFP-Nup43 revealed their distal localization compared with the inner centromeres labeled with the CREST serum, and a slightly less distal position than CENP-F, an outer kinetochore protein (Rattner et al., 1993).
Figure 6.
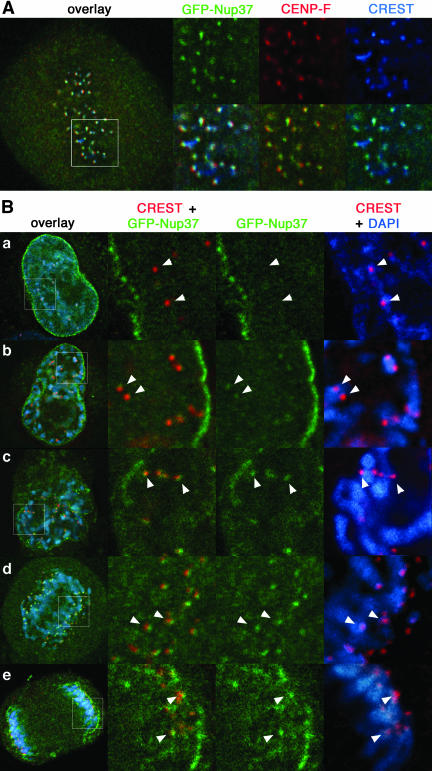
GFP-Nup37 is localized at kinetochores from prophase to anaphase. (A) Deconvolved images of fixed metaphase HeLa cells expressing GFP-Nup37 (shown in green) and labeled with anti-CENP-F antibody (shown in red) and the CREST serum (shown in blue). Insets showing enlargements of the marked area reveal that GFP-Nup37 lies distal to the centromeres labeled with the CREST serum, and slightly more central than CENP-F, an outer kinetochore protein. (B) Fixed HeLa cells expressing GFP-Nup37 were labeled with the CREST serum and DAPI. Deconvolved images of interphase (a), prophase (b), late prophase/prometaphase (c), early anaphase (d), and late anaphase (e) are presented. The insets show enlargements of the marked area. Arrows point to the position of centromeres. Note the weak kinetochore labeling of GFP-Nup37 in early prophase and the persistence of this staining until late anaphase.
In addition, analysis of cells at various stages of mitosis revealed for both GFP-Nup37 and GFP-Nup43 a faint kinetochore labeling in prophase (Figure 6B, a and b and Supplemental Figure S2 B, a and b), whereas no signal could be detected in interphase (a). This staining was enhanced during late prophase/prometaphase (Figure 6B, c), persisted until early anaphase (Figure 6B, d and Supplemental Figure S2 B, c), and started to fade in late anaphase (Figure 6B, e and Supplemental Figure S2 B, d). Accordingly, both the localization within kinetochores and the kinetics of appearance of these nucleoporins to these mitotic structures is indistinguishable from the one observed for Nup133 and Nup107 (Belgareh et al., 2001).
Nup96 Is Tightly Associated with the Nup107-160 Complex during the Entire Cell Cycle and Is Localized at Kinetochores in Mitosis
While this study was in progress, Nup96 and Sec13 were reported to be distributed differently throughout the cell during mitosis: whereas a major fraction of Nup96 was found to colocalize with tubulin on the spindle apparatus, Sec13 did not colocalize with the spindle apparatus and was reportedly distributed in a diffuse pattern throughout the cell (Enninga et al., 2003).
To further characterize the behavior of Nup96 during mitosis, we first performed immunoprecipitation experiments from interphase or mitotic HeLa cell extracts by using affinity-purified anti-Nup133 antibodies. As shown in Figure 7A, Nup96 was very efficiently codepleted with Nup133 from both interphase and mitotic supernatants and recovered in the corresponding immune pellets, indicating that most, if not all, Nup96 is stably associated with the Nup107-160 complex during the entire cell cycle. Unlike Nup96, Nup85 and Nup37 were less efficiently depleted from these extracts (Figure 7A), a result that most likely reflects a less robust interaction of these nucleoporins with the rest of the Nup107-160 complex under our immunoprecipitation conditions.
Figure 7.
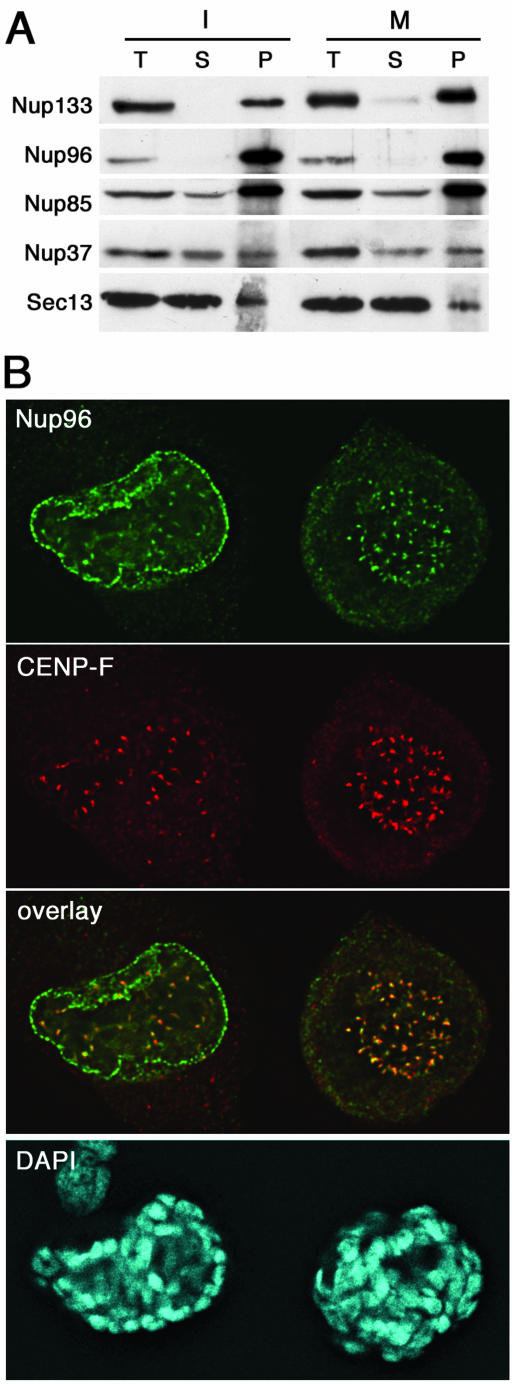
Nup96 stably interacts with the Nup107-160 complex in mitosis and is localized at kinetochores in mitotic HeLa cells. (A) Immunoprecipitation of interphase (I) or mitotic (M) HeLa S3 cell extracts by using affinity-purified anti-Nup133 antibodies. Equivalent amounts of total extracts (T) and depleted supernatants (S) and fivefold equivalents of the immune pellets (P) were analyzed by immunoblot by using antibodies directed against Nup133, Nup96, Nup85, Nup37, and Sec13. Similar coimmunoprecipitation efficiencies were obtained with these various nucleoporins from both interphase and mitotic extracts. Note the very efficient codepletion and coprecipitation of Nup96 together with Nup133. (B) Deconvolved images of HeLa cells double-labeled with affinity-purified anti-Nup96 and monoclonal anti-CENP-F antibodies. DNA was stained with DAPI. Note that Nup96 is clearly detected at kinetochores in prophase (cell on the left) and metaphase.
We then analyzed the subcellular localization of Nup96 in mitotic HeLa cells, by using an affinity-purified antibody directed against Nup96 that recognizes both Nup96 and its splice variant p87 (Fontoura et al., 1999). Although a spindle labeling was clearly detected when using the nonpurified serum, the affinity-purified serum no longer recognized the spindle and instead revealed a clear labeling of kinetochores in metaphase (Figure 7B, right). Similar results were also obtained using another anti-Nup96 serum (Hase and Cordes, 2003) (our unpublished data). As shown in Figure 7B, the Nup96/p87 labeling partially overlapped the staining obtained with anti-CENP-F and was juxtaposed to the labeling obtained with the CREST serum that labels constitutive constituents of centromeres (our unpublished data). As also previously observed with other constituents of the Nup107-160 complex, a specific labeling of Nup96 at kinetochores was already observed during prophase, when Nup96 is mainly localized at the nuclear envelope (Figure 7B, left).
A Fraction of Sec13 Associates with Kinetochores
For GFP3-Sec13, most of its labeling reflected the expected localization of this COPII constituents at ER exit sites and vesicular transport structures (Tang et al., 2000). A faint signal could also be observed at the NE in both live and fixed interphase HeLa cells (Figure 8, A and B). This dual ER/NE localization is in agreement with the biochemical interaction of Sec13 with both Sec31, a constituent of the COPII complex (Tang et al., 2000; see also Figure 3), and the various constituents of the Nup107-160 NPC subcomplex (Vasu et al., 2001; Enninga et al., 2003; see also Figure 3). In addition, a very faint signal also could be observed at kinetochores in live mitotic cells (Figure 8A, inset). To further characterize this labeling, GFP3-Sec13-transfected HeLa cells were analyzed by immunofluorescence by using the human autoimmune CREST serum and affinity-purified anti-Nup133 antibodies. As shown in Figure 8B, deconvolved images of such cells revealed the nearly perfect colocalization of the GFP3-Sec13 and Nup133 labeling at kinetochores, and their partial overlap with the CREST signal. The remaining mitotic GFP3-Sec13 signal, including peripheral vesicular structures, overlapped with the signal obtained with the Sec31 antibody (Supplemental Figure S3, arrows). Noteworthy, no NE or kinetochore labeling could be detected with the Sec31 antibody (Supplemental Figure S3). This result correlates with the lack of Sec31 in the GFP-Nup43 immunepellet that contains the constituents of the Nup107-160 complex (Figure 3), suggesting that other COPII components are not colocalizing with the Nup107-160 complex.
As an alternative approach to localize Sec13, we also used affinity-purified anti-Sec13 antibodies. As shown in Figure 8C, this study confirmed the specific localization of a fraction of endogenous Sec13 at both the nuclear pores and kinetochores. Note, as already observed for other members of the Nup107-160 complex (see Figure 6, Supplemental Figure S2, and Belgareh et al., 2001) that the labeling of Sec13 at kinetochores persists in early anaphase.
DISCUSSION
Seh1 Behaves as a Constituent of the Nup107-160 Complex
The vertebrate Nup107-160 NPC subcomplex was so far demonstrated to be composed of at least six distinct nucleoporins, including the WD-repeat containing Sec13 protein. In this study, we aimed to identify the additional WD-repeat nucleoporins that belong to this vertebrate NPC subcomplex. Although the S. cerevisiae WD-repeat nucleoporin Seh1 is a stable constituent of the ScNup84 complex, we could not convincingly demonstrate the presence of human Seh1 in the Nup107-160 complex by using biochemical approaches. Yet, both GFP-Seh1 and myc6-Seh1 were properly targeted to the NPC during interphase. Comparison of the human and S. cerevisiae Seh1 protein sequences predicts a more basic pKi for human Seh1 (8.97) compared with ScSeh1 (theoretical pKi = 5.58). With the exception of Nup43 (theoretical pKi = 7.8), all the other constituents of the Nup107-160 complex have pKi in the range of 5-5.6. Whether this feature could explain the extremely weak interaction of GFP-Seh1 and myc6-Seh1 with the Nup107-160 complex in our immunoprecipitation experiments remains to be determined. The fact that human Seh1 may be lost during our purification processes without affecting the assembly of the other constituents of this complex, suggests that this WD nucleoporin is likely to be a peripheral constituent of the vertebrate Nup107-160 complex. Noteworthy, S. cerevisiae Nup133 was not initially identified in the ScNup84 complex, and its interaction with components of this complex could only be demonstrated in vitro (Lutzmann et al., 2002) or by immunoprecipitations performed after enrichment of the complex on Nup100-coated beads (Allen et al., 2002). In contrast, vertebrate Nup133 seems to be stably associated with the Nup107-160 complex. These data thus likely reflect species-dependent behavior of individual constituents within the evolutionarily conserved ScNup84/vertebrate Nup107-160 complex.
The link between Seh1 and the Nup107-160 complex was further strengthened by the fact that Seh1 knockdown by RNAi induces phenotypes reminiscent to those induced by partial depletion of Nup133, Nup107, or Nup85 (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003a), most prominently the reduction of many nucleoporins at NPCs. The consequences of Seh1 depletion on the delocalization of various nucleoporins from the NE was less pronounced compared with the effect of Nup133 or Nup107 siRNAs, suggesting a minor contribution of Seh1 to the overall function of the Nup107-160 complex in human cells. Yet, this phenotype was unexpected, because deletion of the Seh1 ortholog in S. cerevisiae, while affecting cell growth particularly at lower temperatures, does not induce any of the phenotypes observed for other constituents of the ScNup84 complex, namely, NPC clustering and growth defect at 37°C (Siniossoglou et al., 1996). While a decreased mAb414 labeling was recently reported in gp210-depleted cells (Cohen et al., 2003), such defects were not observed with siRNA duplexes targeting Tpr or Nup358/RanBP2 (Hase and Cordes, 2003; Salina et al., 2003). Although no alteration or even a modest increase of Nup107 labeling was observed at 3-4 d after Nup153 depletion (Hase and Cordes, 2003), these studies showed a late decrease of Nup107 NE staining intensity concomitant to morphological alterations of the Nup153-deficient nuclei. In addition, we noticed a cytoplasmic accumulation of SC-35 foci that correlated with a minor increase of G1-like cells in some Nup133-, Nup107- (Walther et al., 2003a; our unpublished data), Seh1-, but also Nup153-depleted cells. These common late phenotypes might thus reflect the interaction between Nup153 and the Nup107-160 complex (Vasu et al., 2001) and/or their common involvement in NPC assembly at the end of mitosis (Walther et al., 2003b). Alternatively, and as recently described in the case of gp210 depletion, these late phenotypes could be less specific and reflect a population of sub-G1 cells at an early stage of apoptosis (Cohen et al., 2003).
Noteworthy, the kinetic of GFP-Seh1 turnover at the NPCs is indistinguishable from the ones determined for the other constituents of the Nup107-160 complex, including Nup133 and Nup107 (Belgareh et al., 2001), Nup85, Sec13, Nup37, and Nup43 (Rabut, Doye, and Ellenberg, unpublished data). In particular, despite some common phenotypes induced by their depletion, the extremely stable association of Seh1 with NPCs clearly differs from the dynamic behavior of Nup153 or gp210 at NPCs (Rabut, Doye, and Ellenberg, unpublished data). Finally, as the other constituents of the Nup107-160 complex, and unlike most other nucleoporins, a fraction of GFP-Seh1 is specifically localized at kinetochores during mitosis. Together, these features strongly suggest that Seh1 behaves as a constituent of the Nup107-160 complex.
The Human Nup107-160 Complex Contains Two Higher Eukaryote-specific WD-Repeat Nucleoporins, Nup37 and Nup43
Our study further revealed the stable interaction of two recently identified eukaryote-specific WD nucleoporins, Nup37 and Nup43, with the Nup107-160 complex. These data at first strengthen the identity of these two proteins as bona fide nucleoporins, which was so far based on their copurification with enriched NPCs and their punctuate nuclear rim colocalization with the mAb414 antibody at the nuclear periphery (Cronshaw et al., 2002). We further show that the entire cellular fraction of Nup37 comigrates with other constituents of the Nup107-160 complex on sucrose gradient and upon gel filtration and is as efficiently coprecipitated with Nup133 as other previously characterized constituents of this complex, such as Nup85. This indicates that most, if not all, Nup37, is associated with this NPC subcomplex. The interaction of these two-higher eukaryote specific nucleoporins with the Nup107-160 complex initially raised the hypothesis that they could belong to distinct complexes, that would specify for instance a cytoplasmic versus a nuclear form of the Nup107-160 complex. This is, however, not the case as Nup37 and Nup43 efficiently coprecipitate with each other as well as with Sec13. Our data thus provide strong evidence for the existence of a single complex that contains three, and most likely four (including Seh1) various distinct WD-repeat nucleoporins. This means that among the so far 30 nucleoporins identified in vertebrates, nearly one-third can now be assigned to the Nup107-160 complex that thus represents a major building block of the vertebrate NPC. The lack of coimmunoprecipitation of endogenous Nup107 with GFP-Nup107 (Figure 1C) and of endogenous Nup133, Nup85, and Nup37, with their corresponding GFP-tagged variant (our unpublished data), indicate that the Nup107-160 complex likely contains a single copy of each of these nucleoporins.
In S. cerevisiae, Sec13 and Seh1 were, respectively, found to interact directly with Nup145-C (the S. cerevisiae Nup96 counterpart) and Nup85. Recently, it also was demonstrated that mammalian Sec13 interacts with Nup96 (Enninga et al., 2003). Although it thus seems likely that vertebrate Seh1 associates with Nup85, the specific constituents of the Nup107-160 complex that directly interact with Nup37 and Nup43 remain to be identified. WD repeat propeller structures have been proposed to create a stable platform that can form reversible complexes with several proteins, thus coordinating sequential and/or simultaneous interactions (Smith et al., 1999). Although a possible function of these nucleoporins could be to stabilize interactions within the Nup107-160 complex, they may also interact with other nucleoporins that do not belong to this complex, or with specific transport factors. Whereas potential orthologs of Nup37 and Nup43 can be clearly identified in the Drosophila genome database (as the products of CG11875 and CG7671, respectively; our unpublished data), these two nucleoporins have no identifiable yeast orthologs (Cronshaw et al., 2002), a feature that suggests a function restricted to higher eukaryotic NPCs. In view of the recently described role of the Nup107-160 complex in vertebrates, this specific function could be NPC disassembly and reassembly at the end of mitosis. Yet, the fact that no orthologs of these two nucleoporins could so far be identified in C. elegans (Galy et al., 2003) that displays an open mitosis make this hypothesis less likely. siRNA approaches and two hybrid screens aimed at identifying the various partners of these nucleoporins may help to further unravel the function of these higher eukaryotes specific WD-repeat nucleoporins in the assembly or function of the Nup107-160 complex.
The Nup107-160 Complex Seems to be Targeted as One Entity to Kinetochores
The Nup107-160 complex, purified from interphase extract, retains its integrity in mitosis. Indeed, we previously demonstrated that four of its constituents, namely, Nup133, Nup107, Nup160, and Nup96 are stably associated when purified from mitotic cells (Belgareh et al., 2001). In addition, Nup85 and Sec13 were reported previously to coimmunoprecipitate with the other members of the Nup107-160 complex in Xenopus egg extract that are arrested at a metaphase-like stage (Vasu et al., 2001; Harel et al., 2003). In this study, we could further demonstrate that human Nup85, Nup37, and a fraction of Sec13 are as stably associated with the Nup107-160 complex when purified from interphase or mitotic extracts.
In addition to their biochemical interaction in mitotic extracts, we could demonstrate that all Nup107-160 constituents are associated with kinetochores. In the case of Sec13, the kinetochore signal was rather difficult to observe over the cytoplasmic background, a feature that was anticipated if one considers that only the small fraction of Sec13 that belongs to the Nup107-160 complex is targeted to kinetochores. The targeting of these GFP-tagged nucleoporins to kinetochores was specific, because it was not detected with Aladin, the unique WD-repeat nucleoporin that does not belong to the Nup107-160 complex. Similarly, no detectable signal could be observed at kinetochores with GFP-Nup35. Such a signal could have been expected because the S. cerevisiae ortholog of this nucleoporin, Nup53, associate with Mad1 and Mad2 in yeast (Iouk et al., 2002), and vertebrates Mad1 localizes to kinetochores in mitosis (Campbell et al., 2001).
The constituents of this complex further display a similar kinetics of targeting to this structure: they could all be already detected from prophase until anaphase (Figures 6, 7, and 8C, and Supplemental Figure S2; our unpublished data). Accordingly, the Nup107-160 subcomplex is likely to be targeted as one entity to kinetochores. The persistence of this signal during anaphase, a feature previously reported for Nup133, distinguishes this subset of NPC/kinetochore constituents from checkpoint markers such as Mad1 and Mad2 that also associate with NPCs during interphase.
Because it is anticipated that ∼16 copies of this symmetric subcomplex are present at each NPC (Cronshaw et al., 2002), and considering that the kinetochore signal is at least as bright, and frequently brighter than the signal recorded for one individual NPC (G. Rabut and J. Ellenberg, unpublished data), one can expect the presence of multiple Nup107-160 complexes per kinetochore. Accordingly, the Nup107-160 complex may represent a significant building block of vertebrate kinetochores. Knock-down of Nup358, another protein that displays a dual localization at both NPCs and kinetochores recently revealed a function of this nucleoporin in kinetochore assembly and in microtubule-kinetochore interactions (Salina et al., 2003; Joseph et al., 2004). Whether a similar function might be assigned to the Nup107-160 complex, or whether its kinetochore localization could be somehow linked to its early recruitment to mitotic chromatin in a Xenopus extracts (Walther et al., 2003a,b) and reflect mechanisms involved in the coordination between chromosome segregation and NPC reassembly during mitosis remains to be investigated.
Supplementary Material
Acknowledgments
We thank Janet Cronshaw and Mike Matunis (Johns Hopkins Bloomberg School of Public Health, Baltimore, MD), Benjamin Glick (University of Chicago, Chicago, IL), Beatriz Fontoura (University of Miami School of Medicine, Miami, FL), Bor Luen Tang and Hong Wanjin (Institute of Molecular and Cell Biology, Singapore), Volker Cordes (Karolinska Institute, Stockholm, Sweden), Ulf Nehrbass (Institut Pasteur, Paris, France) and Franck Perez (Institut Curie) for generous gifts of plasmids, siRNAs, and antibodies; Lucien Cabanié, Séverine Morizur, and Vincent Fraisier (Institut Curie) for help in antibody purification, sucrose gradient and gel filtration experiments, and deconvolution microscopy; and F. Perez and the members of V. Doye, J. Ellenberg, and M. Bornens laboratories for helpful advice and critical reading of the manuscript. V.D. was founded by the Institut Curie, the Centre National dela Recherche Scientifique, the Association pour la Recherche contre le Cancer, and la Ligue Contre le Cancer (Comité deParis). A.A. and G.R. were, respectively, supported by fellowships through the Ministère délégué à la Recherche et aux nouvelles Technologies and the European Molecular Biology Laboratory International Ph.D. Programme. J.E. acknowledges support from the German Research Council (DFG, EL 246/1-1) and the Human Frontiers Science Program (RGP0031/2001-M).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0878. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-12-0878.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Allen, N.P.C., Patel, S.S., Huang, L., Chalkley, R.J., Burlingame, A., Lutzmann, M., Hurt, E.C., and Rexach, M. (2002). Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1, 930-946. [DOI] [PubMed] [Google Scholar]
- Askjaer, P., Galy, V., Hannak, E., and Mattaj, I.W. (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans Embryos. Mol. Biol. Cell 13, 4355-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, J.R., Jeganathan, K.B., Baker, D.J., Wu, X., Kang-Decker, N., and Van Deursen, J.M. (2003). Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160, 341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., et al. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer, T., Enninga, J., Dales, S., Blobel, G., and Zhong, H. (2003). Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 100, 981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, M., Chan, G., and Yen, T. (2001). Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114, 953-963. [DOI] [PubMed] [Google Scholar]
- Cohen, M., Feinstein, N., Wilson, K.L., and Gruenbaum, Y. (2003). Nuclear pore protein gp210 is essential for viability in HeLa cells and Caenorhabditis elegans. Mol. Biol. Cell 14, 4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin, J., Hartnell, L., Imreh, G., Hallberg, E., Lippincott-Schwartz, J., and Ellenberg, J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye, V., and Hurt, E. (1997). From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9, 401-411. [DOI] [PubMed] [Google Scholar]
- Elbashir, S., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Enninga, J., Levay, A., and Fontoura, B.M. (2003). Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Cell. Biol. 23, 7271-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., Blobel, G., and Matunis, M.J. (1999). A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-Kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144, 1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., Mattaj, I.W., and Askjaer, P. (2003). C. elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell 14, 5104-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Dang, T., Pane, N., Shevchenko, A., Mann, M., Forbes, D., and Hurt, E. (1997). Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell 8, 2017-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, A.T., and Glick, B.S. (2000). Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11, 3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S., Bechert, K., Tuschl, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- Harel, A., Orjalo, A.V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., Zimmerman, E., Elbaum, M., and Forbes, D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11, 853-864. [DOI] [PubMed] [Google Scholar]
- Hase, M.E., and Cordes, V.C. (2003). Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14, 1923-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui, A.E., Furuya, K., Yanagida, M., and Matsumoto, T. (2002). Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115, 1603-1610. [DOI] [PubMed] [Google Scholar]
- Iouk, T., Kerscher, O., Scott, R.J., Basrai, M.A., and Wozniak, R.W. (2002). The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159, 807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M., Schallhorn, A., and Wurm, F.M. (1996). Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., Liu, S.T., Jablonski, S.A., Yen, T.J., and Dasso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611-617. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Tan, S., Karpova, T., McNally, J., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, N., Sandblad, L., Hase, M., Hunziker, A., Hergt, M., and Cordes, V. (2002). The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111, 236-255. [DOI] [PubMed] [Google Scholar]
- Lennon, G.G., Auffray, C., Polymeropoulos, M., and Soares, M.B. (1996). The I. M. A. G. E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33, 151-152. [DOI] [PubMed] [Google Scholar]
- Lutzmann, M., Kunze, R., Buerer, A., Aebi, U., and Hurt, E. (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21, 387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Wu, J.A., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizak, C., Martin-Lluesma, S., Moutel, S., Roux, A., Kreis, T.E., Goud, B., and Perez, F. (2003). Recombinant antibodies against subcellular fractions used to track endogenous Golgi protein dynamics in vivo. Traffic 4, 739-753. [DOI] [PubMed] [Google Scholar]
- Pritchard, C.E., Fornerod, M., Kasper, L.H., and van Deursen, J.M. (1999). RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 145, 237-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner, J.B., Rao, A., Fritzler, M.J., Valencia, D.W., and Yen, T.J. (1993). CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskeleton 26, 214-226. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., Enarson, P., Rattner, J.B., and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield, P., Garrard, S., and Derewenda, Z. (1999). Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34-39. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., Lutzmann, M., Santos-Rosa, H., Leonard, K., Mueller, S., Aebi, U., and Hurt, E. (2000). Structure and assembly of the Nup84p complex. J. Cell Biol. 149, 41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Wimmer, C., Rieger, M., Doye, V., Tekotte, H., Weise, C., Emig, S., Segref, A., and Hurt, E.C. (1996). A novel complex of nucleoporins, wich includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84, 265-275. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181-185. [DOI] [PubMed] [Google Scholar]
- Spector, D.L., Fu, X.-D., and Maniatis, T. (1991). Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S.R. (2003). Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775-789. [DOI] [PubMed] [Google Scholar]
- Tang, B.L., Peter, F., KrijnseLocker, J., Low, S.H., Griffiths, G., and Hong, W.J. (1997). The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol. Cell. Biol. 17, 256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B.L., Zhang, T., Low, D.Y.H., Wong, E.T., Horstmann, H., and Hong, W. (2000). Mammalian Homologues of Yeast Sec31p. An ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J. Biol. Chem. 275, 13597-13604. [DOI] [PubMed] [Google Scholar]
- Teixeira, M.T., Siniossoglou, S., Podtelejnikov, S., Benichou, J.C., Mann, M., Dujon, B., Hurt, E., and Fabre, E. (1997). Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16, 5086-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, D.L., and Weintraub, H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434-1447. [DOI] [PubMed] [Google Scholar]
- Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W.H., and Forbes, D.J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155, 339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., et al. (2003a). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113, 195-206. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Askjaer, P., Gentzel, M., Habermann, A., Griffiths, G., Wilm, M., Mattaj, I.W., and Hetzer, M. (2003b). RanGTP mediates nuclear pore complex assembly. Nature 424, 689-694. [DOI] [PubMed] [Google Scholar]
- Wang, X., Babu, J.R., Harden, J.M., Jablonski, S.A., Gazi, M.H., Lingle, W.L., de Groen, P.C., Yen, T.J., and van Deursen, J.M. (2001). The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLEBS-containing proteins. J. Biol. Chem. 276, 26559-26567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.