Abstract
In animal and yeast cells, the mitotic spindle is aligned perpendicularly to the axis of cell division. This ensures that sister chromatids are separated to opposite sides of the cytokinetic actomyosin ring. In fission yeast, spindle rotation is dependent upon the interaction of astral microtubules with the cortical actin cytoskeleton. In this article, we show that addition of Latrunculin A, which prevents spindle rotation, delays the separation of sister chromatids and anaphase promoting complex-mediated destruction of spindle-associated Securin and Cyclin B. Moreover, we find that whereas sister kinetochore pairs normally congress to the spindle midzone before anaphase onset, this congression is disrupted when astral microtubule contact with the actin cytoskeleton is disturbed. By analyzing the timing of kinetochore separation, we find that this anaphase delay requires the Bub3, Mad3, and Bub1 but not the Mad1 or Mad2 spindle assembly checkpoint proteins. In agreement with this, we find that Bub1 remains associated with kinetochores when spindles are mispositioned. These data indicate that, in fission yeast, astral microtubule contact with the medial cell cortex is monitored by a subset of spindle assembly checkpoint proteins. We propose that this checkpoint ensures spindles are properly oriented before anaphase takes place.
INTRODUCTION
In eukaryotes, the separation of sister chromatids is mediated by the interaction of spindle microtubules with specialized regions of chromosomes known as kinetochores. At the start of prometaphase, kinetochores are not attached to microtubules. The kinetochore of one sister chromatid then captures a microtubule nucleated from a spindle pole. Once its sister kinetochore has captured microtubules from the other pole, the chromosome becomes bioriented. During metaphase, all bioriented chromosomes move to the equatorial plane, known in animal cells as the metaphase plate, in a process called chromosome congression (Rieder and Salmon, 1994). At anaphase, cohesion is lost, allowing sister chromatids to separate to their respective poles. Finally, the cytokinetic actomyosin ring contracts perpendicularly to, and at the site of, the spindle midzone to ensure that each set of sister chromatids is separated to daughter cells.
Cell cycle progression in all eukaryotic cells is monitored by a series of checkpoints that ensure both the fidelity and the temporal and spatial order of cell cycle events (Hartwell and Weinert, 1989). One of the best studied of these is a checkpoint that monitors the assembly of the mitotic spindle (Yu, 2002; Zhou et al., 2002; Cleveland et al., 2003). Components of this checkpoint, which is often referred to as the spindle assembly checkpoint (SAC), were first identified in budding yeast and include the Mad1, Mad2, Mad3, Bub1, Bub3, and Mps1 proteins (Li and Murray, 1991; Hoyt et al., 1991; Weiss and Winey, 1996). Structural and functional homologues of these proteins have been identified in all other eukaryotes so far examined, including fission yeast (He et al., 1997, 1998; Bernard et al., 1998; Ikui et al., 2002; Millband and Hardwick, 2002). In response to microtubule-disrupting agents, these proteins translocate to kinetochores and inhibit the anaphase promoting complex (APC) (Chen et al., 1996; Li and Benezra, 1996), an E3 ubiquitin ligase that is responsible for the destruction not only of Cyclin B but Securin, an inhibitor of Separase (Funabiki et al., 1996b; Zachariae and Nasmyth, 1999). Separase cleaves Scc1/Rad21, a component of the Cohesin complex that holds sister chromatids together (Uhlmann et al., 1999; Tomonaga et al., 2000; Uhlmann et al., 2000). Microtubule-disrupting agents thus block anaphase onset by inhibiting the activation of Separase.
The molecular nature of the defect that is sensed at kinetochores is not clear. Two models have been put forward. In the first (attachment model), anaphase is initiated when all kinetochores are bound to a bipolar spindle (Rieder et al., 1994, 1995). In the second (tension model), anaphase is initiated only when balanced tension is exerted upon kinetochores when chromosomes congress to the metaphase plate (McIntosh, 1991, Li and Nicklas, 1995; Stern and Murray, 2001). Originally, it was thought that Mad2 was the key effector of the SAC, because Mad2 interacts with and inhibits Cdc20 (Fizzy/Slp1), an activator of the APC (Hwang et al., 1998; Kim et al., 1998). However, the situation now seems more complex. Although heterodimeric complexes of Mad1-Mad2, BubR1(Mad3)-Bub3, and Bub1-Bub3 exist in interphase cells, higher order complexes are formed in mitosis and in response to microtubule-disrupting agents (Chen et al., 1999; Hardwick et al., 2000; Millband and Hardwick, 2002). In particular, a mitotic checkpoint complex has been purified from human cells that contains the Mad2, Cdc20, BubR1(Mad3), and Bub3 proteins and is a significantly more potent inhibitor of APC in vitro than Mad2 alone (Sudakin et al., 2001; Fang, 2002). A similar complex, containing Mad2, Cdc20, Mad3, and Bub3, has been identified in yeast (Hardwick et al., 2000; Millband and Hardwick, 2002). Independently, Tang et al. (2001) have isolated a distinct inhibitory complex that contains Cdc20, BubR1, and Bub3 but not Mad2. Intriguingly some spindle assembly checkpoint proteins, including Bub1, BubR1(Mad3), and Bub3 are present at kinetochores as chromosomes congress at the metaphase plate, whereas Mad1 and Mad2 are not (Waters et al., 1998; Hoffman et al., 2001). One possibility is that a distinct set of SAC components monitor bipolar attachment of kinetochores to spindle poles and chromosome congression.
We previously proposed that, in fission yeast, a spindle orientation checkpoint (SOC) delays anaphase onset when astral microtubule interaction with the medial cell cortex and thereby spindle rotation is disturbed (Gachet et al., 2001, 2004). Recently, Rajagopalan et al. (2004) have suggested, by analysis of a mutant in the kendrin-like protein Pcp1, that metaphase spindle alignment is monitored at the spindle pole by the Bub1 and Mph1 but not Mad1, Mad2, Mad3, or Bub3 checkpoint proteins. This is curious because first, both Mad3 and Bub3 are necessary for Mps1(Mph1) to impose a metaphase arrest in budding yeast (Hardwick et al., 1996), and second, Bub3 is required both for the association of Bub1 to kinetochores and for the checkpoint function of Bub1 in yeast and mammalian cells (Farr and Hoyt, 1988; Taylor et al., 1988; Warren et al., 2002; Gillett et al., 2004). In this article, we have reassessed the mechanism by which spindle position is monitored in fission yeast by analyzing kinetochore dynamics when contact of astral microtubules with the medial cell cortex is disturbed.
MATERIALS AND METHODS
Cell Culture
Media, growth, and maintenance of strains were as described previously (Moreno et al., 1991). Strains used in this study are listed in Table 1. Cell cultures were grown at 28°C in YES unless otherwise stated. Latrunculin A was purchased from Molecular Probes (Eugene, OR), dissolved in dimethyl sulfoxide at a stock concentration of 1 mg/ml. All strains were leu1-32 ura4-D18 unless otherwise stated. ade6- is either ade6-M210 or ade6-M216. Cultures of nmt1-gfp-atb2 cells were grown at 28°C in minimal medium, and thiamine was added at a concentration of 3.75g/l to partially repress expression of nmt1-gfp-atb2. In these conditions, nmt1-gfp-atb2 cells grew at a rate indistinguishable from wild type.
Table 1.
Strains used in this study
| Strain no. | Genotype | Source |
|---|---|---|
| JM2418 | h+ plo1-GFP:kanR | D. McCollum |
| YG 309 | h− GFP-nmt1-atb2(lys1) | D-Q. Ding |
| JM2591 | h− ndc80-GFP:kanR | J. Kilmartin |
| JM2590 | h+ cdc13-GFP(LEU2) | M. Yanagida |
| JM2564 | h+ cut2-364:cut2-GFP(LEU2)ura4+ | M. Yanagida |
| JM2379 | h+ mad1 :: ura4 | T. Matsumoto |
| AE148 | h− mad2 :: ura4 | T. Matsumoto |
| JM2324 | h− mad3 :: ura4 | K. Hardwick |
| JM2323 | h− bub1 :: ura4 | K. Hardwick |
| JM2325 | h− bub3 :: ura4 | K. Hardwick |
| SS560 | h− mph1 :: ura4 ade6- | S. Sazer |
| JM2747 | h+ bub1(K762M) lys1-1 his7-366 ade6- | J.-P. Javerzat |
| JM2748 | h+ mad2-GFP(LEU2) ura4-D18 | T. Toda |
| MA239 | h+ bub1-GFP:kanR | T. Toda |
| JM2608 | h+ cdc11-CFP:kanR | This study |
| JM2746 | h+ cut2-GFP(LEU2) cdc11-CFP:kanR | This study |
| JM2763 | h− ndc80-GFP:kanR cdc11-CFP:kanR | This study |
| JM2751 | h− mad2 :: ura4 cdc13-GFP(LEU2) | This study |
| JM2752 | h+ bub1 :: ura4 cdc13-GFP(LEU2) ade6- | This study |
| JM2891 | h− mad1 :: ura4 ndc80-GFP:kanR cdc11-CFP :: kanR | This study |
| JM2749 | h+ mad2 :: ura4 ndc80-GFP:kanR cdc11-CFP:kanR | This study |
| JM2892 | h− mad3 :: ura4 ndc80-GFP:kanR cdc11-CFP :: kanR | This study |
| JM2750 | h+ bub1 :: ura4 ndc80-GFP:kanR cdc11-CFP:kanR | This study |
| JM2889 | h− bub3 :: ura4 ndc80-GFP:kanR cdc11-CFP:kanR | This study |
| JM2890 | h+ mph1 :: ura4 ndc80-GFP:kanR cdc11-CFP :: kanR | This study |
| JM2896 | h− bub1(K762M) ndc80-GFP:kanR cdc11-CFP :: kanR | This study |
| JM2757 | h− mad2-GFP(LEU2) ndc80-CFP:kanR | This study |
| JM2758 | h− mad2-GFP(LEU2) cdc11-CFP:kanR | This study |
| JM2759 | h− bub1-GFP:kanR ndc80-CFP:kanR | This study |
| JM2760 | h+ bub1-GFP:kanR cdc11-CFP:kanR ade6- | This study |
| JM2761 | h+ cps8-188 ndc80-GFP:kanR | This study |
| JM2762 | h− cdc11-123 ndc80-GFP:kanR ade6- | This study |
| JM2808 | h− bub1 :: ura4 cps8-188 ndc80-GFP:kanR | This study |
| JM2809 | h− bub1 :: ura4 cdc11-123 ndc80-GFP:kanR | This study |
All strains are leu1-32 ura4-D18 unless otherwise stated. ade6- is either ade6-M210 or ade6-M216.
Epitope Tagging
Carboxy-terminal green fluorescent protein (GFP) and cyan fluorescent protein (CFP) epitope tagging of Cdc11p and Ndc80p were done by polymerase chain reaction-based gene targeting (Bähler et al., 1998). The growth rates of cdc11-CFP, ndc80-GFP, and cdc11-CFP ndc80-GFP cells were indistinguishable from wild type. Compound tagged and mutant strains were constructed by standard genetic methods (Moreno et al., 1991).
Cell Synchronization
Cell synchrony was achieved either by centrifugal elutriation or lactose gradient size selection. Cells were resuspended in fresh medium at 106 cells/ml and released at 28°C unless otherwise stated. The peak synchrony of septation was >35% in each experiment in the absence of drug.
Cell Fixation
plo1-GFP, ndc80-GFP, ndc80-CFP, cdc11-CFP, mad2-GFP, and bub1-GFP cells were fixed in 3.7% formaldehyde for 10 min at room temperature. cdc13-GFP cells were fixed in 100% cold methanol for 8 min and washed in phosphate-buffered saline. cut2-GFP cells were observed by live imaging. Actin rhodamine-phalloidin staining and tubulin immunofluorescence were performed as described previously (Marks and Hyams, 1985).
Microscopy
Live analysis of cells was performed in an imaging chamber (CoverWell PCI-2.5; Grace Bio-Labs, Bend, OR) filled with 1 ml of 1% agarose in minimal medium with or without 1.25 μm Latrunculin A and sealed with a 22 × 22-mm glass coverslip. Fluorescence microscopy was performed on a Deltavision Spectris system containing a photometrics CH350L liquid cooled charge-coupled device camera and Olympus IX70 inverted microscope with a 100× objective equipped with Deltavision data collection system (Applied Precision, Issaquah, WA). Stacks of six Z-sections (0.35 μm apart) were taken at each time point with exposure times of 1 s for both GFP and CFP. Projected images were made for each time point followed by intensity adjustments and conversion to 24 bit TIFF images. The position of the spindle poles and kinetochores were determined using OpenLab software (Improvision, Coventry, United Kingdom) and downloaded to Microsoft Excel for analysis.
RESULTS
The SOC Is Activated by Selective Disruption of Actin Cables
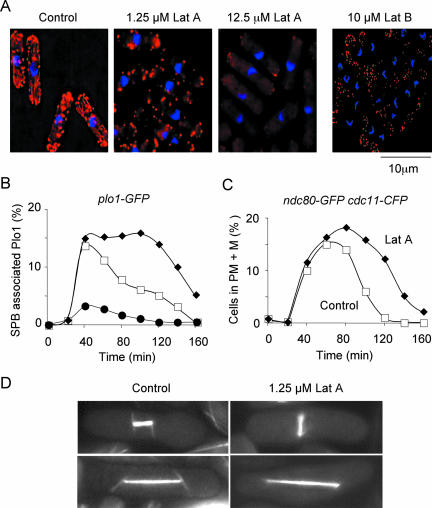
Latrunculin A and Latrunculin B are potent inhibitors of actin polymerization in vivo (Spector et al., 1983; Ayscough et al., 1997). There have been several seemingly contradictory reports of the effects of Latrunculin A and Latrunculin B on cell cycle progression in fission yeast. For example, we have previously shown that addition of 10 μM Latrunculin B to synchronized cultures of wild-type fission yeast cells does not block spindle formation but causes cells to delay in mitosis with unseparated sister chromatids and misoriented spindles (Gachet et al., 2001, 2004). We attributed this effect to activation of a SOC. Conversely, other researchers have shown that addition of 10 μM Latrunculin A blocks the G2/M transition by preventing cell growth (Rupes et al., 2001). To resolve this issue, we examined the effect of various concentrations of Latrunculin A on actin structures and mitotic progression in synchronous wild-type plo1-GFP cells. In the absence of any treatment, both actin cables and actin patches are clearly visible. In the presence of 1.25 μM Latrunculin A, actin cables were absent although actin patches were still evident, particularly at the cell tips (Figure 1A). A similar pattern was observed when cells were treated with 10 μM Latrunculin B, the concentration used in our previous studies (Figure 2A; Gachet et al., 2001, 2004). In the presence of 12.5 μM Latrunculin A, both actin cables and patches were absent (Figure 1A). Whereas addition of 12.5 μM Latrunculin A blocked cell growth and the appearance of Plo1 kinase on spindle poles, addition of 1.25 μM Latrunculin A did not block mitotic entry but delayed cells in mitosis with Plo1 associated to both spindle poles separated by ∼2.0-2.5 μm (Figure 1B). To examine more accurately the mechanism by which spindle misorientation regulates the timing of anaphase A, we constructed a strain that expresses a centromere-associated protein, Ndc80, tagged with GFP and a spindle pole body-associated protein, Cdc11, tagged with CFP (Krapp et al., 2001; Wigge and Kilmartin, 2001). This strain enabled us to measure the period of time cells remained in prometaphase and metaphase (with kinetochores between but not adjacent to separated spindle poles). Addition of 1.25 μM Latrunculin A did not delay separation of spindle poles, confirming that the G2/M transition is not affected under these conditions (Figure 1C). However, in the presence of 1.25 μM Latrunculin A cells remain in prometaphase or metaphase for approximately twice as long than in control cells (as judged by the area under the curve) (Figure 1C). Under these conditions, astral microtubules undergo more erratic phases phase or growth and catastrophe and are thus more difficult to visualize (Figure 1D). We find that, whereas 95% of spindles were aligned within 30° of the longitudinal axis at anaphase onset in control cells, only 51% of spindles were properly oriented in the presence of 1.25 μM Latrunculin A. This confirms our previous observations that disruption of the actin cytoskeleton disrupts spindle rotation (Figure 1D, Gachet et al., 2001; Gachet et al., 2004). We conclude that whereas complete disruption of the actin cytoskeleton blocks cell growth, selective disruption of actin cables inhibits spindle rotation, presumably by inhibiting the formation of the cytokinetic actomyosin ring, and delays anaphase onset without effecting the G2/M transition. These data resolve the discrepancy between previous studies (Gachet et al., 2001; Rupes et al., 2001).
Figure 1.
Selective disruption of actin cables delays the separation of sister chromatids without inhibiting mitotic entry. (A) Log phase cultures of wild-type cells were either untreated (control) or treated with 1.25 μM Latrunculin A, 12.5 μM Latrunculin A, or 10 μM Latrunculin B for 15 min and stained for actin containing structures. (B) plo1-gfp cells were synchronized and incubated in the absence (open squares) or presence of either 1.25 μM Latrunculin A (closed diamonds) or 12.5 μM Latrunculin A (closed circles). The percentage of cells with Plo1-GFP on both SPBs was determined at the times shown (n = 150). (C) Synchronized ndc80-gfp cdc11-cfp cells were incubated in the absence (open squares) or presence of 1.25 μM Latrunculin A (closed diamonds). The percentage of cells in prometaphase and metaphase (PM + M) was determined at the times indicated (n = 150). (D) Representative images of nmt1-atb2-gfp cells in mitosis either in the absence (left) or presence (right) of 1.25 μM Latrunculin A. Cells are either before (top) or after (bottom) sister chromatid separation. Microtubules were visualized by live cell imaging.
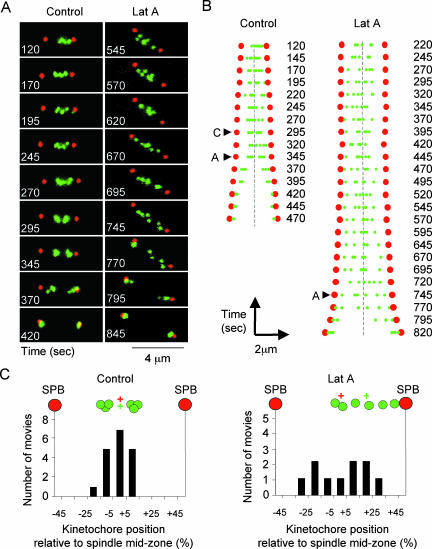
Figure 2.
Latrunculin A prevents congression of centromeres to the spindle midzone. (A) Images from a movie of ndc80-gfp cdc11-cfp cells grown either in the absence (control) (Video 1) or presence of 1.25 μM Latrunculin (Lat A) (Video 2). The localization of kinetochores (green) and spindle pole bodies (red) is shown at the times indicated. (B) Analysis of images from movies of ndc80-gfp cdc11-cfp cells grown either in the absence (control) (Video 1) or presence of 1.25 μM latrunculin (Lat A) (Video 2). The localization of kinetochores (green) and spindle pole bodies (red) is shown at the times indicated. The position of each of the six kinetochores (green circles) relative to the positions of the two spindle pole bodies (red circles) is represented at 25-s intervals both for the untreated cell (control) and that treated with Lat A. The distance between each kinetochores and spindle pole bodies is plotted on the x-axis and time is shown in the y-axis. The spindle midzone (equi-distant between the poles) is shown as a dotted line. The point of chromosome congression (c) and anaphase onset (a) are shown. (C) Measurement of the central position of kinetochores (green cross) relative to the spindle midzone (red cross) taken 50 s before the onset of anaphase in movies of ndc80-gfp cdc11-cfp cells in the absence (control, left) or presence (Lat A, right) of 1.25 μM Latrunculin A.
To circumvent the need to attain a critical cell size, Rajagopalan et al. (2004) analyzed the effect of 50 μM Latrunculin A on cell cycle progression in cdc25-22 cells and concluded that, under these conditions, Latrunculin A specifically activates the SOC. This prompted us to carry out a careful study of the effect of various concentrations of Latrunculin A on mitotic entry and progression in these cells. First, cdc25-22 cells blocked at the G2/M transition were treated with a range of Latrunculin A concentrations and then stained with phalloidin. Whereas both actin cables and actin patches were clearly visible in the absence of drug, addition of 2.5 μM Latrunculin A disrupted actin cables, although actin patches were still visible (Supplementary Figure 1A). Addition of 5 μM or higher Latrunculin A disrupted both actin cables and actin patches (Supplementary Figure 1A). To examine the effect of the various concentrations of Latrunculin A on mitotic entry and progression, cdc25-22 plo1-GFP cells were arrested in late G2 and released to the permissive temperature. Plo1 binds to spindle pole bodies (SPBs) only as cells enter mitosis and this requires activation of the Cdc2/Cdc13 kinase (Mulvihill et al., 1999). Addition of 12.5 μM Latrunculin A delayed chromosome separation (Supplementary Figure 1B) but did not affect association of Plo1 to spindle poles (Supplementary Figure 1C). However, at higher concentrations, Latrunculin A blocks the association of Plo1 to spindle poles and mitotic entry (Supplementary Figure 1, B and C). The IC50 value for this effect is 43 μM, >10 times greater than the IC50 value of Latrunculin A for the disruption of actin structures in vivo. These data strongly suggest that at high concentrations, Latrunculin A inhibits mitotic entry by a mechanism that is unrelated to its effects as an inhibitor of actin polymerization.
Latrunculin Impedes Congression of Centromeres to the Spindle Midzone
To examine specifically the role of spindle mispositioning on mitotic progression, all further experiments were performed with 1.25 μM Latrunculin A. We previously showed that separation of chromosome arms, as judged by examining the separation of a GFP marker integrated 30 kb from centromere 1 (lys1:lacO his7:GFP-NLS-lacI), is inhibited by addition of 10 μM Latrunculin B (Gachet et al., 2001). To investigate the effect of 1.25 μM Latrunculin A on mitotic progression, we filmed kinetochore dynamics during mitosis in ndc80-gfp cdc11-cfp cells. Single cell analysis revealed that cells remained in prometaphase and metaphase for 11 ± 2 min, and this was extended to 20 ± 3 min in the presence of 1.25 μM Latrunculin A (Figure 2). In the absence of drug, kinetochores make rapid oscillatory movements between the spindle poles during phase 2 (Figure 2A and Video 1). The position of each kinetochore was measured by fluorescence intensity and mapped relative to the position of the two spindle pole bodies for each frame of the movie (Figure 2B and Video 1), showing that the six kinetochores congress into two bunches of three either side of the spindle midzone ∼50 s before the onset of anaphase A (Figure 2, A and B). The position of the kinetochores at congression was within the central 30% of the mitotic spindle in 17 of 18 cells filmed (Figure 2C). When anaphase takes place, each set of three sister chromatids arrive at their respective spindle pole bodies simultaneously. In the presence of Latrunculin A, kinetochores continue to oscillate between the spindle poles, suggesting that kinetochores are still under tension and no congression to the spindle midzone was observed before anaphase onset (Figure 2, A and B, and Video 2). In addition the spindle was longer (3.5 μm) when anaphase took place. In 10 independent movies, kinetochores were found to separate at more diverse positions on the mitotic spindle (Figure 2C). These results demonstrate that centromeres congress to the spindle midzone before anaphase onset, and this is disrupted when the actin cytoskeleton is disturbed.
Latrunculin Inhibits Degradation of Spindle-associated Cyclin B and Securin
In a previous study, we found that 10 μM Latrunculin B delays the degradation of Rad21, a component of the cohesin complex, suggesting that activation of Separase (Cut1) is delayed (Gachet et al., 2001). However, Latrunculin B did not inhibit the bulk degradation of either Cut2 or Cdc13 (Cyclin B), as judged by Western blots of cell extracts (Gachet et al., 2001). This led us to believe that Latrunculin B does not inhibit the action of APC. However, we could not discount the possibility that a small population of Cdc13 and Cut2 may remain undegraded during the delay or that Cdc13 and Cut2 relocalize to an insoluble fraction during mitosis. To address this question we analyzed Cdc13 and Cut2 levels microscopically using synchronized cdc13-CFP and cut2-GFP strains (Tatebe et al., 2001). In late G2 a strong signal of Cdc13 is observed in both chromatin and nucleolar domains, as observed previously (Alfa et al., 1990; Tatebe et al., 2001). As cells enter mitosis, the strong nucleolar staining diminishes, leaving a weaker nuclear signal (Figure 3, A and B). At the same time, Cdc13 concentrates at the unseparated spindle pole bodies. Cdc13 remains associated with chromatin and on the spindle and spindle poles until just before anaphase when it disappears, suggesting that Cdc13-GFP is effectively ubiquitinated by APC at this time (Alfa et al., 1990; Tatebe et al., 2001; Figure 3, A and B). In the presence of 1.25 μM Latrunculin A, early relocalization and degradation of Cdc13 was not affected, although a proportion of Cdc13 remained associated to the spindle and spindle pole bodies for a longer period than in the control (Figure 3, A and B). Quantitative analysis indicates that <20% of the total Cdc13-GFP observed in G2 cells remains on the spindle and spindle pole bodies during metaphase. In movies of single cells, we also observed that Cdc13 remains associated to the spindle for longer in the presence of Latrunculin A (18 min) than in the control (8 min) (Supplementary Figure 2). Cut2 is located in the nucleus of G2 cells and associates only transiently to the spindle in M phase, as observed previously (Funabiki et al., 1996a; Tatebe et al., 2001; Figure 3, C and D). Addition of Latrunculin A resulted in an extended metaphase delay with Cut2 associated to the mitotic spindle (Figure 3, C and D). Cut9 is a core component of the APC in fission yeast (Funabiki et al., 1996b). We find that Cdc13 and Cut2 associate strongly to mitotic spindles in cut9-366 cells at the restrictive temperature, suggesting that this is an important site of APC action (our unpublished data). These results suggest that Latrunculin A inhibits sister chromatid separation by delaying activation of the APC.
Figure 3.
Latrunculin A delays the destruction of spindle associated Cdc13 (Cyclin B) and Cut2 (Securin). (A) Synchronized cdc13-gfp cells were incubated in the absence (open squares) or the presence of 1.25 μM Latrunculin A (closed diamonds). The percentage of cells displaying spindle associated Cdc13 was determined (n = 150). (B) Representative images of cdc13-gfp cells from A showing the localization of Cdc13 at the times indicated in the absence or presence of 1.25 μM Latrunculin A. (C) Synchronized cut2-gfp cdc11-cfp cells were incubated either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A. The percentage of cells with either separated SPBs (left) or spindle associated Cut2 (right) was determined at the times indicated (n = 150). (D) Representative images of cut2-gfp cdc11-cfp cells from (C) showing the localization of Cut2 (green) or spindle pole bodies (red) at the times indicated, either in the absence or presence of 1.25 μM Latrunculin A.
Latrunculin Activates a Bub1-, Bub3-, and Mad3-dependent Anaphase Checkpoint
We previously showed that addition of 10 μM Latrunculin B delays nuclear separation in cells lacking the Mad2 spindle assembly checkpoint protein (Gachet et al., 2001). Because Latrunculin A delays the degradation of spindle-associated Cdc13 and Cut2, we undertook a careful investigation of the role of SAC proteins in delaying anaphase under these conditions. Components of the SAC include Mad1, Mad2, Mad3, Bub1, Bub3, and Mph1 (the homologue of budding yeast Mps1) (He et al., 1997, 1998; Bernard et al., 1998; Ikui et al., 2002; Millband and Hardwick, 2002). Each of these genes was individually deleted in a ndc80-gfp cdc11-cfp strain to visualize centromere and SPB dynamics and, by this means, to assess the time spent in prometaphase and metaphase. In agreement with previous observations, Latrunculin A delayed the onset of anaphase in cells lacking Mad2 to a similar extent to that observed in control cells (Figure 4A). By contrast, addition of 1.25 μM Latrunculin A failed to impose a metaphase delay in cells lacking Bub1 (Figure 4B). To confirm this result, we filmed individual mad2Δ ndc80-GFP cdc11-CFP and bub1Δ ndc80-GFP cdc11-CFP cells in mitosis. In the absence of Mad2 metaphase lasted ∼6 min, and this was extended to 16 min in the presence of Latrunculin A (Supplementary Figure 3A). By contrast, in the absence of Bub1, metaphase lasted 4 min, and this was not extended in the presence of Latrunculin A (Supplementary Figure 3B). To examine whether the failure of bub1Δ cells to delay in metaphase was due to the role of Bub1 in APC inactivation, Cdc13-GFP localization was monitored through mitosis. Whereas addition of Latrunculin A caused Cdc13 to persist on mitotic spindles in the absence of Mad2 (Figure 4C), it failed to do so in the absence of Bub1 (Figure 4D). This suggests Bub1 acts independently of Mad2 to block activation of the APC.
Figure 4.
Latrunculin A inhibits APC and anaphase onset via a Bub1-dependent but Mad2-independent mechanism. (A) Synchronized mad2Δ ndc80-gfp cdc11-cfp and (B) bub1Δ ndc80-gfp cdc11-cfp cells were incubated either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A. The percentage of cells in prometaphase and metaphase (PM + M) was determined at the times indicated (n = 150). Synchronized mad2Δ cdc13-gfp (C) or bub1Δ cdc13-gfp (D) cells were incubated either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A. The percentage of cells showing spindle associated Cdc13 was determined at the times indicated (n = 150). Similar results were obtained from six independent experiments.
We next examined the length of prometaphase and metaphase in bub1(K762M) ndc80-gfp cdc11-cfp cells, which express an inactive Bub1 kinase. We found that Latrunculin A was unable to delay sister chromatid separation in these cells, suggesting that the catalytic activity of Bub1 kinase is required to delay anaphase under these conditions (Figure 5A). Furthermore, no anaphase delay was observed by addition of Latrunculin A to cells lacking either Bub3 (Figure 5B) or Mad3 (Figure 5C). However, addition of Latrunculin A to cells lacking Mad1 delayed the onset of anaphase as in wild-type cells (Figure 5D). We also observed a delay in cells lacking Mph1, but this was reproducibly less pronounced than in wild-type cells or in cells lacking Mad1 or Mad2 (Figure 5E). Note that in these experiments addition of 1.25 μM Latrunculin A completely blocks the appearance of septa (Figure 5, A-E). These results suggest that disruption of the actin cytoskeleton delays activation of the APC via the Bub1, Bub3, and Mad3 spindle assembly checkpoint proteins.
Figure 5.
Anaphase delay induced by Latrunculin A requires Bub3 and Mad3 and the catalytic activity of Bub1. The following strains were synchronized in early G2 and transferred to fresh medium either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A. (A) bub1-K762M ndc80-gfp cdc11-cfp. (B) bub3Δ ndc80-gfp cdc11-cfp. (C) mad3Δ ndc80-gfp cdc11-cfp. (D) mad1Δ ndc80-gfp cdc11-cfp. (E) mph1Δ ndc80-gfp cdc11-cfp cells. The percentage of cells with septa (left) or in prometaphase and metaphase (PM + M) (right) was determined at the times indicated (n = 150). Similar results were obtained from at least five independent experiments.
Latrunculin Prolongs the Association of Bub1 with Kinetochores
The differential requirement for Mad2 and Bub1 in monitoring spindle mispositioning prompted us to examine their cellular location relative to centromeres. To do this, we constructed mad2-GFP ndc80-CFP and bub1-GFP ndc80-CFP strains. In control cells, Mad2 is observed throughout the nucleus in G2 and then, as cells enter mitosis, relocates to a region underlying the unseparated spindle pole that colocalizes with kinetochores, and it remains in this location as the spindle forms. Mad2, however, does not colocalize with kinetochores during metaphase (Figure 6A). By contrast, Bub1 remains associated with all kinetochores in prometaphase and metaphase, and the Bub1-GFP signal only diminishes a few minutes before anaphase onset (Figure 6B). To confirm these observations, we constructed mad2-GFP cdc11-CFP and bub1-GFP cdc11-CFP strains and examined the localization of Mad2 and Bub1 relative to the position of spindle poles. We found that Mad2 colocalizes with centromeres only in early mitosis and then is seen most prominently with only one of the separated SPBs (Supplementary Figure 4). Bub1, however, is broadly nuclear in G2, localizes to a region underlying the unseparated SPB in early mitosis, and then is found between the two SPBs, which are separated by a <2.5-μm spindle until it disappears shortly before anaphase (Supplementary Figure 4). Mad2 localization was monitored in mad2-gfp cdc11-cfp cells during an unperturbed mitosis or in the presence of 1.25 μM Latrunculin A or a sub-lethal dose of the microtubule-depolymerizing drug benomyl. Although Mad2 remained bound to both the SPB and kinetochores for longer in the presence of benomyl, the localization of Mad2 was unaffected by addition of Latrunculin A (Figure 6C). By contrast, treatment of synchronous populations of bub1-gfp ndc80-cfp cells with 1.25 μM Latrunculin A caused Bub1 to remain associated with kinetochores for longer than in control cells (Figure 6D). This was confirmed by live imaging of individual bub1-gfp cdc11-cfp cells (Figure 6E). Whereas we observed a maximum of two clusters of Bub1 dots in control cells, we frequently observed cells with up to 6 Bub1 dots for an extended period in the presence of Latrunculin A (Figure 6E). These data suggest that the Bub1 kinase is required at kinetochores to prevent anaphase onset when the actin cytoskeleton is disturbed.
Figure 6.
Bub1 associates to kinetochores when the SOC is activated. (A) Images of log phase cultures of mad2-gfp ndc80-cfp cells in either G2 (i), prometaphase (ii), metaphase (iii), anaphase (iv), or telophase (v) showing the localization of Mad2 (green) and Ndc80 (red). Colocalization is seen in yellow. (B) Images of log phase cultures of bub1-gfp ndc80-cfp cells in either G2 (i), various stages between prometaphase and metaphase (ii-iv), and telophase (v) showing the localization of either Bub1 (green) and Ndc80 (red). Colocalization is seen in yellow. (C) Synchronized mad2-gfp cdc11-cfp were transferred to fresh medium either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A or with 3 μM benomyl (open triangles). The percentage of cells displaying SPB associated Mad2 was determined at the times indicated (n = 150). (D) Synchronized bub1-gfp ndc80-cfp were transferred to fresh medium either in the absence (open squares) or the presence (closed diamonds) of 1.25 μM Latrunculin A or with 3 μM benomyl (open triangles). The percentage of cells displaying kinetochore associated Bub1 was determined at the times indicated (n = 150). (E) Images from movie of bub1-gfp cdc11-cfp cells incubated either in the absence (control) or the presence (Lat A) of 1.25 μM Latrunculin A. Time zero is when the spindle became 2 μm in length. Localization of Bub1 at various times is shown (green).
The SOC is activated by disruption of astral microtubule contact with the cortical actin cytoskeleton
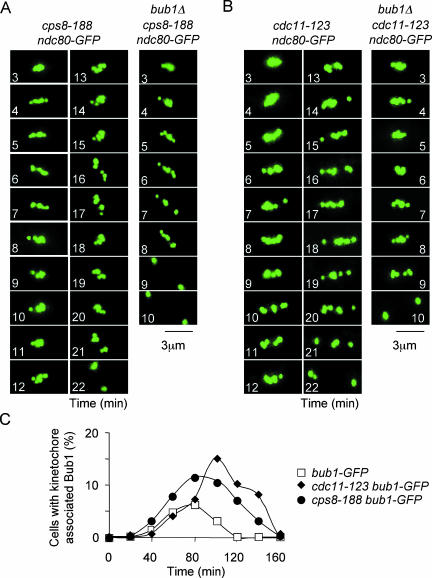
In previous studies, we showed that disruption of astral microtubule interaction with the cell cortex delays the timing of chromosome separation (Gachet et al., 2001, 2004). To examine the delay in anaphase A onset more accurately, we monitored centromere dynamics in the actin mutant cps8-188 and in cdc11-123 cells, in which astral microtubule attachment to the spindle pole and spindle rotation are compromised (Ishiguro and Kobayashi, 1996; Krapp et al., 2001; Gachet et al., 2004). Single cell analysis revealed that cps8-118 ndc80-gfp cells remained in prometaphase and metaphase for 20 ± 3 min (Figure 7A), longer than in wild-type cells at the same temperature, but similar to that observed in wild-type cells in the presence of 1.25 μM latrunculin A (Figure 2), consistent with our previous observations (Gachet et al., 2004). By contrast, the length of prometaphase and metaphase was 8 ± 2 min in bub1Δ cps8-118 ndc80-gfp cells (Figure 7A) but unchanged in mad2Δ cps8-118 ndc80-gfp cells (our unpublished data). Similarly, the length of prometaphase and metaphase in cdc11-123 cells was longer than in wild-type cells at the same temperature, and this was abolished by inactivation of Bub1 (Figure 7B; Gachet et al., 2004). We find that Bub1 only binds kinetochores for ∼4 min in wild-type cells. For this reason, Bub1 is bound to kinetochores in a small percentage of cells even in highly synchronous populations (Figure 7C). Nevertheless, we find that Bub1 colocalizes kinetochores for longer in cps8-188 and cdc11-123 cells than in control cells at the same temperature (Figure 7C). These data strongly argue that Bub1 imposes an anaphase delay at kinetochores when astral microtubule contact with the cell cortex, and thus spindle position, is disturbed.
Figure 7.
Bub1 monitors astral microtubule contact with the cell cortex. (A) Images from a movie of cps8-188 ndc80-gfp cells or bub1Δ cps8-188 ndc80-gfp cells in mitosis. The localization of kinetochores (green) is shown at the times indicated. Time zero is the beginning of phase 1 when kinetochores are first seen to separate. (B) Images from a movie of cdc11-123 ndc80-gfp cells or bub1Δ cdc11-123 ndc80-gfp cells in mitosis. The localization of kinetochores (green) is shown at the times indicated. Time zero is the beginning of phase 1 when kinetochores are first seen to separate. (C) Synchronized bub1-gfp (open squares), cps8-188 bub1-gfp (closed diamonds) or cdc11-123 bub1-gfp (closed circles) cells were incubated in fresh medium and the percentage of cells displaying Bub1 at kinetochores was then determined at the times indicated (n = 150). Similar results were obtained from three independent experiments.
DISCUSSION
We previously proposed that addition of Latrunculin B activates a SOC in fission yeast that dictates the timing of chromosome separation (Gachet et al., 2001). In agreement with this, we have found that astral microtubules interact with the cytokinetic actomyosin ring and that disruption of this process both prevents spindle alignment and delays anaphase onset (Gachet et al., 2004). In this article, we confirm by live imaging of ndc80-gfp cdc11-cfp cells that addition of Latrunculin A, at concentrations that specifically disrupt actin cables, delays the separation of sister chromatids. This effect is also observed cps8-188 cells, in which the cortical actin cytoskeleton is disturbed, and in cdc11-123 cells, in which nucleation of astral microtubules is partially disrupted (Gachet et al., 2004). These results provide further evidence that the onset of anaphase in fission yeast is regulated by a checkpoint that is activated when interaction of astral microtubules with the cortical actin cytoskeleton is disturbed. In this article, we have examined in more detail the mechanism by which anaphase is delayed under these conditions.
Chromosomes are held together in interphase and early mitosis by a complex that includes the Rad21(Scc1) protein (Uhlmann et al., 1999; Tomonaga et al., 2000). We previously showed that Latrunculin B delays the degradation of Rad21 but does not inhibit the degradation of either Cut2 or Cdc13 (Cyclin B), as judged by Western blot of cell extracts (Gachet et al., 2001). This led us to believe that Latrunculin B blocks sister chromatid separation without inhibiting the activation of APC. We reexamined this conclusion by visually examining Cdc13 and Cut2 levels in cdc13-gfp and cut2-gfp cells treated with Latrunculin. Although we observe a substantial reduction (>80%) in Cdc13-GFP fluorescence in early mitosis, a proportion of Cdc13 remains strongly associated with short misoriented spindles and spindle pole bodies when cells enter mitosis. Importantly, spindle-associated Cdc13 disappears just before sister chromatid separation, suggesting that it is effectively ubiquitinated by APC at this time. It is conceivable that a proportion of Cdc13 is degraded by APC in early mitosis, as is Cyclin A in mammalian cells (Geley et al., 2001). However, we note that Cdc13 degradation is observed in temperature-sensitive mutants of the APC, raising the possibility that a distinct degradation pathway acts on Cdc13 in early M phase (Chang et al., 2001). Regardless, we also find that Cut2 is stabilized on mitotic spindles in the presence of Latrunculin, and this also disappears before anaphase onset. These results indicate that, in contrast to our previous conclusion (Gachet et al., 2001), the APC is inhibited when astral microtubule contact with the medial actin cytoskeleton is disturbed.
It is well known that sister chromatid separation is inhibited by addition of microtubule-disrupting agents to cells. This causes the recruitment of Mad1, Mad2, Mad3, Mph1, Bub1, and Bub3 proteins to unattached kinetochores and inhibition of APC (Yu, 2002; Zhou et al., 2002; Cleveland et al., 2003). Activation of the spindle assembly checkpoint and recruitment of Mad2 to kinetochores is also observed in mutants, such as alp7/mia1, in which the stability of both astral and spindle microtubules is perturbed. In these cells, Mad2 is required to maintain cell viability (Oliferenko and Balasubramanian, 2002; Sato et al., 2003). By contrast, we found that in response to Latrunculin A, both the separation of sister chromatids and the degradation of spindle associated Cdc13 and Cut2 are delayed in cells lacking Mad1 or Mad2, but not in cells lacking Mad3 or Bub3. In accordance with this, we find that Bub1 colocalizes with kinetochores when spindles are misoriented but Mad2 does not. Instead, Mad2 localizes to a region underlying the unduplicated spindle pole body in early mitosis and colocalizes with only one pole during spindle elongation, as observed by others (Garcia et al., 2002). It remains to be determined whether this is because Mad2 remains closely associated only to the old spindle pole. Importantly, anaphase onset and the disappearance of Bub1 from kinetochores is delayed in cps8-188 and cdc11-123 cells and the anaphase delay in these mutants is dependent on Bub1 but not Mad2. These results suggest that spindle assembly and spindle orientation are monitored by distinct checkpoints that inhibit APC by an overlapping but nonidentical mechanism (Figure 8). We note that in budding yeast Bub1 and Bub3 bind kinetochores every cell cycle, whereas Mad1 and Mad2 only associate to kinetochores when spindles are damaged, indicating that components of the SAC are nonequivalent (Gillett et al., 2004). Intriguingly, a complex containing Cdc20, BubR1(Mad3), and Bub3, but lacking Mad2, has been isolated from tissue culture cells as a potent inhibitor of APC activity in vitro (Tang et al., 2001). We are currently generating reagents to examine whether a similar complex inhibits APC in fission yeast when spindles are mispositioned.
Figure 8.
Spindle assembly and orientation checkpoints in fission yeast. The spindle assembly checkpoint inhibits the APC via the Mad1, Mad2, Mph1, Mad3, Bub1, and Bub3 proteins in response to a lack of bipolar attachment of kinetochores to spindle poles. The spindle orientation checkpoint inhibits APC by a pathway that requires Bub1, Bub3, and Mad3 but not Mad1 or Mad2. Microtubule depolymerizing agents activate both spindle assembly and spindle orientation checkpoints. Actin depolymerizing drugs only disrupt astral microtubule interaction with the cell cortex and activate only the spindle orientation checkpoint.
Recently, Rajagopalan and colleaques have suggested, by analysis of a mutant in the kendrin-like spindle pole component Pcp1 [pcp1(Δ400-900)], that metaphase spindle position is monitored by the Bub1 but not Mad1 or Mad2 spindle assembly checkpoint proteins (Rajagopolan et al., 2004). However, we find that, by analyzing kinetochore dynamics, the delay imposed by disrupting astral microtubule contact with the cell cortex is dependent not only on Bub1 but also on the Bub3 and Mad3 spindle assembly checkpoint proteins. Furthermore, whereas Rajapolan and colleagues find that disruption of mph1 abolishes the anaphase delay in pcp1(Δ400-900) cells, we find that addition of Latrunculin A imposes an anaphase delay in cells lacking Mph1, although this is reproducibly less pronounced than in wild-type cells. One possibility is that the metaphase delay in pcp1(Δ400-900) cells is due to a mitotic defect other than spindle misorientation, because spindle alignment is apparently not affected in this mutant (Rajagopalan et al., 2004). Alternatively, because mitotic progression in pcp1(Δ400-900) cells was judged only by staining of nuclei and spindles, the timing of sister chromatid separation may have been masked by an additional delay over anaphase B (spindle elongation). We suggest that a careful analysis of kinetochore dynamics in pcp1(Δ400-900) cells, such as that performed in this study, may help to resolve this issue. We point out that first, both Mad3 and Bub3 are necessary for Mps1(Mph1) to impose a metaphase arrest in budding yeast (Hardwick et al., 1996), and second, Bub3 is required both for the association of Bub1 to kinetochores and for the checkpoint function of Bub1 (Farr and Hoyt, 1988; Taylor et al., 1988; Warren et al., 2002; Gillett et al., 2004). In this regard, the localization of Bub1 in metaphase arrested pcp1(Δ400-900) cells will be of interest. In other experiments, we have found that addition of 50 μM Latrunculin A to cdc25-22 cells inhibits mitotic onset by a mechanism that is independent of its effects as an inhibitor of actin polymerization (Supplementary Figure 1). Thus, the delay in anaphase onset observed by Rajagopalan and colleagues may be due, at least in part, to a delay in mitotic entry. This may also explain why these researchers failed to identify a role for Mad3 and Bub3 in their experiments (Rajagopalan et al., 2004).
In this and previous articles, we have shown that anaphase is delayed when the medial actin cytoskeleton is disturbed (Gachet et al., 2001, 2004). We attributed this effect to activation of an SOC because spindle rotation is blocked and also there is no known role for actin at the kinetochore. Despite this, it is still unclear whether the SOC monitors astral microtubule contact with the cell cortex, the integrity of the cortical actin cytoskalston or spindle angle itself. Recently, Rajagopolan and colleagues proposed that spindle position is monitored by a tension checkpoint at the spindle pole (Rajagopalan et al., 2004). However, we find that sister kinetochores continue to undergo dynamic movement along the mitotic spindle when spindles are mispositioned, suggesting that the spindle is still under tension. So what do the Bub1, Bub3, and Mad3 checkpoint proteins sense? Intriguingly, we have found that astral microtubules often interact with the cytokinetic actomyosin ring shortly before the onset of anaphase, an event that is necessary to align the mitotic spindle perpendicularly to the axis of cell division (Gachet et al., 2004). One attractive possibility is that the SOC monitors attachment of astral microtubules with the cytokinetic actomyosin ring. This would be analogous to the role of SAC components in monitoring attachment of spindle microtubules to the kinetochore. If this is the case, a signal must be transmitted from the cell cortex to the kinetochore, because Bub1 binds kinetochores in SOC-activated cells. Clearly, further experimentation will be needed to test this and other possibilities.
In mammalian cells, chromosomes align on a metaphase plate that is formed equidistant between the centrosomes and at the site of the future cleavage furrow (Rieder and Salmon, 1994). Similarly, in budding yeast, kinetochores congress to the spindle midzone as the spindle is positioned across the bud neck, suggesting that a pseudometaphase plate also exists in fungi (Pearson et al., 2001). In agreement with this, we find that centromeres congress to the spindle midzone just before sister chromatid separation in fission yeast. Because anaphase onset follows shortly after the establishment of a metaphase plate, it is thought that chromosome congression is monitored by a checkpoint that dictates the timing of anaphase. Notably, some SAC components such as Bub1, BubR1(Mad3), and Bub3 bind kinetochores at the metaphase plate, whereas Mad1 and Mad2 do not (Waters et al., 1998; Campbell and Hardwick, 2003). We find that in the presence of Latrunculin A, or in mutants in which astral microtubule contact with the cell cortex is disturbed, chromosome congression does not take place. This may be a secondary consequence of an anaphase delay. Alternatively, a subset of spindle assembly checkpoints, including Bub1, Bub3, and Mad3, may ensure both that chromosomes congress to the spindle midzone (metaphase plate) and that the spindle is positioned perpendicularly to the cytokinetic actomyosin ring before anaphase onset. If this is the case, the spindle orientation checkpoint in fission yeast may be indistinguishable from the checkpoint that ensures formation of a metaphase plate in animal cells.
Supplementary Material
Acknowledgments
We thank Jean-Paul Javerzat, John Kilmartin, Tomohiro Matsumoto, Dan McCollum, Viesturs Simanis, Takashi Toda, Vincent Vanoosthuyse, and Mitsuhiro Yanagida for supplying strains, and, in particular, Kevin Hardwick for strains and communicating results before publication. We also thank Stamatis Pagakis and Kate Sullivan (Microscopy facility, National Institute for Medical Research) and Céline Reyes (University of Toulouse, Toulouse, France) for technical assistance and Lee Johnston (National Institute for Medical Research) and Sara Mole (University College London) for advice and encouragement. S.T. is supported by a postdoctoral fellowship from the Association of International Cancer Research. J.S.H. and J.B.A.M. are supported by grants from the Wellcome Trust, Association of International Cancer Research, the Leverhulme Trust, University College London, and the Medical Research Council.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0256. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0256.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Alfa, C.E., Ducommun, B., Beach, D., and Hyams, J.S. (1990). Distinct nuclear and spindle pole body population of cyclin-cdc2 in fission yeast. Nature 347, 680-682. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A 3rd, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Bernard, P., Hardwick, K., and Javerzat, J.-P. (1998). Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143, 1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, L., and Hardwick, K.G. (2003). Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. J. Cell Sci. 116, 617-628. [DOI] [PubMed] [Google Scholar]
- Chang, L., Morrell, J.L., Feoktistova, A., and Gould, K.L. (2001). Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol. Cell. Biol. 21, 6681-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., Brady, D.M., Smith, D., Murray, A.W., and Hardwick, K.G. (1999). The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10, 2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., Waters, J.C., Salmon, E.D., and Murray, A.W. (1996). Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274, 242-246. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- Fang, G. (2002). Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13, 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, K.A., and Hoyt, M.A. (1988). Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 18, 2738-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., Kumada, K., and Yanagida, M. (1996a). Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 15, 6617-6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., Yamano, H., Kumada, K., Nagao, K., Hunt, T., and Yanagida, M. (1996b). Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381, 438-441. [DOI] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Millar, J.B.A., and Hyams, J.S. (2001). A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 412, 352-355. [DOI] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Millar, J.B.A., and Hyams, J.S. (2004). Mechanism controlling perpendicular alignment of the spindle to the axis of cell division in fission yeast. EMBO J. 23, 1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.A., Koonrugsa, N., and Toda, T. (2002). Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21, 6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley, S., Kramer, E., Gieffers, C., Gannon, J., Peters, J.M., and Hunt, T. (2001). Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153, 137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett, E.S., Espelin, C.W., and Sorger, P.K. (2004). Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164, 535-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K.G., Johnston, R.C., Smith, D.L., and Murray, A.W. (2000). MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148, 871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K.G., Weiss, E., Luca, F.C., Winey, M., and Murray, A.W. (1996). Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273, 953-956. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., and Weinert, T.A. (1989). Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629-634. [DOI] [PubMed] [Google Scholar]
- He, X., Patterson, T.E., and Sazer, S. (1997). The Schizosaccharomyces pombe spindle checkpoint protein Mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94, 7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Jones, M.H., Winey, M., and Sazer, S. (1998). Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell Sci. 111, 1635-1647. [DOI] [PubMed] [Google Scholar]
- Hoffman, D.B., Pearson, C.G., Yen, T.J., Howell, B.J., and Salmon, E.D. (2001). Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12, 1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., Totis, L., and Roberts, B.T. (1991). S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66, 507-517. [DOI] [PubMed] [Google Scholar]
- Hwang, L.H., Lau, L.F., Smith, D.L., Mistrot, C.A., Hardwick, K.G., Hwang, E.S., Amon, A., and Murray, A.W. (1998). Budding yeast Cdc 20, a target of the spindle checkpoint. Science 279, 1041-1044. [DOI] [PubMed] [Google Scholar]
- Ikui, A.E., Furuya, K., Yanagida, M., and Matsumoto, T. (2002). Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115, 1603-1610. [DOI] [PubMed] [Google Scholar]
- Ishiguro, J., and Kobayashi, W. (1996). An actin point-mutation neighboring the `hydrophobic plug' causes defects in the maintenance of cell polarity and septum organization in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 392, 237-241. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Lin, D.P., Matsumoto, S., Kitazono, A., and Matsumoto, T. (1998). Fission yeast Slp 1, an effector of the Mad2-dependent spindle checkpoint. Science 279, 1045-1047. [DOI] [PubMed] [Google Scholar]
- Krapp, A., Schmidt, S., Cano, E., and Simanis, V. (2001). S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11, 1559-1568. [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray, A.W. (1991). Feedback control of mitosis in budding yeast. Cell 66, 519-531. [DOI] [PubMed] [Google Scholar]
- Li, Y., and Benezra, R. (1996). Identification of a human mitotic checkpoint gene: hsMAD2. Science 274, 246-248. [DOI] [PubMed] [Google Scholar]
- Li, X., and Nicklas, R.B. (1995). Mitotic forces control a cell-cycle checkpoint. Nature 373, 630-632. [DOI] [PubMed] [Google Scholar]
- Marks, J., and Hyams, J.S. (1985). Localisation of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur. J. Cell Biol. 39, 27-32. [Google Scholar]
- McIntosh, J.R. (1991). Structural and mechanical control of mitotic progression. Cold Spring Harbor Symp. Quant. Biol. 56, 613-619. [DOI] [PubMed] [Google Scholar]
- Millband, D.N., and Hardwick, K.G. (2002). Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22, 2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Mulvihill, D.P., Petersen, J., Ohkura, H., Glover, D.M., and Hagan, I.M. (1999). Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell 10, 2771-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko, S., and Balasubramanian, M.K. (2002). Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat. Cell Biol. 4, 816-820. [DOI] [PubMed] [Google Scholar]
- Pearson, C.G., Maddox, P.S., Salmon, E.D., and Bloom, K. (2001). Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152, 1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, S., Bimbo, A., Balasubramanian, M.K., and Oliferenko, S. (2004). A potential tension-sensing mechanism that ensures timely anaphase onset upon metaphase spindle orientation. Curr. Biol. 14, 69-74. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and Salmon, E.D. (1994). Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 124, 223-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., Schultz, A., Cole, R., and Sluder, G. (1994). Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127, 1301-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., Cole, R.W., Khodjakov, A., and Sluder, G. (1995). The checkpoint delaying anaphase in response to chromosome mono-orientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes, I., Webb, B.A., Mak, A., and Young, P.G. (2001). G2/M arrest caused by actin disruption is a manifestation of the cell size checkpoint in fission yeast. Mol. Biol. Cell 12, 3892-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Koonrugsa, N., Toda, T., Vardy, L., Tournier, S., and Millar, J.B.A. (2003). Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat. Cell Biol. 5, 764-766; author reply 766. [DOI] [PubMed] [Google Scholar]
- Spector, I., Shochet, N.R., Kashman, Y., and Groweiss, A. (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493-495. [DOI] [PubMed] [Google Scholar]
- Stern, B.M., and Murray, A.W. (2001). Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11, 1462-1467. [DOI] [PubMed] [Google Scholar]
- Sudakin, V., Chan, G.K., and Yen, T.J. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe, H., Goshima, G., Takeda, K., Nakagawa, T., Kinoshita, K., and Yanagida, M. (2001). Fission yeast living mitosis visualized by GFP-tagged gene products. Micron. 32, 67-74. [DOI] [PubMed] [Google Scholar]
- Tang, Z., Bharadwaj, R., Li, B., and Yu, H. (2001). Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1, 227-237. [DOI] [PubMed] [Google Scholar]
- Taylor, S.S., Ha, E., and McKeon, F. (1988). The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, T., et al. (2000). Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., Lottspeich, F., and Nasmyth, K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37-42. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., Wernic, D., Poupart, M.A., Koonin, E.V., and Nasmyth, K. (2000). Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375-386. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., Brady, D.M., Johnston, R.C., Hanna, J.S., Hardwick, K.G., and Spencer, F.A. (2002). Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13, 3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, J.C., Chen, R.H., Murray, A.W., and Salmon, E.D. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141, 1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E., and Winey, M. (1996). The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132, 111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., and Kilmartin, J.V. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. (2002). Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14, 706-714. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., and Nasmyth, K. (1999). Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13, 2039-2058. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Yao, J., and Joshi, H.C. (2002). Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115, 3547-3555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.