Abstract
Purpose: Genetic polymorphisms in CYP3A4 can change its activity to a certain degree, thus leading to differences among different populations in drug efficacy or adverse drug reactions. Methods: The study was intended to validate the genetic polymorphisms in CYP3A4 in Uygur Chinese population, we sequenced and screened for genetic variants including 5’UTR, promoters, exons, introns, and 3’UTR region of the whole CYP3A4 gene in 100 unrelated, healthy. Results: Twenty-one genetic polymorphisms in CYP3A4, and nine of them were novel. We detected CYP3A4*8, a putative poor-metabolizer allele, with the frequency of 0.5% in Uygur population. Tfsitescan revealed that the density of transcription factor varied in the different promoter regions, among which some were key regions for transcription factor binding. Conclusion: our results provide basic information about CPY3A4 alleles in Uygur and suggest that the enzymatic activities of CPY3A4 may differ among the diverse ethnic populations of China.
Keywords: CPY3A4, genetic polymorphism, haplotype, Uygur, ethnic difference
Introduction
The cytochrome P450 (CYP450) super-gene family codes more than 500 kinds of enzymes can mainly be found in the liver and small bowel in vivo. CYP450 enzymes exist many isozymes which can be divided into a number of genes/subgenes families, the enzymes of the same family own similar functions [1-3]. Notably, CYP450 enzymes show extensive structural differences due to genetic polymorphisms in the corresponding genes, thus giving rise to different enzymatic activities and leading to great intra- and inter-population differences in drug efficacy and adverse reactions.
CYP3A4, a subfamily of CYP450, expanding on chromosome 7q22.1, included 13 exons and 12 introns [4]. It is the most important drug metabolizing enzymes in the liver which accounts for 30-40% of total P450 enzymes and involved in metabolism of more than 50% of clinical commonly used drugs [5,6]. CYP3A4 is a polymorphic enzyme, the expression of CYP3A4 among individuals varied as much as 40 times, to understand the expression of CYP3A4 may determine drug efficacy and safety, thus helping people make the right due to dose [7,8].
Uygur is an ethnic with a population of 10,069,347 (according to the sixth population survey of China in 2011). They live mostly in the Xinjiang Autonomous Region. Places of residence are relatively stable in the Uygur population and there is little migration [9]. We systematically screened the whole CYP3A4 genes in 100 healthy, unrelated Uygurs for polymorphisms, hoping to find corresponding phenotypes and offer recommendations pertaining to the drug substrates of CYP3A4 in the Uygur population.
Materials and methods
Subjects
We recruited a random sample of 100 healthy, unrelated Uygurs (including 50 males and 50 females) between October 2010 and December 2011 from Xinjiang University in Xinjiang Autonomous Region for population genetics research. All of the chosen subjects were Uygur Chinese living in the Xinjiang Autonomous Region of China and had at least three generations of paternal ancestry in their ethnic. We used detailed recruitment and exclusion criteria excluding subjects with chronic diseases involving vital organs (heart, lung, liver, kidney, and brain) and other related diseases. The purpose of the exclusion procedures was to minimize the known environmental and therapeutic factors that influence genetic variation in the gene CYP3A4.
We informed all of the participants about the experimental procedures and the purpose of the study. The Human Research Committee of the Xinjiang University and Northwest University for Approval of Research Involving Human Subjects approved the use of human blood in this study. We also obtained signed informed consent from each study participant.
PCR and DNA sequencing
We used GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Ltd.) according to the manufacture’s protocol to extract genomic DNA from peripheral blood. Our PCR primers were designed using primer-Blast (http://frodo.wi.mit.edu/primer3/) to amplify about 900bp of the 5’ flanking regions, all exons, all introns, and 3’UTR of the CYP3A4 gene. We performed the PCR in a total volume of 10 μl containing 1 μl genomic DNA (20 ng/μl), 5 μl Hot Star Taq Master Mix, 0.5 μl each primer pair (5 μM), and 3 μl deionized water. The cycling protocol consisted of denaturation at 95°C for 15 min, followed by 35 cycles of 95°C for 30 s, 55-64°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 3 min to hold. The PCR products were purified by incubating them with 0.5 μl shrimp alkaline phosphate (Roche, Basel, Switzerland), 1.5 μl deionized water, and 8 μl HotStar PCR product, for a total volume of 10 μl, at 37°C for 30 min, followed by heat inactivation at 80°C for 15 min. We directly sequenced the purified PCR products using an ABI Prism BigDye Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems), on an ABI Prism 3100 sequencer (Applied Biosystems).
Data analysis
We used the Squencher 4.10.1 (http://www.genecodes.com/) software for analysis of the sequences including base calling, fragment assembly, and detection of SNPs, insertions, and deletions. We named the CYP3A4 variants based on the nucleotide reference sequence AY545216 and CYP allele nomenclature (http://www.cypalleles.ki.se/). We assessed linkage disequilibrium (LD) and Hardy-Weinberg equilibrium for each genetic variant using HAPLOVIEW 4.1 (http://broad.mit.edu/mpg/haploview) [10]. We constructed haplotypes from the selected tag SNPs and derived the haplotype frequencies for the Uygur population.
Transcriptional prediction
We analyzed variants in the CYP3A4 promoter region to predict their potential effects on transcription. We used the online tool Tfsitescan (www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) and the transcription factor binding sites database to investigate the effects that promoter-region variants have on transcription [11]. We analyzed the wild-type and allelic variants separately. Depending on the metabolic activity of CYP3A4, we divided the subjects into three phenotypic groups: poor metabolizer (PM), extensive metabolizer (EM), and ultra-rapid metabolizer (UM), according to CYP allele nomenclature (http://www.cypalleles.ki.se/) [12].
Functional prediction of nonsynonymous mutation
We analyzed nonsynonymous mutations in the CYP3A4 exon region to predict their potential effects on protein function. We used online tool SIFT (http://sift.bii.a-star.edu.sg/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) to evaluate the effect on exon-region variants have on protein function [13-15]. The results of SIFT prediction can be classified in four categories: tolerant (0.201-1.00), borderline (0.101-0.20), potentially intolerant (0.051-0.10), and intolerant (0-0.05). The results of PolyPhen-2 can be classified in five categories: probably benign (0-0.999), bordline (1.000-1.249), potentially damaging (1.250-1.499), possibly damaging (1.500-1.999) and probably damaging (≥2).
Results
Genetic variants
We successfully sequenced CYP3A4 from 100 volunteer subjects, including 50 males and 50 females. We identify a total of twenty-one CYP3A4 polymorphisms in the current Uygur population, among which nine of the polymorphisms are not previously reported in the NCBI database nor the Human Cytochrome P450 Allele Nomenclature Committee tables, most variants have the frequency of less than 5%, besides four variants in introns showed relatively high frequencies of 74% (15977T > C, intron7), 11.5% (16613C > T, intron7), 11.5% (20230G > A, intron10) and 13% (25721A > G, intron12) (Table 1). One of the novel polymorphisms is within the promoter region, three are in exons among which one is nonsynonymous mutations, four are in the introns, and one is in 3’UTR region. We did not find any CYP duplications or deletions. The primers of Promoter, 13 exons and 3’UTR region were seen in Table S1.
Table 1.
Polymorphisms and frequency distribution of CYP3A4 in Uyghur population
| # | SNP ID | Position | Region | Allele | Effect | Flanking Sequence | MAF |
|---|---|---|---|---|---|---|---|
| 1 | / | -747C > G | Promoter | CYP3A4*1F | / | TGTACAGCAC S CTGGTAGGGA | 0.025 |
| 2 | / | -667C > T | Promoter | CYP3A4*1W | / | GGGAAACAGG Y GTGGAAACAC | 0.005 |
| 3 | new1 | -549C > G | Promoter | / | AGGAAAGACT S TAAGAGAAGG | 0.005 | |
| 4 | rs2740574 | -392A > G | Promoter | CYP3A4*1B | / | GACAAGGGCA R GAGAGAGGCG | 0.015 |
| 5 | / | -62C > A | 5’UTR | CYP3A4*1D | / | CATAGCCCAG M AAAGAGCAAC | 0.005 |
| 6 | rs55913187 | 3858C > T | Intron1 | Not translated | TAATCATTGC Y GTCAGAGTTA | 0.015 | |
| 7 | new2 | 5939A > T | Intron5 | Not translated | TTTTCATCCC W ATTAGAGGCA | 0.005 | |
| 8 | / | 13908G > A | Exon5 | CYP3A4*8 | 130R > Q | AAGAGATTAC R ATCATTGCTG | 0.005 |
| 9 | new3 | 15380C > T | Intron6 | Not translated | TCACACCCAG Y GTAGGGCCAG | 0.015 | |
| 10 | / | 15727G > C | Intron7 | CYP3A4*1P | Not translated | TAAGTATGTG S ACTACTATTT | 0.015 |
| 11 | / | 15753T > C | Intron7 | T15781G* | Not translated | TATTTATCTT Y CTCTCTTAAA | 0.025 |
| 12 | / | 15977T > C | Intron7 | C78013T* | Not translated | AACTTTCTGC Y TCTATGGATT | 0.740 |
| 13 | 16613C > T | Intron7 | C78649T* | Not translated | ACTGCTGTAG Y GGTGCTCCTT | 0.115 | |
| 14 | new4 | 16934A > T | Exon8 | 264T > S | CCTCGAAGAT W CACAAAAGGT | 0.005 | |
| 15 | new5 | 17864A > C | Intron9 | Not translated | AGAGGCATGC M GGCATAGATA | 0.005 | |
| 16 | rs2242480 | 20230G > A | Intron10 | CYP3A4*1G | Not translated | TGAGTGGATG R TACATGGAGA | 0.115 |
| 17 | rs4986910 | 23172T > C | Exon12 | CYP3A4*3 | 445M > T | TGCATTGGCA Y GAGGTTTGCT | 0.005 |
| 18 | new7 | 25721A > G | Intron12 | Not translated | ATTACTCCAT R GAGATCAGAA | 0.130 | |
| 19 | new8 | 25925C > T | Exon13 | 499T > T | GGGATGGCAC Y GTAAGTGGAG | 0.010 | |
| 20 | new9 | 25931T > C | Exon13 | 501S > S | GCACCGTAAG Y GGAGCCTGAA | 0.005 | |
| 21 | new10 | 26759G > C | 3’UTR | / | TGGTGGACTC S CCTGTAATCT | 0.005 |
MAF: minor allele frequency.
Allele frequency and genotype frequency
We detected nine CYP3A4 alleles in the Uygur population (Table 2). The CYP3A4*1A represented the wild type CYP3A4 allele and had the highest frequency (81%), followed by the CYP3A4*1G (11.5%), CYP3A4*1F (2.5%), CYP3A4*1B (1.5%) and CYP3A4*1P (1.5%) allele. The rest alleles: 3A4*1W, *1D, *3 and *8; were relatively rare with frequencies of 0.5%. We also compared the allle frequency in different populations and we found that the allele frequency of CYP3A4*1B in the current study was 1.5%, which was as much as 82% in African population (Table 3).
Table 2.
Alleles and frequencies of CYP3A4 in Uyghur population
| Allele | Alleles (N) | Frequency (%) | Enzyme Activity |
|---|---|---|---|
| CYP3A4*1A | 162 | 81% | Normal |
| CYP3A4*1F | 5 | 2.5% | / |
| CYP3A4*1W | 1 | 0.5% | / |
| CYP3A4*1B | 2 | 1.5% | / |
| CYP3A4*1D | 1 | 0.5% | / |
| CYP3A4*1P | 3 | 1.5% | / |
| CYP3A4*1G | 23 | 11.5% | / |
| CYP3A4*3 | 1 | 0.5% | Decreased |
| CYP3A4*8 | 1 | 0.5% | Decreased |
Table 3.
Comparison of CYP3A4 alleles in different populations
| Ethnicity | N | *1B | *3 | *4 | *5 | *6 | *8 | *11 | *13 | *14 | *15 | *16 | *17 | *18 | *21 | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uyghur | 100 | 1.5% | 0.5% | 0.5% | / | |||||||||||
| Han Chinese | 100 | 0.5% | 0.5% | 1% | 1% | 0.5% | [19] | |||||||||
| Chinese Han, She, Dong | 60 | 1% | 1% | 7% | 16% | 2% | 1% | [18] | ||||||||
| Taiwan Chinese | 102 | 2.9% | 1.9% | 0.5% | [32] | |||||||||||
| Japanese | 416 | 0.1% | 0.2% | 1.4% | 2.8% | [33] | ||||||||||
| Malaysian | 121 | 0% | 0% | 2.1% | [28] | |||||||||||
| European | 95 | 4% | 2% | [34] | ||||||||||||
| African | 95 | 82% | 0% | [34] |
We identified ten CYP3A4 genotypes in the Uygur population, with frequencies ranging from 1% to 67%, listed in Table 4. Eight of the genotypes have normal enzyme activity and two have decreased enzyme activity.
Table 4.
Genotype and frequency distribution in Uyghur population
| Genotype | Subjects (N) | Frequency (%) | Phenotype |
|---|---|---|---|
| *1A/*1A | 66 | 67% | Normal |
| *1A/*1G | 19 | 19% | Normal |
| *1A/*1F | 5 | 5% | Normal |
| *1A/*1P | 3 | 3% | Normal |
| *1A/*1W | 1 | 1% | Normal |
| *1G/*1D | 1 | 1% | Normal |
| *1G/*1B | 1 | 1% | Normal |
| *1A/*1B | 1 | 1% | Normal |
| *1G/*3 | 1 | 1% | PM |
| *1G/*8 | 1 | 1% | PM |
LD analysis
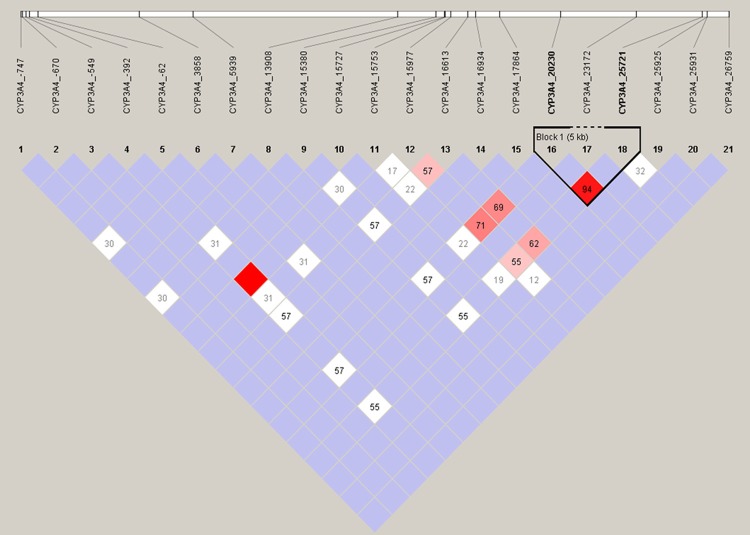
We performed LD analysis using Haploview with confidence intervals to define blocks. By default, our method ignores markers with MAF < 0.05; SNPs with lower frequencies have little power to detect LD (Figure 1). We found the twenty-one genetic polymorphisms in CYP3A4 rendered a relatively weak linkage relationship since the D’ value (a measure of the extent of LD for each pair of SNPs) on the square is relatively small. We identified one LD block comprised of three haplotypes of GA, AG and GG with the frequencies of 87%, 11% and 2% respectively.
Figure 1.
Linkage disequilibrium analysis of CYP3A4. LD is displayed by standard color schemes, with bright red for very strong LD (LOD > 2, D’ = 1), pink red (LOD > 2, D’ < 1) and blue (LOD < 2, D’ = 1) for intermediate LD, and white (LOD < 2, D’ < 1) for no LD.
Genetic variants in the transcription factor binding site
We found four polymorphisims including -747C > G, -667C > T, -549C > G, and -392A > G in promoter region. The search for transcription factor binding domains by Tfsitescan revealed that the -747C > G, -667C > T and -392A > G mutation altered the original binding sites of transcription factors thus potentially affecting the binding proteins and resulting in alteration of the corresponding gene expression, the new polymorphism -549C > G mutation makes no effect on the binding of the transcription factors (Figure 2).
Figure 2.
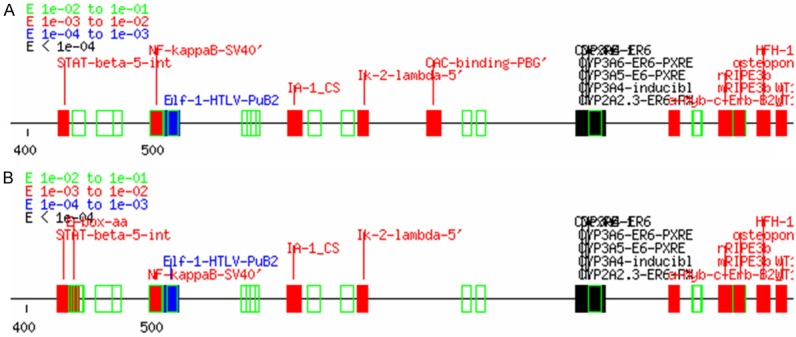
Transcription factor binding sites prediction of genetic variants in promoter region. A. Prediction map of TfsiteScan with the original sequence. B. Prediction map of TfsiteScan with the mutantre sequence.
Nonsynonymous mutation in the functional change of protein function
We found three polymorphisims including16934A > T, 13908G > A and 23172T > C caused nonnsynonymous mutation in exon region. SIFT and PolyPhen showed high consistency that novel variant 16934A > T had little effect on the change of protein function, but for 13908G > A and 23172T > C, the function were changed to a damaging direction (Table 5A, 5B).
Table 5A.
Results of SIFT predictions of nsSNPS
| SNPs | Coordinates | Codons | Ensemble Transcript ID | Substitution | dbSNP | Score* | Prediction |
|---|---|---|---|---|---|---|---|
| 13908 G > A | 7,99367788 C/T | CCA CaA | ENST000 00336411 | R130Q | novel | 0 | DAMAGING |
| 16934 A > T | 7,99364762 T/A | ACA tCA | ENST000 00424826 | T180S | novel | 0.51 | TOLERATED |
| 23172 T > C | 7,99358524 A/G | ATG AcG | ENST000 00424826 | M361T | novel | 0 | DAMAGING |
Table 5B.
Results of PolyPhen predictions of nsSNPS
| RS# | Position | SNPs | HumVar | HumDiv | Prediction |
|---|---|---|---|---|---|
| / | Exon5 | 13908G > A R130Q | 1.000 (sensitivity:0.00; specificity:1.00) | 1.000 (sensitivity:0.00; specificity:1.00) | Probably damaging |
| new | Exon8 | 16934A > T T264S | 0.001 (sensitivity:0.99; specificity:0.09) | 0.000 (sensitivity:1.00; specificity:0.00) | Benign |
| rs4986910 | Exon12 | 23172T > C M445T | 0.909 (sensitivity:0.69; specificity:0.90) | 0.987 (sensitivity:0.73; specificity:0.96) | Probably damaging |
Discussion
Our study, for the first time, systematically screened for variants of CYP3A4 by direct sequencing among the Uygur population and compared the results with other ethnic populations around the world. We found twenty-one genetic variants including nine novel polymorphisms, nine alleles and ten genotypes. One of the novel genetic variants within the promoter region and makes no effect on the binding of transcription factors. Among the three variants within exons resulted in nonsynonymous mutation, the novel one makes little effect on protein function, but the other two changed the protein function to a damaging direction. In conclusion, our results provide a basic profile of CYP3A4 in the Uygur population, and thus can be used to inform optimal dosage recommendations and individualized medicine.
CYP3A4 is an important member of phase I drug metabolism enzymes cytochrome P450 (CYP450) and plays an important role in the metabolism of exogenous compounds and the transformation of internal compounds. It involved in more than 50% commonly clinically used drug [5,7]. The metabolic activity of CYP3A4 exhibit up to 40-folds individual differences, and the metabolism ability of substrate drugs expands from 10 to 20-folds, such a large difference was the co-function of genetic polymorphism, regulation of gene expression and the surrounding environment of the drug or chemical substance interaction, among which 90% of the difference was caused by inherited factors [7,8,16,17].
The 15977T > C, 16613C > T and 25721A > G variants were reported in Chinese population for the first time, enriching the contents of CYP3A4 polymorphisms in China. Especially for the variant 15977T > C, with the frequency of as high as 74%, we have not found such a high frequency in other CYP3A4 polymorphisms. The 20230G > A (CYP3A4*1G) frequency in the current study was 11.5%, lower than two studies centered on Chinese Han polymorphisms of CYP3A4 with the frequency of 20230G > A (CYP3A4*1G) was 25% and 37% [18,19].
As for allele, we for the first time identified CYP3A4*1B in Chinese population. CYP3A4*1B (A-392G) was considered to be the most commonly see mutations around the world with the frequency of 4%, 6%, 6.9%, 12.5%, 40% and 72% in Caucasian, Chilean, Jordanian, Central Americans, African Americans and Africans respectively [20-25]. The frequency of CYP3A4*1B in the current study is only 1.5%. The activity of CYP3A4*1B was found to be controversial in previous studies, some found the allele had the decreased activity while some were normal [26,27]. Whether the function of the allele was normal or not deserved further validation.
We for the first time identified CYP3A4*8, a decreased-activity metabolizers in Asia. The allele was the 13908G > A mutation in Exon5, leading to amino change Arg130Gln. It was first identified in Caucasus with the frequency of 0.33% [28]. Tamoxifen, metabolized by CYP3A4 and other CYPs, is a common anti-cancer drug used in breast cancer [29,30]. We prompt doctors should cautiously consider the dosage of Tamoxifen and other drugs metabolized by CYP3A4 since CYP3A4*8, the decreased-activity metabolizer, is firstly identified in Asian populations in Uygurs.
The LD distribution provides a basic profile of the genomic structure of CYP3A4 in the Uygur population. A haplotype on a block is set of closely linked genetic markers present on one chromosome that tends to be inherited together. The twenty-one genetic polymorphisms in CYP3A4 rendered a relatively weak linkage relationship and we found only one block comprised of three SNPs. The linkage relationship may be an explanation of why most of the commonly see alleles in CYP3A4 pose no threat on the change of enzyme activity.
For the analysis of novel genetic variants in the promoter region, -747C > G, -667C > T, and -392A > G altered the transcription binding site in promoter region. The -667C > T mutation, leading to the loss of BRE’ binding site and appearance of a series new transcription binding sites such as E-box-aa, E47_CS’, PTF-7SK-RNA_PSE’, E2A_CS, E2A_site_consen’, E47-MyoD_CS’, MyoD-MCK-right_’, C/EBP-SV40-1’, AP-3_CS and so on. This is an indicator that the mutation makes great effect on the regulation of promoter region and thus explaining why CYP3A4 content differed interindividually. Some studies reported that the -667C > T mutation makes no effect on the CYP3A4 content [7,24,31]. This inconsistency may be explained by the complexity of the regulation network in promoter regions.
SIFT and PolyPhen prediction in coding region revealed that the novel variant 16934A > T makes no effect on the construction and function of the protein, but for 13908G > A (CYP3A4*8) and 23172T > C (CYP3A4*3), the prediction were damaging, which was in accordance with the reported results that the two alleles represented decreased-activity enzymes.
Acknowledgements
This work was supported by the National Science and Technology Major Project (No. 2012ZX09506001-007).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.CYP450 genotypes: metabolism and efficacy of the antidepressant mirtazapine. Pharmacogenomics. 2013;14:129. [PubMed] [Google Scholar]
- 2.Siedlinski M, Cho MH, Bakke P, Gulsvik A, Lomas DA, Anderson W, Kong X, Rennard SI, Beaty TH, Hokanson JE, Crapo JD, Silverman EK, Investigators CO, Investigators E. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang QH, Hu JP, Wang BL, Li Y. Effects of capsaicin and dihydrocapsaicin on human and rat liver microsomal CYP450 enzyme activities in vitro and in vivo. J Asian Nat Prod Res. 2012;14:382–395. doi: 10.1080/10286020.2012.656605. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto H, Toide K, Kitamura R, Fujita M, Tagawa S, Itoh S, Kamataki T. Gene structure of CYP3A4, an adult-specific form of cytochrome P450 in human livers, and its transcriptional control. Eur J Biochem. 1993;218:585–595. doi: 10.1111/j.1432-1033.1993.tb18412.x. [DOI] [PubMed] [Google Scholar]
- 5.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 6.van der Weide K, van der Weide J. The influence of the CYP3A4*22 polymorphism on serum concentration of quetiapine in psychiatric patients. J Clin Psychopharmacol. 2014;34:256–260. doi: 10.1097/JCP.0000000000000070. [DOI] [PubMed] [Google Scholar]
- 7.Westlind A, Lofberg L, Tindberg N, Andersson TB, Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: relationship to genetic polymorphism in the 5’-upstream regulatory region. Biochem Biophys Res Commun. 1999;259:201–205. doi: 10.1006/bbrc.1999.0752. [DOI] [PubMed] [Google Scholar]
- 8.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Simayi M, Wushouer Q, Xia Y, He Y, Yan F, Zhang J, Cui S, Wen H. Association between polymorphisms in ADAM33, CD14, and TLR4 with asthma in the Uygur population in China. Genet Mol Res. 2014;13:4680–4690. doi: 10.4238/2014.June.18.11. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Duan P, Zhang Y, Han X, Liu J, Yan W, Xing Y. Effect of neuronal induction on NSE, Tau, and Oct4 promoter methylation in bone marrow mesenchymal stem cells. In Vitro Cell Dev Biol Anim. 2012;48:251–258. doi: 10.1007/s11626-012-9494-z. [DOI] [PubMed] [Google Scholar]
- 12.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–1671. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 13.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 16.Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, Kashuba AD. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–388. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson GR. Genetic variability in cytochrome P450 3A5 and in vivo cytochrome P450 3A activity: some answers but still questions. Clin Pharmacol Ther. 2004;76:99–103. doi: 10.1016/j.clpt.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Du J, Xing Q, Xu L, Xu M, Shu A, Shi Y, Yu L, Zhang A, Wang L, Wang H, Li X, Feng G, He L. Systematic screening for polymorphisms in the CYP3A4 gene in the Chinese population. Pharmacogenomics. 2006;7:831–841. doi: 10.2217/14622416.7.6.831. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Q, Yu X, Shu C, Cai Y, Gong W, Wang X, Wang DM, Hu S. Analysis of CYP3A4 genetic polymorphisms in Han Chinese. J Hum Genet. 2011;56:415–422. doi: 10.1038/jhg.2011.30. [DOI] [PubMed] [Google Scholar]
- 20.Paris PL, Kupelian PA, Hall JM, Williams TL, Levin H, Klein EA, Casey G, Witte JS. Association between a CYP3A4 genetic variant and clinical presentation in African-American prostate cancer patients. Cancer Epidemiol Biomarkers Prev. 1999;8:901–905. [PubMed] [Google Scholar]
- 21.Roco A, Quinones L, Agundez JA, Garcia-Martin E, Squicciarini V, Miranda C, Garay J, Farfan N, Saavedra I, Caceres D, Ibarra C, Varela N. Frequencies of 23 functionally significant variant alleles related with metabolism of antineoplastic drugs in the chilean population: comparison with caucasian and asian populations. Front Genet. 2012;3:229. doi: 10.3389/fgene.2012.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousef AM, Bulatova NR, Newman W, Hakooz N, Ismail S, Qusa H, Zahran F, Anwar Ababneh N, Hasan F, Zaloom I, Khayat G, Al-Zmili R, Naffa R, Al-Diab O. Allele and genotype frequencies of the polymorphic cytochrome P450 genes (CYP1A1, CYP3A4, CYP3A5, CYP2C9 and CYP2C19) in the Jordanian population. Mol Biol Rep. 2012;39:9423–9433. doi: 10.1007/s11033-012-1807-5. [DOI] [PubMed] [Google Scholar]
- 23.Sinues B, Vicente J, Fanlo A, Vasquez P, Medina JC, Mayayo E, Conde B, Arenaz I, Martinez-Jarreta B. CYP3A5*3 and CYP3A4*1B allele distribution and genotype combinations: differences between Spaniards and Central Americans. Ther Drug Monit. 2007;29:412–416. doi: 10.1097/FTD.0b013e31811f390a. [DOI] [PubMed] [Google Scholar]
- 24.Wandel C, Witte JS, Hall JM, Stein CM, Wood AJ, Wilkinson GR. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5’-promoter region polymorphism. Clin Pharmacol Ther. 2000;68:82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 25.Cavaco I, Reis R, Gil JP, Ribeiro V. CYP3A4*1B and NAT2*14 alleles in a native African population. Clin Chem Lab Med. 2003;41:606–609. doi: 10.1515/CCLM.2003.091. [DOI] [PubMed] [Google Scholar]
- 26.Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90:1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 27.Ando Y, Tateishi T, Sekido Y, Yamamoto T, Satoh T, Hasegawa Y, Kobayashi S, Katsumata Y, Shimokata K, Saito H. Re: Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1999;91:1587–1590. doi: 10.1093/jnci/91.18.1587. [DOI] [PubMed] [Google Scholar]
- 28.Ruzilawati AB, Suhaimi AW, Gan SH. Genetic polymorphisms of CYP3A4: CYP3A4 *18 allele is found in five healthy Malaysian subjects. Clin Chim Acta. 2007;383:158–162. doi: 10.1016/j.cca.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Majidzadeh AK, Gharechahi J. Plasma proteomics analysis of tamoxifen resistance in breast cancer. Med Oncol. 2013;30:753. doi: 10.1007/s12032-013-0753-y. [DOI] [PubMed] [Google Scholar]
- 30.Cho YA, Lee W, Choi JS. Effects of curcumin on the pharmacokinetics of tamoxifen and its active metabolite, 4-hydroxytamoxifen, in rats: possible role of CYP3A4 and P-glycoprotein inhibition by curcumin. Pharmazie. 2012;67:124–130. [PubMed] [Google Scholar]
- 31.Ball SE, Scatina J, Kao J, Ferron GM, Fruncillo R, Mayer P, Weinryb I, Guida M, Hopkins PJ, Warner N, Hall J. Population distribution and effects on drug metabolism of a genetic variant in the 5’ promoter region of CYP3A4. Clin Pharmacol Ther. 1999;66:288–294. doi: 10.1016/S0009-9236(99)70037-8. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh KP, Lin YY, Cheng CL, Lai ML, Lin MS, Siest JP, Huang JD. Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos. 2001;29:268–273. [PubMed] [Google Scholar]
- 33.Fukushima-Uesaka H, Saito Y, Watanabe H, Shiseki K, Saeki M, Nakamura T, Kurose K, Sai K, Komamura K, Ueno K, Kamakura S, Kitakaze M, Hanai S, Nakajima T, Matsumoto K, Saito H, Goto Y, Kimura H, Katoh M, Sugai K, Minami N, Shirao K, Tamura T, Yamamoto N, Minami H, Ohtsu A, Yoshida T, Saijo N, Kitamura Y, Kamatani N, Ozawa S, Sawada J. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat. 2004;23:100. doi: 10.1002/humu.9210. [DOI] [PubMed] [Google Scholar]
- 34.Garsa AA, McLeod HL, Marsh S. CYP3A4 and CYP3A5 genotyping by Pyrosequencing. BMC Med Genet. 2005;6:19. doi: 10.1186/1471-2350-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.