Abstract
microRNAs (miRNAs) are a class of small non-coding RNAs that play important roles in a variety of biological process. It has been reported that dysregulation of miRNA is always associated with cancer progression and development, and miR-378 aberrant expression has been found in some types of cancers. However, the association of miR-378 and glioma has not been evaluated. In this work, we measured the expression of miR-378 in glioma tissues and non-neoplastic brain tissues was measured using real-time PCR, and found that miRNA-378 expression level was significantly lower in glioma tissues compared with non-neoplastic brain tissues. Patients with lower miR-378 expression level had significantly poorer overall survival. Multivariate Cox regression analysis showed that miR-378 expression was an independent prognostic factor for 5-year overall survival. Over-expression of miR-378 inhibits glioma cell migration and invasion. In conclusion, our results indicated that miR-378 may serve as a tumor suppressor and play an important role in inhibiting tumor migration and invasion. Our work implicates the potential effect of miR-378 on the prognosis of glioma.
Keywords: miR-378, glioma, prognosis, cell migration and invasion
Introduction
Glioma is one of the most common malignant brain tumors and represents about 30% of all intrinsic neoplasms of the central nervous system. It have been reported that glioma has a poor prognosis because of the characteristic progressive overgrowth and diffuse invasion. The World Health Organization (WHO) classification divides glioma into four grades (I to IV), however, this criteria may not sufficient to estimate patient prognosis [1-3]. Therefore, investigating the pathogenesis and biological features of glioma is crucial to enhance early detection and treatment.
MicroRNAs (miRNAs) are a class of small noncoding RNAs with the size range of 21-25 nucleotides, and it plays important role at post-transcriptional regulation levels [4]. It can lead to mRNA degradation or inhibition of translation through imperfect hybridization to target 3’-untranslated regions (UTR). Many evidences suggest that miRNAs play important roles in various biological processes, including cell proliferation and differentiation [5,6]. In addition, studies have de-monstrated that dysregulation of miRNA is al-ways associated with the tumorigenesis and progression of various types of tumors, and a number of miRNAs have been reported to regulate tumor metastasis [7,8]. Many miRNAs may serve as markers for cancer diagnosis and prognosis.
Dysregulation of miRNA expression have been documented to play crucial role in tumorigenesis and cancer progression [9]. Recently, the expression of miR-378 was shown to be des-regulated in several human cancers, such as colorectal cancer and oral carcinoma [10,11]. However, the underlying molecular mechanism and clinical significance of miR-378 in glioma remains unclear. In this work, we explored the expression difference between glioma tissues and non-neoplastic brain tissues, and evaluated the clinical significance of miR-378 in glioma patients. Furthermore, we examined the effects of miR-378 on glioma cells migration and invasion, which implicates the potential effects of miR-378 on glioma prognosis.
Materials and methods
Patients and tissue samples
Surgical specimens of glioma tissues and adjacent non-neoplastic brain tissues were obtained from patients with a diagnosis of glioma who underwent surgery at the Wuxi Second People’s Hospital (Jiangsu Province, China) between April 2006 and June 2012. All patients had complete five-year follow-up, and informed written consents were obtained from all patients. None of the patients had received radiotherapy before surgery excision. The tumor tissue and adjacent non-neoplastic brain tissues were divided by tissue laser microdissection. All tissue samples were immediately frozen in liquid nitrogen and stored at -80°C until further use.
RNA extraction and RT-PCR
The total RNA was extracted using the TRIzol Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. Quantitative RT-PCR was performed using All-in-OneTM mi-RNA quantitative RT-PCR Detection Kit (GeneCopoeia, Rockville, MD). The primer se-quences of miR-378 and U6 were purchased from Applied Biosystems (ABI, Foster City, CA, USA). RT-PCR was performed using TaKaRa SYBR Green PCR Kit (TaKaRa), and measured in a LightCycler 480 system (Roche, Basel, Switherland). Relative quantification of miR-378 expression was calculated by using the 2-ΔΔCT method.
Cell culture and transfection of miRNA
Four human glioma cell lines (U87, U251, MT330 and SJ-G2) and the normal skin fibroblasts (HF) were obtained from the American Type Culture Collection and cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum, 100 µ/ml penicillin and 100 mg/ml streptomycin, at 37°C in a humidified atmosphere containing 5% CO2. The pre-miR miRNA-378 (Pre-miR-378), pre-miR negative control (Pre-miR-nc) were purchased from Ribobio (China). A final concentration of 2 × 105 cells were seeded into each well of a 6-well plate and transfected for 48 hours using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s protocol.
In vitro migration and invasion assays
The cell migration and invasion abilities were assessed using transwell inserts with 8 µm pores (BD Biosciences, San Jose, CA, USA). For transwell migration assay, 2 × 105 cells were added into the top chamber lined with a noncoated membrane. For invasion assay, 2 × 105 cells were placed to each upper compartment of the chamber pre-coated with matrigel matrix. In both assays, cells were suspended in medium without growth or serum factors, and medium containing 10% fetal bovine serum was used as the chemoattractant in the lower chamber. After the incubation for 48 hours and stained with hematoxylin for 10 minutes, the non-invaded cells were removed from the top chambers. The number of cells on the lower surface of the membrane was counted under a microscope in five random fields.
Statistical analysis
The data presented in this work have been repeated at least three independent experiments. The differences between groups were measured using the nonparametric test (Mann-Whitney U test) and categorical data were evaluated using Chi-square test. Kaplan-Meier method was used to calculate the overall survival rate. The Cox proportional hazards model was employed for the multivariate analysis. All statistical analyses were performed using SPSS version 16.0 (SPSS, Chicago, IL, USA). All of the P values were two-sided, and the differences were considered to be statistically significant at P-value < 0.05.
Results
Expression of miR-378 in glioma tissues and cell lines
At first, we explored the expression levels of miR-378 in 100 pairs of glioma tissues and adjacent non-neoplastic brain tissues from 100 glioma patients by qRT-PCR. The result showed that miR-378 expression level was significantly lower in glioma tissues (2.3 ± 0.77) compared with adjacent non-neoplastic brain tissues (4.5 ± 1.04, P < 0.0001, Figure 1A). Further analysis of miR-378 expression in cell lines showed that miR-378 expressions in glioma cell lines (U87, U251, MT330 and SJ-G2) were significantly lower compared with normal skin fibroblast cell line (Figure 1B).
Figure 1.
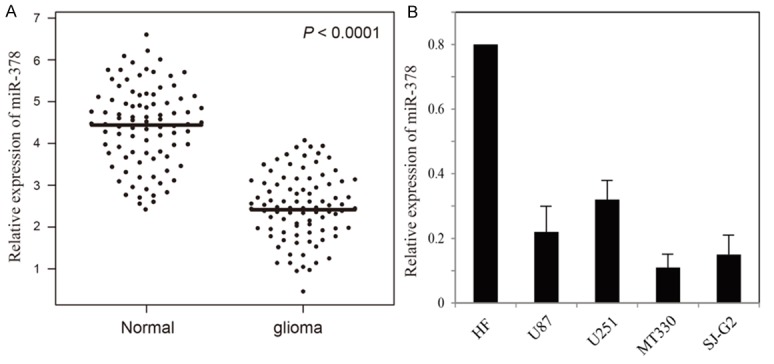
Relative expression levels of miR-378 in glioma tissues and cell lines. A. The relative expression of miR-378 in glioma tissues and adjacent non-neoplastic brain tissues. The bars represent the means of the relative expression of miR-378. B. The relative expression of miR-378 in glioma cell lines.
Clinical significance of miR-378 expression in glioma
To measure the association between miR-378 expression and clinicopathological characteristics, the 100 patients with glioma were divided into two groups according to the median value of miR-378 expression level, including high-expression group (n = 50) and low-expression group (n = 50), respectively. The correlation between miR-378 expression and pathological factors was evaluated (Table 1), and the result showed that low expression of miR-378 was significantly associated with tumor size (P = 0.012), KPS (P = 0.005) and WHO grade (P < 0.001). No significant differences about gender, age were found. In the Kaplan-Meier survival analysis, the patients with lower miR-378 expression level had a significantly poorer prognosis than those with high miR-378 expression level (P = 0.001, Figure 2). Univariate proportional hazard model showed that the WHO grade and miR-378 expression level were prognostic predictors. Those parameters which significantly associated with overall survive in the univariate analysis were further evaluated by multivariate analysis. The result showed that WHO grade (P = 0.001) and miR-378 expression level (P = 0.001, Table 2) were independent prognostic factors for overall survival.
Table 1.
Clinicopathological associations of miR-378 expression in glioma
| miR-378 expression | |||
|---|---|---|---|
| Variable | Low (n = 50) | High (n = 50) | P-value |
| Ages (years) | 0.18 | ||
| < 50 | 19 | 24 | |
| ≥ 50 | 31 | 26 | |
| Gender | 0.98 | ||
| Male | 28 | 26 | |
| Female | 22 | 24 | |
| Tumor size | 0.012 | ||
| < 5 cm | 32 | 37 | |
| ≥ 5 cm | 18 | 13 | |
| KPS | 0.005 | ||
| < 90 | 35 | 28 | |
| ≥ 90 | 15 | 22 | |
| WHO grade | < 0.001 | ||
| I | 0 | 18 | |
| II | 1 | 15 | |
| III | 20 | 12 | |
| IV | 29 | 5 | |
Figure 2.
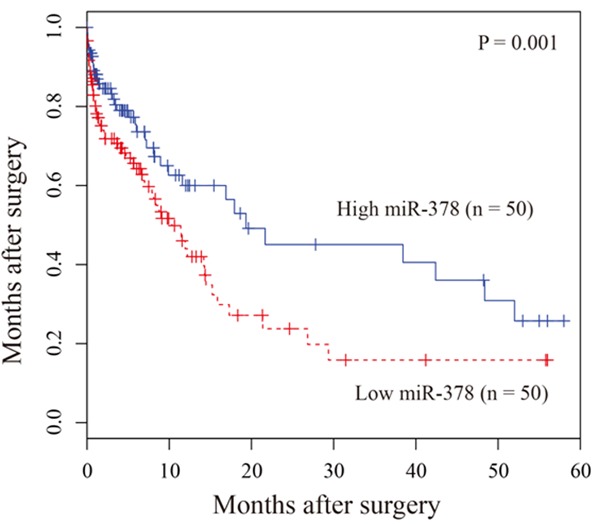
Kaplan-Meier survival curves of patients with glioma based on miR-378 expression status. Patients with low expression group have significantly poorer prognosis than those in high expression group.
Table 2.
Univariate and multivariate analyses of prognostic factors in glioma
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk | 95% CI | P-value | Risk | 95% CI | P-value | |
| Age (Years) | 1.06 | 0.96-1.22 | 0.59 | 1.01 | 0.92-1.22 | 0.55 |
| Gender | 1.16 | 0.82-1.48 | 0.44 | 1.09 | 0.88-1.44 | 0.43 |
| KPS | 1.41 | 0.76-2.55 | 0.11 | 1.21 | 0.74-2.11 | 0.09 |
| Tumor size | 1.08 | 0.96-1.24 | 0.12 | 1.06 | 0.96-4.68 | 0.26 |
| WHO grade | 2.52 | 1.22-4.47 | < 0.001 | 1.78 | 1.11-3.68 | 0.001 |
| miR-378 | 2.26 | 1.16-3.05 | < 0.001 | 1.68 | 1.14-5.96 | 0.001 |
In vitro effect of miR-378 on glioma cell migration and invasion
To investigate whether miR-378 regulates glioma cell migration and invasion, we performed in vitro glioma cell migration and invasion assays by transfecting pre-miR-378 or pre-miR-nc into U87 cell. The results sho-wed that overexpression of miR-378 significantly inhibited glioma cell migration and invasion (Figure 3).
Figure 3.
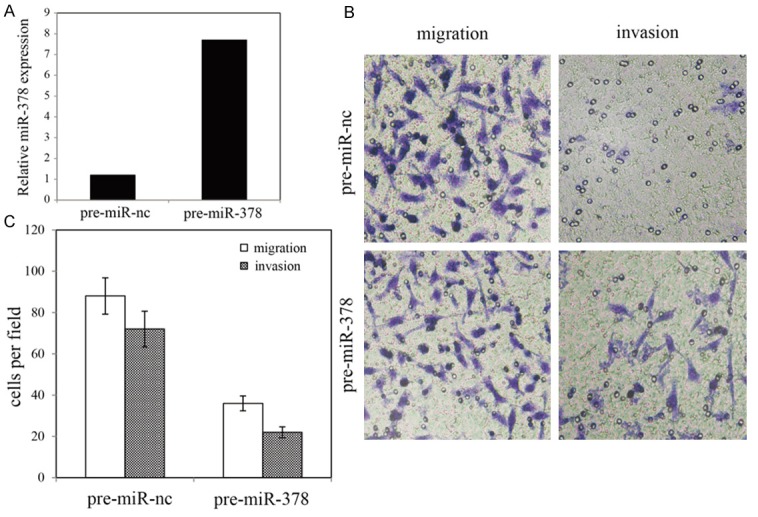
Effects of miR-378 on migration and invasion of U87 cell line. A. qRT-PCR analysis of miR-378 in U87 cells. B. Invasion and migration assay of U87 cells and representative fields of migration and invasive cells. C. Average number of invasive and migration cells per field from three independent experiments.
Discussion
Glioma is one of the most common tumors with a relative low survival rate. Prognostic factor detection in glioma is essential to predict patients’ survival and determine optimal therapeutic strategies. Up to date, many efforts have been devoted to explore specific biological markers with prognosis in glioma [12]. As a novel biomarker, the potential rules of miRNAs in predicting prognosis has been extensively identified. Until now, nearly 1900 human miRNAs have been identified, which can regulate 60%~80% of the genes in humans [13]. Due to the crucial role of miRNA regulation at the post-transcription level, miRNAs are involved in many important biological functions, such as differentiation, proliferation. It has also been implemented in diverse pathological processes in cancers [14-16].
It has been reported that dysregulation of miR-378 performed an important function in diverse cancer types, such as oral carcinoma and colorectal cancer [10,11,17,18]. However, there are still no studies on the association between miR-378 and glioma. In this work, we investigated the expression level of miR-378 in glioma and adjacent non-neoplastic brain tissues. Moreover, we determined whether miR-378 expression could predict the outcome of glioma patients. The results showed that the expression of miR-378 was extensively down-regulated in glioma tissues and cell lines. Further multivariate survival analysis showed that miR-378 might be involved in glioma and could be used as a potential prognostic biomarker for glioma. Subsequent work showed that overexpression of miR-378 could significantly inhibit cell migration and invasion, which indicates that miR-378 might function as a tumor suppressor in glioma.
In conclusion, our present work showed that miR-378 expression was decreased in glioma and is tightly associated with cancer cell migration and invasion. Our data implicated for the first time that miR-378 expression was an independent prognostic factor of glioma patients.
Disclosure of conflict of interest
None.
References
- 1.Dellaretti M, Reyns N, Touzet G, Dubois F, Gusmão S, Pereira JL, Blond S. Diffuse brainstem glioma: prognostic factors. J Neurosurg. 2012;117:810–4. doi: 10.3171/2012.7.JNS111992. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau A, Mokhtari K, Duyckaerts C. The 2007 WHO classification of tumors of the central nervous system-what has changed? Curr Opin Neurol. 2008;21:720–7. doi: 10.1097/WCO.0b013e328312c3a7. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DR, Galanis E. Incorporation of prognostic and predictive factors into glioma clinical trials. Curr Oncol Rep. 2013;15:56–63. doi: 10.1007/s11912-012-0279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–12. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, Yang XQ, Zhang GH, Xu XC, Zhu T, Wu Q. Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC Cancer. 2012;12:51. doi: 10.1186/1471-2407-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23:1229–34. doi: 10.1177/039463201002300427. [DOI] [PubMed] [Google Scholar]
- 12.Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Ann Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12:319–21. doi: 10.1016/S1470-2045(11)70067-5. [DOI] [PubMed] [Google Scholar]
- 15.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 16.Liu A, Tetzlaff MT, Vanbelle P, Elder D, Feldman M, Tobias JW, Sepulveda AR, Xu X. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–27. [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JK, Kiet TK, Blansit K, Ramasubbaiah R, Hilton JF, Kapp DS, Matei D. MiR-378 as a biomarker for response to anti-angiogenic treatment in ovarian cancer. Gynecol Oncol. 2014;133:568–74. doi: 10.1016/j.ygyno.2014.03.564. [DOI] [PubMed] [Google Scholar]
- 18.Yin JY, Deng ZQ, Liu FQ, Qian J, Lin J, Tang Q, Wen XM, Zhou JD, Zhang YY, Zhu XW. Association between mir-24 and mir-378 in formalin-fixed paraffin-embedded tissues of breast cancer. Int J Clin Exp Pathol. 2014;7:4261–7. [PMC free article] [PubMed] [Google Scholar]


