Abstract
Breast cancer is the most common cancer in women around the world. However, the molecular mechanisms underlying breast cancer pathogenesis are only partially understood. Here, in this study, we found that miR-1228 was up-regulated in breast cancer cell lines and tissues. Ectopic expression of miR-1228 mimics leads to promoted cell growth, invasion and migration. Using bioinfomatic analysis and 3’UTR luciferase reporter assay, we determined SCAI can be directly targeted by miR-1228, which can down-regulate endogenous SCAI protein level. Furthermore, our findings demonstrate that SCAI was down-regulated in breast cancer cell lines and tissues. Rescue experiment demonstrated that miR-1228 promoted cell growth is attenuated by over-expression of MOAP1 and miR-1228 promoted cell invasion and migration can be attenuated by over-expression of SCAI. Taken together, this study provides evidences that miR-1228 serves as an oncogene to promote breast cancer proliferation, invasion and migration, which may become a critical therapeutic target for breast cancer treatment.
Keywords: Breast cancer, cell proliferation, cell invasion, cell migration, miR-1228, SCAI
Introduction
Breast cancer is the most common cancer in women around the world [1]. It is a complex disease, characterized by heterogeneity of genetic alterations and influenced by several environmental factors. Oncogene amplification or dysregulation usually occurs late in tumor progression and correlates well with aggressiveness of tumor [2]. This may lead to the spreading of tumor cells from the primary neoplasm to distant sites [3,4]. Many proteins including proteases, adhesion molecules, angiogenesis, and growth factor are involved in and proliferation and metastasis [5]. Therefore, understanding the gene and protein expression changes in breast cancer progression may aid in early diagnosis and therapeutic intervention.
MicroRNAs (miRNAs) are ~22 nucleotide non-coding single-stranded RNAs that regulate gene expression through translational repression and/or transcript cleavage, based on specific binding to the complementary sequence in the coding or noncoding region of mRNA transcripts [6-8]. It has been demonstrated that miRNAs can function in a variety of biological processes, including cellular proliferation [9], metastasis [10], apoptosis [11], differentiation [12,13] and metabolism [14]. Aberrant miRNA expression has been found to be associated with the development and progress of some cancers [15]. Furthermore, based on microarray analysis of global miRNA expression profiles in cancer tissues, researchers have revealed that miRNA profiles can discriminate malignancies of the breast [16], lung [17], pancreas [18] and liver [19,20] from their counterparts.
miR-1228 is located in chromosome 12 and housed within the LRP1 gene, which has diverse functions in the cell and has been implicated to play a role in atherosclerosis and Alzheimer’s disease [21]. It appears to be phylogenetically restricted to primates, with some presenting conserved hairpin structures in human/rhesus/chimp [22]. Previous studies indicated that miR-1228 expression is dysregulated in many tumors such as malignant mesothelioma, lung adenocarcinoma, breast cancer. Treatment with an antitumor agent, Resveratrol, can lead to a significant reduction in miR-1228 expression in human non-small cell lung cancer cells. Enforced miR-1228 expression can sensitize cells to stress-induced apoptosis through targeting MOAP1 protein, suggesting that miR-1228 is a crucial regulator of cellular apoptosis [23].
In the present study, we have we found that miR-1228 was up-regulated in breast cancer cell lines and tissues. Ectopic expression of miR-1228 mimics leads to promoted cell growth, invasion and migration. Using bioinfomatic analysis, our findings demonstrated that the 3’UTR of SCAI contains a putative binding site for miR-1228. We then determined SCAI can be directly targeted by miR-1228 using 3’UTR luciferase reporter assay. Enforced expression of miR-1228 represses the endogenous expression of SCAI. Furthermore, our findings demonstrated that SCAI was down-regulated in breast cancer cell lines and tissues. Rescue experiment demonstrated that miR-1228 promoted cell growth is attenuated by over-expression of MOAP1 and miR-1228 promoted cell invasion and migration can be attenuated by over-expression of SCAI. Taken together, this study provides evidences that miR-1228 serves as an oncogene to promote breast cancer proliferation, invasion and migration, which may become a critical therapeutic target for breast cancer treatment.
Materials and methods
Patient samples
Breast cancer specimens and adjacent normal tissues were collected in Department of General Surgery, The Fourth Affiliated Hospital of Harbin Medical University. All the patients recruited into the present study did not receive radiotherapy or chemotherapy or any other treatment before and after operation. Surgical specimens of the tumor resection were collected, and lumps of tumors as well as adjacent normal tissues, which were at least 2 cm distal to tumor margins, were snap-frozen in liquid nitrogen for detection of miR-1228 and SCAI expression. Written informed consent was obtained from all study participants. The use of tissue samples were approved by the ethical committees of the Department of General Surgery, The Fourth Affiliated Hospital of Harbin Medical University.
Cell culture and transfection
The breast cancer cell lines (MDA-MB-468, MCF-7, MCF-7), and non-malignant breast epithelial cell (MCF-10A) were obtained from the ATCC and maintained in RPMI 1640 or Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% antibiotics (Invitrogen, USA). Transfection of the cells with miR-1228 mimics, miR-1228 inhibitors (anti-miR-1228), pcDNA3-SCAI or pcDNA3-MOPA1 (Genepharma, China) was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Detection of cell phenotypes
The effect of miR-1228 on proliferation of breast cancer cells was evaluated by MTT and colony formation assays. MCF-7 and MDA-MB-468 cells were plated in 96-well culture plates (3×103 per well). After 24 h incubation, the cells were transfected with miR-1228 mimics, anti-miR-1228 and their controls for 12, 24, 36 and 48 hours. Then the MTT (0.5 mg/ml; Sigma-Aldrich, USA) was added to each well (20 μl/well). After 4 hours of additional incubation, MTT solution was discarded and 200 ml of DMSO (Sigma, USA) was added and the plates shaken gently. The absorbance was measured on an ELISA reader at a wavelength of 570 nm. For colony formation assay, cells were counted and seeded in 12-well plates (in triplicate) at 100 cells per well. Fresh culture medium was replaced every 3 days. The number of viable cell colonies was determined after 14 days and colonies were fixed with methanol, stained with crystal violet, photographed and counted. Each experiment was performed in triplicate.
Tumor cell invasion assay
Invasion assay was performed with the Transwell chamber with 8 μm pores (Corning, USA). Fifty microliters diluted matrigel (2 mg/ml, BD Biosciences, Bedford, MA) was placed on the inner surface. Cells were transfected for 24 h and isolated to make a final concentration at 2×105/ml, which then placed on the top chamber. RMPI1640 with 20% FBS was added to the bottom chamber. After 24 h, non-invading cells were removed from the top of the Matrigel with a cotton-tipped swab. Invading cells at the bottom of the Matrigel were fixed in methanol and stained with Crystal violet. The invasiveness was determined by counting the penetrated cells under a microscope at ×200 magnification of 5 random fields in each well. Each experiment was performed in triplicate.
Wound assay to assess cell migration
Cells were transfected for 24 h and then isolated and plated in twelve-well plates (3×105/well) for 24 h. When the cells reached 90% confluence, sterile pipette tips was used to scratch the wound uniformly. Cell motility was assessed by measuring the movement of cells into a scraped wound. The speed of wound closure was monitored after 72 h by measuring the distance of the wound from 0 h. Each experiment was conducted in triplicate.
Western blotting
Western blotting was performed to determine protein expression of MOAP1 or SCAI. Cells were washed twice with cold PBS and total cellular protein was extracted using a modified RIPA buffer with 0.5% sodium dodecyl sulfate (SDS) in the presence of proteinase inhibitor cocktail (Complete mini, Roche). The protein concentration in the supernatants was determined using Bradford protein dye reagent (Bio-Rad, Hercules, CA) and equal amounts of protein lysates were separated on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked with 5% non-fat milk, followed by incubation with antibodies against MOAP1 (1:1000, Millipore) or SCAI (1:1000, Millipore). As a secondary antibody, horseradish peroxidase (HRP) conjugated secondary antibody was used, then visualized with enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia) according to the manufacturer’s protocol. GAPDH (1:1000, Santa Cruz, CA) was used as an internal control.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) from breast cancer tissues and adjacent non-tumor tissues for detection the expression of miR-1228 or SCAI. For quantitative real-time PCR, RNA was first reversely transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). After that, qPCR was performed with an SYBR Green I real-time PCR kit (GenePharma, Shanghai, China) according to the manufacturer’s instructions with the ABI 7300 Real-Time PCR System. GAPDH mRNA was used as internal control. The amplification protocol was as follows: an initial 95°C for 5 min and 50 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. The analysis was performed with the three independent preparations of total RNA samples harvested from each sample group. The expression level of SCAI mRNA was normalized to that of GAPDH mRNA, and the change in expression level was calculated using the 2-ΔΔCt method. A two-tailed t-test (P<0.05 was identified to indicate a statistically significant difference) was performed to identify the differentially expressed miRNAs. The following primers were used: miR-1228 sense: 5-CTTGACATGATTAGCTGGCATGATT-3; antisense: 5-CCTGTGCAATATGCCGTGTAGA-3; U6 sense: 5-CTTGACATGATTAGCTGGCATGATT-3; antisense: 5-CTTGACATGATTAGCTGGCATGATT-3; SCAI sense: 5-GAGTGCTCGCAGCTCATACCT-3; antisense: 5-CCTCACGGCCTGGGATTT-3; GAPDH sense: 5-CCAAAATCAGATGGGGCAATGCTGG-3; antisense: 5-TGATGGCATGGACTGTGGTCATTCA-3.
Construction of 3’UTR reporter plasmid and luciferase assay
The human SCAI wild-type 3’UTR (WT-3’UTR) harboring miR-1228 target sequence as well as the seed-sequence mutated version (Mutant-3’UTR) were synthesized by GenPharm (Shanghai, China), The SCAI 3’UTR reporter was generated by inserting the entire WT-3’UTR or Mutant-3’UTR of human SCAI mRNA into XhoI/NotI sites of psiCHECK-2 vector (Promega) downstream of the Renal luciferase gene. For the luciferase assay, 1×105 cells were transfected along with the SCAIWT-3’UTR or Mutant-3’UTR reporter and the miR-1228 mimics in a 24-well plate using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 24 h, firefly and Renilla luciferase activities were measured consecutively using Dual Luciferase Assay (Promega).
Apoptosis analysis
For cell apoptosis assay, MCF-7 cells were transfected with miR-1228 mimics or MDA-MB-231 cells were transfected with MDA-MB-231 for 48 h. Then the cells apoptotic rate was determined by using Annexin V-FITC and PI staining flow cytometry kit (KeyGEN BioTECH, China) according to manufacturer’s instruction. Briefly, the cells in different transfection groups were harvested and washed with PBS for twice. After that, cells were resuspended in 500 ml binding buffer provided by the kit. 5 ml Annexin V and 5 ml propidium iodide (PI) were added to the cells and then incubated at room temperature for 15 minutes in dark. Cells apoptotic rate was then tested by flow cytometry within 1 h.
Data analysis
A Student’s test was performed to analyze the significance of differences between the sample means obtained from three independent experiments. Differences were considered statistically significant at P<0.05.
Results
The expression status of miR-1228 in breast cancer cell lines and tissues
To investigate the role of miR-1228 in breast cancer pathogenesis, here we used quantitative real-time PCR (qRT-PCR) to measure the expression levels of miR-1228 in breast cancer cell lines and tissues. MDA-MB-468, MCF-7, MDA-MB-231 and a non-malignant breast epithelial cell MCF-10A were used. Compared to low metastatic MCF-7 cells, the expression of miR-1228 were obviously increased in highly metastatic cells, MDA-MB-231 (3.1-fold) and MDA-MB-468 (3.4-fold) cells; while the expression of miR-1228 was significantly decreased in non-malignant breast epithelial cell MCF-10A (Figure 1A).
Figure 1.
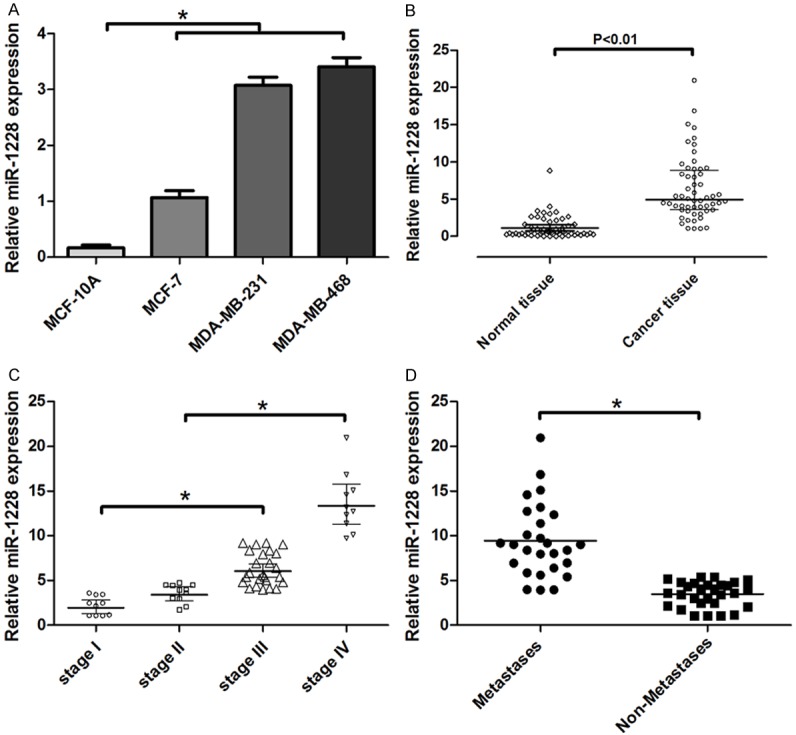
Relative expression of miR-1228 in breast cancer cells and tissues. A: miR-1228 with increased expression in MCF-10A, MCF-7, MDA-MB-231 and MDA-MB-468 cells. B: Relative expression of miR-1228 in breast cancer tissues and adjacent normal tissues. C: Relative expression of miR-1228 with breast cancer staging. D: Relative expression of miR-1228 in metastases and non-metastases tissues.
To further confirm the role of miR-1228 during breast cancer progression, we determined the expression of miR-1228 in breast cancer tissues and adjacent normal tissues by using qRT-PCR. We observed that miR-1228 expression was significantly increased in breast cancer tissue compared with adjacent normal breast tissues (Figure 1B). Tumors expressed increasing levels of miR-1228 with malignancy stage (Figure 1C); Furthermore, we observed that the expression of miR-1228 in metastasis tissues was 9-fold compared with non-metastases tissues (Figure 1D).
miR-1228 promotes cell growth of breast cancer cell lines
To explore the role of miR-1228 in regulating cell growth, a miR-1228 mimic or anti-miR-1228 was transfected into MCF-7 or MDA-MB-231 cells, respectively. Compared with the control group, the transfection of miR-1228 mimic markedly increased the level of miR-1228 in MCF-7 cells, while the level of miR-1228 was significantly decreased in MDA-MB-231 cells (Figure 2A). Next, we tested the effects of miR-1228 mimic or anti-miR-1228 on cellular growth. The MTT and colony formation assays results showed that miR-1228 introduction can promote the cell growth in MCF-7 cells (Figure 2C and 2E), while blocking the expression of miR-1228 by anti-miR-1228 attenuated the proliferation of MDA-MB-231 cells (Figure 2D and 2F). Furthermore, Annexin V assay showed that miR-1228 mimics caused a significant decrease in cell apoptosis compared with the control group (Figure 2G), while Anti-miR-1228 induced obvious increase of MDA-MB-231 cells apoptosis. We also confirmed that miR-1228 mimics repressed the expression of MOAP1, while anti-miR-1228 induced the protein expression of MOAP1 (Figure 2I), which was consistent with previous results illustrating MOPA1 was a direct target of miR-1228 [23]. Taken together, these results revealed a functional role for miR-1228 in regulating breast cancer cells proliferation.
Figure 2.
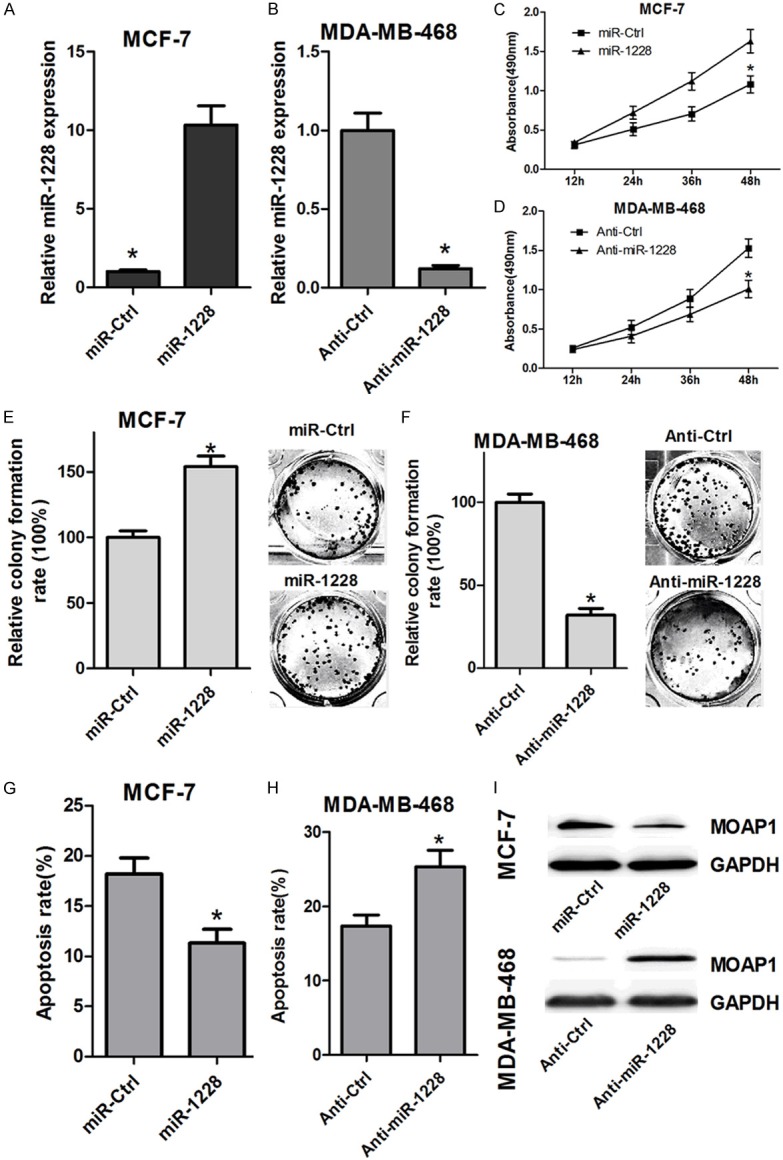
miR-1228 promoted breast cancer cell proliferation and reduces cellular apoptosis. A and B: The efficiency of miR-1228 and anti-miR-1228 were detected with RT-PCR in MCF-7 and MDA-MB-468 cells, respectively. C and D: Cell viability was determined for 12 h, 24 h, 36 h, and 48 h using MTT assay with MCF-7 cells transfected with miR-1228 or MDA-MB-468 transfected with anti-miR-1228. E and F: Cells long-term proliferation capacity was determined by colony formation assay with MCF-7 cells transfected with miR-1228 or MDA-MB-468 cells transfected with anti-miR-1228. G and H: The cells apoptosis was detected using Annexin V assay with MCF-7 cells transfected with miR-1228 or MDA-MB-468 cells transfected anti-miR-1228. I: Western blot was performed to detect the effect of miR-1228 and anti-miR-1228 on the expression of MOAP1 in MCF-7 and MDA-MB-468 cells, respectively.
miR-1228 promotes cell invasion and migration of breast cancer cell lines
To further investigate whether miR-1228 affects cell metastasis, transwell invasion assay and wound healing assay were performed. Compared with the control group, miR-1228 mimics promoted the invasion of MCF-7 cells (Figure 3A), while anti-miR-1228 inhibited MDA-MB-231 cells invasion (Figure 3B). Furthermore, we used wound healing assay to detect the function of miR-1228 on cell migration. As shown in Figure 3C and 3D, the miR-1228 mimics promoted the potential of MCF-7 cells migration, while anti-miR-1228 inhibited the migration potential of MDA-MB-231 cells. These results suggested that miR-1228 can promote breast cancer cells invasion and migration.
Figure 3.
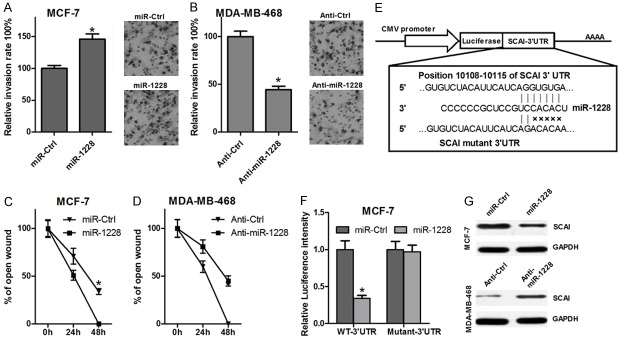
miR-1228 promotes cell metastasis and targets SCAI. A and B: Transwell invasion assay was performed in MCF-7 cells transfected with miR-1228 or MDA-MB-468 cells transfected with anti-miR-1228. C and D: Wound healing assay was performed in MCF-7 cells transfected with miR-1228 or MDA-MB-468 cells transfected with anti-miR-1228. E: miR-1228 seed sequence and its complementary binding site in MOAP1 3’UTR (Position: 10108-10115 bp) are highlighted. F: 3’UTR luciferase reporter assay was performed in MCF-7 cells co-transfected with miR-1228 plus WT-3’UTR or miR-1228 plus mutant-3’UTR. G: The protein expression of SCAI was determined using western blot with MCF-7 cells transfected with miR-1228 or MDA-MB-468 cells transfected with anti-miR-1228, GAPDH was detected as loading control.
SCAI emerges as a direct target of miR-1228
We then investigated the potential mechanism of the function of miR-1228 on cell invasion and migration. Based on the bioinfomatic data using three computational algorithms, TargetScan, miRDB and miRanda, SCAI (suppressor of cancer cell invasion), a previously identified protein that regulates invasive cell migration, was predicted as a potential target of miR-1228.
It is well known that miRNAs cause mRNA cleavage or translational repression by forming imperfect base pairing with the 3’UTR of target genes. SCAI mRNA carries a binding site for miR-1228 (Figure 3E), suggesting that SCAI transcript might be a direct target of miR-1228. We therefore used 3’UTR luciferase reporter assay to verify whether miR-1228 directly targets SCAI. As shown in Figure 3F, miR-1228 mimics dramatically suppressed the luciferase activity of the wild-type SCAI 3’-UTR in MCF-7 cells, whereas the profound inhibition was abolished when the seed sequences of the miR-1228 target sequences were mutated in the SCAI 3’-UTR vector (Figure 3E and 3F). Then the effect of miR-1228 on the expression of SCAI protein was evaluated in MCF-7 and MDA-MB-231 cells transfected with miR-1228 mimics or inhibitor. Compared with the control group, transfection with miR-1228 mimics lead to a marked reduction of SCAI expression in MCF-7 cells, whereas anti-miR-1228 induced a significant increase in SCAI protein expression (Figure 3G). These results provide evidence that miR-1228 inhibits SCAI protein expression through directly targeting the 3’UTR of SCAI mRNA.
miR-1228 promotion of cell growth and metastasis is significantly attenuated by MOAP1 and SCAI overexpression
Since SCAI has been confirmed as a direct target of miR-1228, we further evaluated the expression of SCAI in breast cancer cell lines and tissues. qRT-PCR was used and measure the mRNA expression levels of SCAI in breast cancer cell lines, which showed that miR-1228 had decreasing expression from MCF-10A, MCF-7, MDA-MB-231 to MDA-MB-468 cells (Figure 4A); Furthermore, we observed that SCAI mRNA and protein expression was significantly decreased in breast cancer tissue compared with adjacent normal breast tissues (Figure 4B and 4C).
Figure 4.
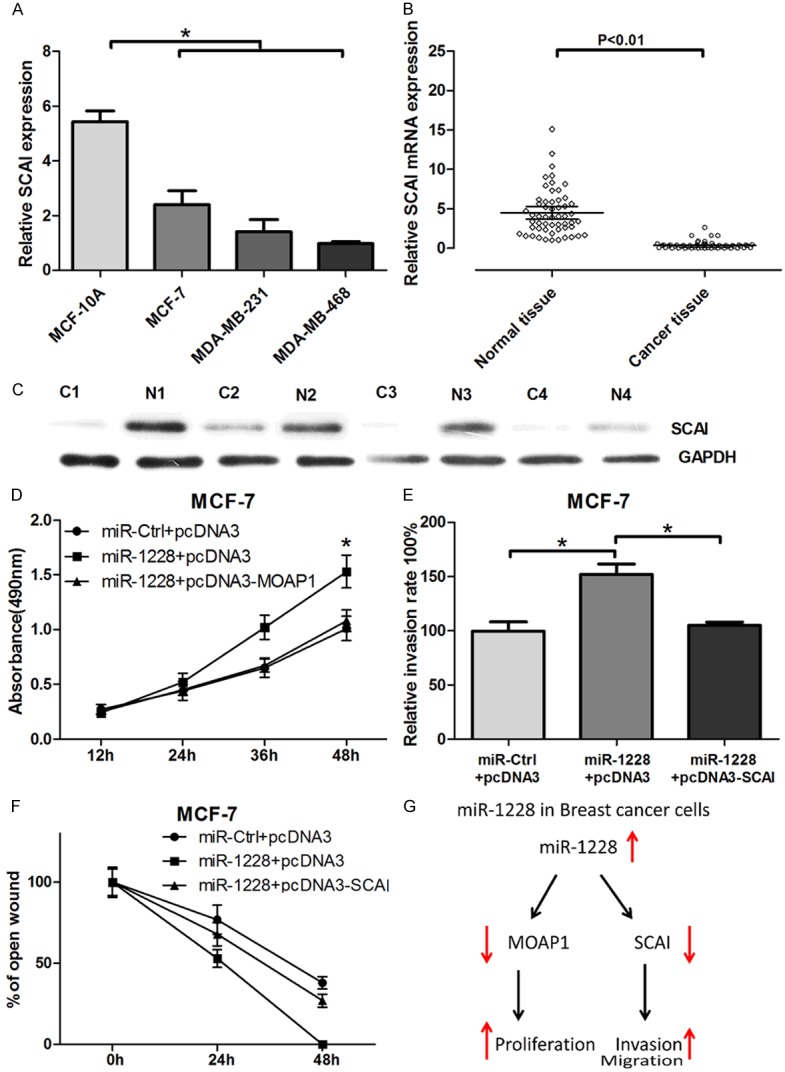
MOAP1 and SCAI can reverse miR-1228 induced cell proliferation, invasion and migration. A: Relative expression of SCAI in MCF-10A, MCF-7, MDA-MB-231 and MDA-MB-468 cells. B: Relative mRNA expression of SCAI in breast cancer tissues and adjacent normal tissues. C: The protein expression of SCAI in n breast cancer tissues and adjacent normal tissues. D: The effect of overexpression MOAP1 on miR-1228 induced cell viability detected with MTT. E and F: The effect of overexpression SCAI on miR-1228 induced cell invasion and migration detected with transwell and wound healing assay. G: An illustration of miR-1228 mechanism on breast cancer cells proliferation, invasion and migration.
To address whether miR-1228 effects on cellular proliferation, invasion and migration are indeed due to the suppression of MOAP1 and SCAI, a rescue experiment was then performed. As shown in Figure 4D, miR-1228 promoted MCF-7 cells proliferation was rescued by the transfection of pcDNA3-MOAP1. Furthermore, we confirmed that transfection of pcDNA3-SCAI in MCF-7 cells ablated the miR-1228 mimics induced cell invasion and migration (Figure 4E and 4F). These results suggested that miR-1228-induced breast cancer cell proliferation, invasion and migration were mediated by MOAP1 and SCAI, as illustrated in Figure 4G.
Discussion
MicroRNAs can modulate a wide variety of biological processes. It has been demonstrated that miRNAs can play specific roles in cancer cell proliferation, differentiation, migration and metastasis. Numerous miRNAs have been reported to be differentially expressed in breast cancer cells and tissues, suggesting their involvement in breast cancer pathogenesis [20]. However, the roles played by miRNAs in the pathogenesis of these diseases remained largely unknown. In this study, we focused on the miR-1228 potential effectiveness in breast cancer. We found the expression of miR-1228 were obviously increased in highly metastatic cells MDA-MB-231 and MDA-MB-468 cells, while the expression of miR-1228 was significantly decreased in non-malignant breast epithelial cell MCF-10A. Breast cancer tissues with the highest malignancy stage expressed the highest levels of miR-1228, and the expression of which was higher in metastases tissues compared with non-metastases tissues. Therefore we inferred that miR-1228 may have proliferation- and metastasis-promoting function on breast cancer pathogenesis. The hypothesis was further confirmed by our in vitro analysis in breast cancer cells with MTT, Transwell and wound healing assays.
The finding that miR-1228 targeting the protein MOAP1 to repress cellular apoptotic signaling has been described in HeLa cells [23]. Here we confirmed the result that miR-1228 can inhibit MCF-7 and MDA-MB-468 apoptosis. Furthermore, we verified that miR-1228 can repress the expression of MOAP1 in breast cancer cells. To explore other possible targets of miR-1228, different computational algorithms were used and we found SCAI may be a potential target of miR-1228. With the 3’UTR luciferase reporter assay, SCAI was identified as the direct target of miR-1228. The increase in miR-1228 expression is accompanied by down-regulation of SCAI expression. This fact may partly explain the down-regulation of SCAI during breast cancer pathogenesis, which was observed that SCAI was down-regulated in breast cancer tissues, and non-malignant breast epithelial cell MCF-10A expressed the highest level of SCAI, while highly metastatic cells (MDA-MB-231 and MDA-MB-468) expressed relatively lower SCAI expression.
SCAI (suppressor of cancer cell invasion) is downregulated in human tumors, implying that it has tumor suppressor characteristics [24,25]. SCAI has been demonstrated as a cofactor for human transcriptional coactivator MAL. Depletion of MAL proteins inhibits motility and invasive behavior [26,27]. Knockdown of SCAI by RNA-mediated interference causes a drastic upregulation of β1-integrin gene expression levels, leading to a striking increase in the ability of invasive cell migration [27], which was consistent with the promotion, which was consistent with invasion and migration induction ability in breast cancer cells and further confirmed our results that miR-1228 can target SCAI.
In summary, this study revealed that SCAI was a direct target of miR-1228. Enforced miR-1228 expression can reduce SCAI expression. Moreover, miR-1228 promoted breast cancer proliferation, invasion and migration was mediated by MOAP1 and SCAI. Therefore, these results can provide new insights into the pathogenesis and therapeutics of breast cancer.
Acknowledgements
This work was supported by Heilongjiang Province Science Fund for Distinguished Young (JC201203); National Natural Science Foundation of China (81372612); Heilongjiang Province Natural Science Foundation of China (H201425).
Disclosure of conflict of interest
None.
References
- 1.Ismail NI, Kaur G, Hashim H, Hassan MS. S100A4 overexpression proves to be independent marker for breast cancer progression. Cancer Cell Int. 2008;8:12. doi: 10.1186/1475-2867-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Olopade OI. MYC in breast tumor progression. Expert Rev Anticancer Ther. 2008;8:1689–1698. doi: 10.1586/14737140.8.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorusso G, Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol. 2008;130:1091–1103. doi: 10.1007/s00418-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 4.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson NM, Yigit MV. The role of microRNA in resistance to breast cancer therapy. Wiley Interdiscip Rev RNA. 2014;5:823–33. doi: 10.1002/wrna.1248. [DOI] [PubMed] [Google Scholar]
- 6.Gromak N. Intronic microRNAs: a crossroad in gene regulation. Biochem Soc Trans. 2012;40:759–761. doi: 10.1042/BST20120023. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S, Teal CB, Man YG, Brem RF, Fu SW. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple negative breast cancer. Breast Cancer Res. 2014;16:435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Shang L, Bai S, Chen J, He X, Martin-Trevino R, Chen S, Li XY, Meng X, Yu B, Wang X, Liu Y, McDermott SP, Ariazi AE, Ginestier C, Ibarra I, Ke J, Luther T, Clouthier SG, Xu L, Shan G, Song E, Yao H, Hannon GJ, Weiss SJ, Wicha MS, Liu S. microRNA100 inhibits self-renewal of breast cancer stem-like cells and breast tumor development. Cancer Res. 2014;74:6648–60. doi: 10.1158/0008-5472.CAN-13-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34:3240–50. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by LincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–68. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 11.Sandhu R, Rein J, D’Arcy M, Herschkowitz JI, Hoadley KA, Troester MA. Overexpression of miR-146a in basal-like breast cancer cells confers enhanced tumorigenic potential in association with altered p53 status. Carcinogenesis. 2014;35:2567–75. doi: 10.1093/carcin/bgu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong C, Nie Y, Qu S, Liao JY, Cui X, Yao H, Zeng Y, Su F, Song E, Liu Q. miR-21 Induces Myofibroblast Differentiation and Promotes the Malignant Progression of Breast Phyllodes Tumors. Cancer Res. 2014;74:4341–4352. doi: 10.1158/0008-5472.CAN-14-0125. [DOI] [PubMed] [Google Scholar]
- 13.Waning DL, Mohammad KS, Guise TA. Cancer-associated osteoclast differentiation takes a good look in the miR(NA)ror. Cancer Cell. 2013;24:407–409. doi: 10.1016/j.ccr.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abba M, Patil N, Allgayer H. MicroRNAs in the Regulation of MMPs and Metastasis. Cancers (Basel) 2014;6:625–645. doi: 10.3390/cancers6020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 20.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/ tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 21.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan B, Zhao JL. miR-1228 prevents cellular apoptosis through targeting of MOAP1 protein. Apoptosis. 2012;17:717–724. doi: 10.1007/s10495-012-0710-9. [DOI] [PubMed] [Google Scholar]
- 24.Brandt DT, Baarlink C, Kitzing TM, Kremmer E, Ivaska J, Nollau P, Grosse R. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol. 2009;11:557–568. doi: 10.1038/ncb1862. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Hu W, Xie B, Gao H, Xu C, Chen J. Downregulation of SCAI enhances glioma cell invasion and stem cell like phenotype by activating Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;448:206–211. doi: 10.1016/j.bbrc.2014.04.098. [DOI] [PubMed] [Google Scholar]
- 26.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliano R. SCAI blocks MAL-evolent effects on cancer cell invasion. Nat Cell Biol. 2009;11:540–542. doi: 10.1038/ncb0509-540. [DOI] [PubMed] [Google Scholar]


