Abstract
Circulating microRNAs (miRNAs) were recognized to be potential non-invasive biomarkers for colorectal cancer (CRC) detection and prediction. Meanwhile, the association of the expression of plasma miRNAs with the risk of CRC patients has rarely been analyzed. Therefore, we conducted this study to evaluate the value of plasma miRNAs for CRC diagnosis and risk estimation. Fasting blood samples from 100 CRC patients and 79 cancer-free controls were collected. Plasma miR-106a, miR-20a, miR-27b, miR-92a and miR-29a levels were detected by RT-qPCR. Sensitivity and specificity were employed to evaluate the diagnostic value of miRNAs for CRC. Univariate and multivariate logistic regression were employed to analyze the association between miRNAs expression and CRC risk. As results, miR-106a and miR-20a were elevated in the patients with CRC. The sensitivity of miR-106a was 74.00% and the specificity was 44.40%, while the cutoff value was 2.03. As for miR-20a, the sensitivity was 46.00% and specificity was 73.42% when employed 2.44 as cutoff value. High expression of plasma miR-106a increased CRC risk by 1.80 -fold. Plasma miR-106a and miR-20a may as noninvasive biomarkers for detecting the CRC. High expression of miR-106a associated with CRC risk.
Keywords: microRNA, colorectal cancer, plasma, diagnosis, risk factor
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females worldwide. Due to the improvements in early detection and treatment, CRC related mortality rates have continued to decrease over the years. Nevertheless, over 1.2 million new cancer cases and 608,700 deaths estimated to have occurred per year [1]. In the United States, the 5-year survival rate is 93.2% for stage I as opposed to only 8.1% for stage IV [2]. Therefore, early diagnosis is of vital importance for the treatment and prognosis of CRC patients. Several clinical examinations, including fecal occult blood tests (FOBT), radiologic tests, and endoscopic examinations, are currently used to screening for CRC [3]. However, these existed methods have limitations due to low efficacy, high cost, or invasive trauma [4-6]. Thus, new markers from non-invasive clinical samples are needed.
MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate expression levels of more than 60% of human gene [7]. Growing evidence indicated that miRNA expression profiles in tissues play an important role in the diagnosis of various malignancies [8-12] including CRC [13,14]. Recent studies has revealed that circulating miRNAs could be detectable in a remarkably stable manner in the blood, plasma, or serum [9,15,16], and several studies have reported the potentially diagnostic utility of circulating miRNAs as blood-based biomarkers [17,18].
In addition to its potential diagnostic value, miRNA can respond for carcinogen exposure by its expression alternation. Circulating miRNAs as accurate biomarkers can reflect the alteration for carcinogenesis induced by chemical carcinogens [19]. Therefore, miRNAs may be considered as factors to evaluate cancer risk, and the association of miRNA with CRC risk has became another new focus for its important roles in carcinogenesis [20]. However, few studies were conducted on the association of circulating miRNA with CRC risk based on present reports.
In this study, we aimed to explore the potential diagnostic value of a panel of five miRNAs (miR-106a, miR-20a, miR-27b, miR-92a, and miR-29a), which were selected according to a comprehensive review of relevant literatures and a previously published microarray data of colorectal tumor tissues [21,22], and to evaluate the association between aberrantly expressed circulating miRNAs and CRC risk.
Materials and methods
Patients and controls
Blood samples of 100 newly diagnosed primary CRC cases were collected before any therapeutic procedures, including surgery, chemotherapy, and radiotherapy after asking the consent of surgeons and patients themselves. All patients were pathologically diagnosed in the Tumor Hospital of Harbin Medical University, from 15 May, 2007 to 1 January, 2008. Clinical data of the CRC patients were derived from medical records. Tumors were staged according to the TNM stage for CRC. Meanwhile, 79 cancer-free controls were recruited from the Second Affiliated Hospital of Harbin Medical University, and their blood samples were collected after asking consent of physician and the patients. The study was approved by Human Research and Ethics Committee Harbin Medical University.
Blood processing and miRNAs extraction
From each participant, 5 to 10 milliliters (ml) of peripheral whole blood was collected into EDTA tube. Plasma was separated from cell components by centrifugation at 2000 g for 10 min at 4°C and 12000 g for another 10 min at 4°C, within eight hours after collection. All the supernatant plasma were separated as aliquots and stored at -80°C until use.
Total RNA was extracted from 250 μl plasma using TRIzol LS reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. Briefly, 750 μl TRIzol LS reagent was added to 250 μl plasma samples, after phase separation by 200 μl chloroform addition and centrifugation. 600 μl 100% isopropanol was added to the aqueous phase and centrifuged to precipitate RNA. RNA pellet was washed by 75% ethanol and air-dried for 5 min, and the final elution volume was 12 μl. The concentrations of all samples were quantified by NanoDrop 2000 spectrophotometer (Thermo, Japan).
Reverse transcription and real-time quantitative PCR
Briefly, 1 μg total RNA from each sample was quantitatively polyadenylated in 20 μl reaction and 3 μl product of polyadenylate was reversely transcribed to cDNA, using polyadenylation polymerase and First-Strand cDNA Synthesis Kit (Tiangen, China) with commercial product reverse primer (Tiangen, China) according to the manufacturer’s protocol. RT- control was included in each batch of reactions. The cDNA product was diluted 1:2 with nuclease-free water and stored at -20°C for further analysis.
Quantification of miRNA expression was detected by real-time quantitative polymerase chain reaction (RT-qPCR) in a finally volume of 10 μl with Roche Lightcycler 480 (Roche, Germany) using SYBR Green PCR Master Mix (Kapa, USA). Each reaction contained 5 μl SYBR Green reagents, 0.2 μl each of 10 μM specific forward primer and universal primer, and 3.6 μl DEPC-treated water. The amplification used the following conditions: 95°C for 5 min, followed by 45 cycles at 95°C for 20 s and 60°C for 34 s. The miRNA-specific forward primers were designed according to the miRNA sequence obtained from the miRBase database (http://microrna.sanger.ac.uk/), which was listed in Table 1. The universal reverse primer was purchased in Tiangen (Tiangen, China). No-template controls for both RT and PCR, which to ensure target specific amplification, were included on each plate and all PCR reactions were carried out in triplicate. In order to validate the specificity of the target PCR product, melting curve analysis was performed at the end of PCR cycles. The comparative cycle threshold (CT) method was applied to quantify the expression levels of miRNAs. The expression levels of miRNAs were normalized to miR-16 which was selected as a reference gene [16]. The relative amount of target miRNAs to miR-16 was calculated using the equation 2-ΔCT, where ΔCT= (ΔC ttarget-ΔCt normalizer). The median of relative amount was chosen to define the cases with high expression or low expression.
Table 1.
The miRNA-specific primer sequences
| MicroRNA | Primer sequence |
|---|---|
| hsa-miR-92a | TAT TGC ACT TGT CCC GGC C |
| hsa-miR-16 | TAG CAG CAC GTA AAT ATT GGC G |
| hsa-miR-29a | GGT AGC ACC ATC TGA AAT CGG TT |
| hsa-miR-106a | GGA AAA GTG CTT ACA GTG CAG GTA G |
| hsa-miR-27b | CGT TCA CAG TGG CTA AGT TCT GC |
| hsa-miR-20a | AAA GTG CTT ATA GTG CAG GTA GGG G |
Statistical analysis
The Mann-Whitney U test was used to assess the differences of miRNA expression between CRC patients and controls. Sensitivity, specificity, Youden index and receiver operating characteristic (ROC) curve analysis were used to evaluate the diagnostic power of the miRNAs. In addition, univariate and multivariate logistic regression were used to evaluate associations between each miRNA and the risk of CRC. Pearson’s X2 test was used to analyze the relationship between differential expression of miRNAs and clinicopathologic characteristics. The statistical analyses were performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL). All tests were two-sided. P-values less than 0.05 were considered statistically significant.
Results
Characteristics and clinical features of subjects
The 100 patients consisted of 60 males and 40 females aged from 27 to 81, with the median age of 58.91. There were 44 males and 35 females included in the cancer free controls. The median age of 79 controls was 60.20, and the range from 27 to 81. The characteristics and clinical features of patients are summarized in Table 2.
Table 2.
The characteristics of subjects
| Parameters | Patients | Controls |
|---|---|---|
| Sex | ||
| Male | 60 | 44 |
| Female | 40 | 35 |
| Age | ||
| <60 | 53 | 38 |
| ≥60 | 47 | 41 |
| Location | ||
| Colon | 35 | |
| Rectum | 63 | |
| Unknown | 2 | |
| Tumor size | ||
| ≤45 mm | 44 | |
| >45 mm | 43 | |
| Unknown | 13 | |
| TNM stage | ||
| I-II | 41 | |
| III-IV | 38 | |
| Unknown | 21 | |
| Lymphnode status | ||
| Positive | 43 | |
| Negative | 51 | |
| Unknown | 6 | |
| Histology | ||
| Adenocarcinoma | 73 | |
| Mucinous adenocarcinoma | 18 | |
| Polypoid adenocarcinoma | 3 | |
| Other | 6 |
Expression of plasma miRNAs in CRC and control
The expression levels of plasma miR-106a and miR-20a were statistically significantly higher in CRC patients compared with controls (miR-106a: 17.13 vs. 5.59, P=0.016; miR-20a: 2.05 vs. 1.25, P=0.038). (Figure 1D and 1E) No significant differences were observed in the plasma levels of miR-92a, miR-29a and miR-27b between cancer patients and controls (miR-92a: 14.59 vs. 15.56, P=0.802; miR-29a: 0.16 vs. 0.15, P=0.548; miR-27b: 0.57 vs. 0.45, P=0.294) (Figure 1A-C; Table 3).
Figure 1.
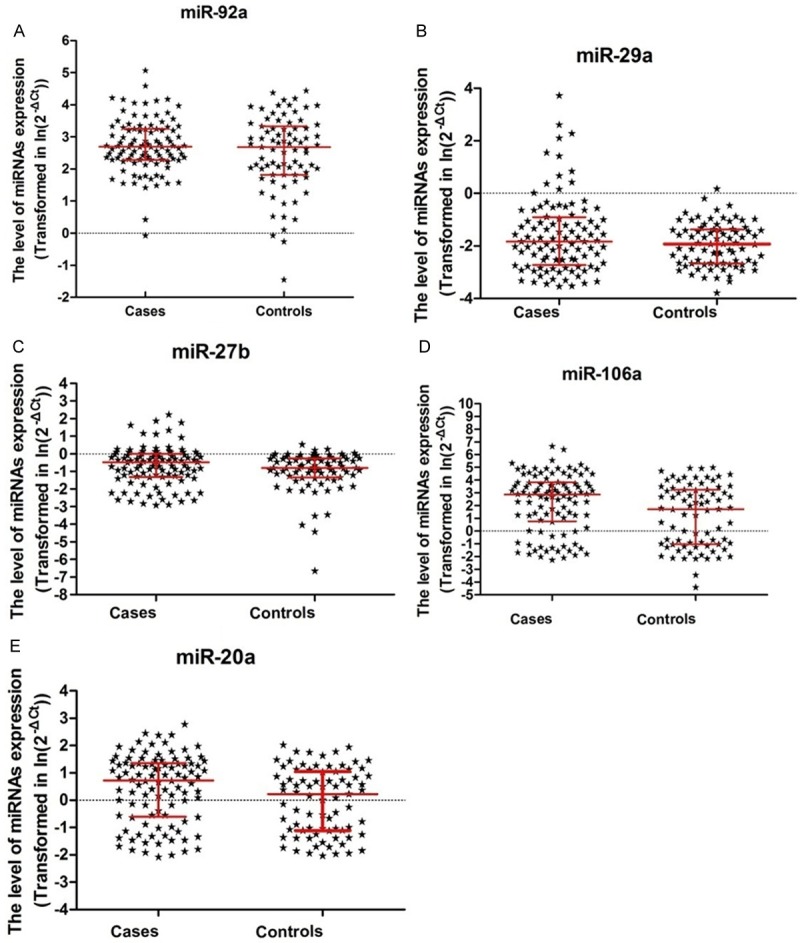
Plasma miR-92a, miR-29a, miR-27b, miR-106a and miR-20a levels in CRC patients and controls.
Table 3.
The expression level of plasma miRNAs between CRC and controls
| miRNAs | Case | Control | P value* | ||
|---|---|---|---|---|---|
|
|
|
||||
| Median Range | Median | Range | |||
| miR-92a | 14.59 | 0.82-157.95 | 15.56 | 0.23-200.05 | 0.802 |
| miR-29a | 0.16 | 0.03-41.36 | 0.15 | 0.02-9.25 | 0.548 |
| miR-27b | 0.57 | 0.03-9.23 | 0.45 | 0.00-12.22 | 0.294 |
| miR-106a | 17.13 | 0.07-776.05 | 5.59 | 0.01-148.40 | 0.016 |
| miR-20a | 2.05 | 0.09-16.08 | 1.25 | 0.13-15.45 | 0.038 |
The significance between CRC and controls was test by Mann-Whitney U test.
Diagnostic value of plasma miR-106a and miR-20a for CRC
When the miR-106a cutoff value was at the first quartile (0.49), the sensitivity and specificity were 80.00% and 31.65%, respectively. While the cutoff value was set at the third quartile (38.59), the corresponding sensitivity and specificity were 29.00% and 81.01%, respectively. The largest Youden Index for miR-106a was 0.184; the corresponding expression level of 2.03 in plasma was as optimal cutoff value. At the cutoff value of 0.36 for miR-20a, the sensitivity was 79.00% and the specificity was 29.11%, while cutoff value elevated to 3.52, the sensitivity and specificity were 32.00% and 83.54%, respectively. When 2.44 was an optimal cutoff value for miR-20a based on the largest Youden Index of 0.194, the corresponding sensitivity and specificity were 46.0% and 73.42%, respectively (Table 4). The area under the ROC (AUC) for miR-106a and miR-20a were 0.605 (95% CI: 0.522-0.688) and 0.590 (95% CI: 0.507-0.674), respectively (Figure 2A and 2B).
Table 4.
The diagnostic value of miR-106a and miR-20a in CRC
| Percentile | miR-106a | miR-20a | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Cutoff | Sensitivity | Specificity | Youden Index | Cutoff | Sensitivity | Specificity | Youden Index | |
| Optimal | 2.03 | 74.0% | 44.40% | 0.184 | 2.44 | 46.00% | 73.42% | 0.194 |
| 25% | 0.49 | 80.0% | 31.65% | 0.117 | 0.36 | 79.0% | 29.11% | 0.081 |
| 50% | 11.08 | 57.0% | 58.23% | 0.152 | 1.65 | 54.0% | 54.43% | 0.084 |
| 75% | 38.59 | 29.0% | 81.01% | 0.100 | 3.52 | 32.0% | 83.54% | 0.155 |
Figure 2.
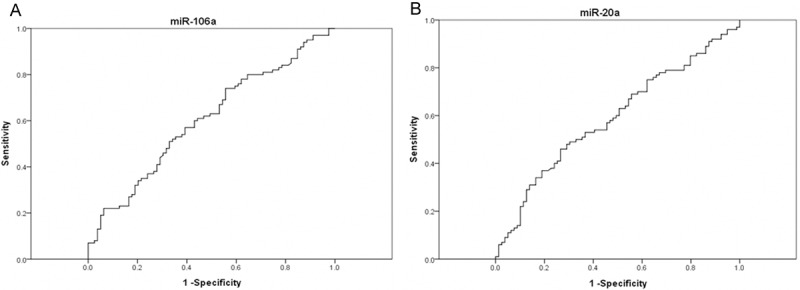
Receiver operating characteristics (ROC) curve analysis using plasma miR-106a (A) and miR-20a (B) for discriminating CRC.
Relationship between plasma miRNAs level and clinical features
Higher level of miR-92a expression was statistically associated with tumor size no more than 4.5 mm (P=0.041). Inversely, lower expression of miR-106a was associated with larger tumor (P=0.013). miR-20a expression was significantly associated with differentiation (P=0.037, Table 5). The expression of miR-29a in adenocarcinoma group was higher than that in other histological subtype group (56.16% vs. 27.78%, 22.22%, P=0.026). No statistically significant associations were observed between the five miRNAs and age, gender, TNM stage, lymph node status, or tumor location (Table 5).
Table 5.
The association of plasma miR-106a and miR-20a with clinical characteristics of CRC patients (No. (%)
| Parameters | miR-106a | miR-20a | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Low | High | P-value | Low | High | P-value | |
| Gender | ||||||
| Male | 26 (44.83) | 32 (55.17) | 0.224 | 29 (50.00) | 29 (50.00) | 1 |
| Female | 24 (57.14) | 18 (42.86) | 21 (50.00) | 21 (50.00) | ||
| Age | ||||||
| <60 | 24 (48.0) | 26 (52.0) | 0.689 | 26 (52.0) | 24 (48.0) | 0.689 |
| ≥60 | 26 (52.0) | 24 (48.0) | 24 (48.0) | 26 (52.0) | ||
| Location | ||||||
| Colon | 20 (57.14) | 15 (42.86) | 0.366 | 20 (57.14) | 15 (42.86) | 0.366 |
| Rectum | 30 (47.62) | 33 (52.38) | 30 (47.62) | 33 (52.38) | ||
| Tumor size | ||||||
| ≤45 mm | 17 (38.64) | 27 (61.36) | 0.013 | 20 (45.45) | 24 (54.55) | 0.334 |
| >45 mm | 28 (65.12) | 15 (34.88) | 24 (55.81) | 19 (44.19) | ||
| TNM stage | ||||||
| I-II | 26 (63.41) | 15 (36.59) | 0.229 | 23 (56.10) | 18 (43.90) | 0.757 |
| III-IV | 19 (50.0) | 19 (50.0) | 20 (52.63) | 18 (47.37) | ||
| Lymphnode status | ||||||
| Positive | 30 (58.82) | 21 (41.18) | 0.157 | 28 (54.9) | 23 (45.1) | 0.418 |
| Negative | 19 (44.19) | 24 (55.81) | 20 (46.51) | 23 (53.49) | ||
| Histology | ||||||
| Adenocarcinoma | 40 (54.79) | 33 (45.21) | 0.278 | 38 (52.05) | 35 (47.95) | 0.796 |
| Mucinous adenocarcinoma | 7 (38.89) | 11 (61.11) | 8 (44.44) | 10 (55.56) | ||
| Other | 3 (33.33) | 6 (66.67) | 4 (44.44) | 5 (55.56) | ||
| Differentiation | ||||||
| Poor | 6 (40.0) | 9 (60.0) | 0.296 | 4 (26.67) | 11 (73.33) | 0.037 |
| Moderate/well | 40 (54.79) | 33 (45.21) | 41 (56.16) | 32 (43.84) | ||
Association between expression of miRNAs and CRC risk
Univariate logistic regression showed the higher expression of miR-106a was significantly associated with an increased risk of CRC (OR=1.23, 95% CI: 1.02-1.49). The higher level of miR-27b and miR-20a also increased the risk of CRC by 1.20 times (for miR-27b, OR=1.20, 95% CI: 0.99-1.45; for miR-20a, OR=1.20, 95% CI: 0.99-1.45, respectively) (Table 6).
Table 6.
The association between miRNAs expression and risk of CRC
| miRNA | Univariable analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| OR | 95% C.I. | P-value | ORadj.* | 95% CI | P-value | |
| miR-92a | 1.14 | 0.95-1.38 | 0.16 | 1.14 | 0.71-1.83 | 0.59 |
| miR-29a | 1.17 | 0.97-1.41 | 0.11 | 1.30 | 0.79-2.14 | 0.30 |
| miR-27b | 1.20 | 0.99-1.45 | 0.06 | 1.55 | 0.95-2.52 | 0.08 |
| miR-106a | 1.23 | 1.02-1.49 | 0.03 | 1.80 | 1.12-2.92 | 0.02 |
| miR-20a | 1.20 | 0.99-1.45 | 0.06 | 1.54 | 0.95-2.50 | 0.08 |
ORadj: adjusted for age and sex.
Further multivariate logistic regression analysis showed that higher expression of miR-106a increased CRC risk by 1.80 times (ORadj=1.80, 95% CI: 1.12-2.92) after adjusting age, sex. High expression of miR-27b and miR-20a also increased the risk of CRC (for miR-27b, ORadj= 1.55, 95% CI: 0.95-2.52, for miR-20a, ORadj= 1.54, 95% CI: 0.95-2.50) (Table 6).
Discussion
MiRNA can exist intact in serum or plasma protected from endogenous RNase activity [9], and its expression profile in human cancer appears to be tissue-specific [15]. Circulating miRNA as a novel biomarker had attracted more attentions on its value of diagnosis, prognosis and treatment. In this study, we analyzed the levels of five miRNAs (miR-92a, miR-29a, miR-27b, miR-106a and miR-20a) in plasma from 100 CRC patients and 79 cancer-free controls. Our results showed that miR-106a and miR-20a were significantly upregulated in the CRC patients comparing with cancer-free controls. Although it had been reported that miR-106a overexpressed in tissue [23,24], and fecal occult blood [25] of colorectal cancer, this is first study that focus on the miR-106a level in plasma formed that it overexpressed in CRC patients compared with cancer-free controls. Dysregulated miR-106a plays a vital role in the process of oncogenesis and development [7]. For example, overexpressed miR-106a promoted gastric cancer cell proliferation and inhibited apoptosis [8]. Feng et al. [26] revealed that miR-106a could inhibit the expression of transforming growth factor-β receptor 2 (TGFBR2), leading to increased CRC cell migration and invasion. MiR-20a was also confirmed to be significantly higher in CRC tissues than in normal tissues [27]. MiR-20a was upregulated by CCAT2 through TCF7L2-mediated transcriptional regulation in colon cancer [28]. MiR-20a can induce cell invasion and migration by targeting the ABL2 in prostate cancer [29] and contribute chemotherapeutic resistance in colorectal adenocarcinoma cell lines [30]. However, its function in CRC formation was not clear.
In our study, miR-92a and miR-29a were not significantly upregulated in CRC patients compared to controls. However, Huang et al. reported miR-92 and miR-29a were upregulated and were identified as plasma diagnostic markers for CRC [16]. At the same time, Cheng H, et al. even found that plasma miR-92 was significantly decreased in the TexGen cohort of colon cancer patients [31]. These conflicting conclusions may because of the genetic variations among different ethnic groups as well as the differences of environmental factors and diets. Zhao ZH, et al. had revealed the profiles of circulating miRNAs in plasma samples from Caucasian American women or that from African American with early breast cancer were inconsistent, and that differentially expressed miRNAs characterized with potential racial difference [32]. Therefore, more studies with well design are required to identify miRNAs in peripheral blood as noninvasive markers for tumor disease.
Our study showed that expression levels of miR-106a and miR-20a in plasma were increased in CRC patients compared with cancer-free controls. The AUC of miR-106a and miR-20a were 0.605 and 0.590, respectively. Compared with CEA, which showed 51.9% sensitivity at 90% specificity with AUC of 0.773, the evaluation of miR-106a and miR-20a showed less sensitivity of 22.0% at 90% specificity [33]. Both miR-106a and miR-20a had relatively lower specificity of 44.49% and 73.42%. In some extent, these two miRNAs only presented potential value for auxiliary diagnosis for CRC patients, and further studies with larger samples are needed.
Accumulating evidence indicated that miRNAs associated with clinical parameters, such as higher serum miRNA expression associated with poorly differentiated gastric cancer [34]. In present study, the expression of plasma miR-92 is lower in the patients with a tumor less than 45 mm in diameter and the miR-106a is high expression in the patients with a tumor less than 45 mm in diameter. Some reports showed that circulating miRNAs were associated with tumor size [35,36], another study did not found the correlation between miRNAs and tumor size [37]. Several reports stated that circulating miRNAs levels were significantly reduced after tumor resection [38,39]. High expression of miR-20a was also associated with poor differentiation (77.3% vs. 26.7%). MiR-20a regulates the Jak-STAT signaling pathways which are closely related to cell differentiation in myeloid leukemia cell [40]. MiR-20a may be a potential factor in the process of CRC progression. Additionally, miR-27b did not show a significant association between their expression levels and clinicopathological characteristics of tumors.
We found that high expression of plasma miR-106a increased the risk of CRC by 1.23 times compared to the low level. After adjusting the age and sex, high expression of miR-106a increased 1.80-fold of CRC risk. High expression miR-27b and miR-20a are the risk of CRC, although they are marginal significance in statistics. More evidence showed that miRNAs play pivotal roles in activating natural killer cells [41], mediating inflammatory response [42], altering the cells microenvironment [43]. Some aberrant expression miRNAs co-existing will increase the predictive value of CRC prognosis [44]. Therefore, we evaluated the combined effects of three miRNAs on CRC risk, and found that the risk effects of CRC noticeably increased with increasing number of putative risk miRNAs. Based on the recent present study, more cohort study will be designed on the association of plasma miRNAs exposure with the cancer risk are needed to confirm our preliminary results.
The limitations of this study should be considered. First, the small sample size caused incredible results by sampling error in case-control study. For example, miR-20a and miR-27b had a marginally statistical significance in CRC risk. Further validations of these markers in large sample in independent studies are necessary. Second, Poly-A method was employed before reverse transcription which would be not completely differ matured miRNA from pre-miRNA. In addition, normalization is a key step for the accurate quantification of miRNA levels with RT-qPCR. A common problem in the circulating miRNA researches is that no consensus endogenous control has been established. We selected miR-16 as endogenous control because of its relatively stable and abundant in plasma/serum [16,45]. However, several reports showed that aberrant expression of miR-16 in plasma/serum was associated with the risk of lymphoma and prostate cancer [46,47].
In conclusion, we observed that expression levels of circulating miR-106a and miR-20a were significantly up-regulated in CRC patients plasma compared with cancer-free controls. The aberrant expression of miR-106a and miR-20a will be a potential biomarker for auxiliary diagnosis. High expression of miR-106a, miR-20a and miR-27b may be increased the CRC risk. High level of plasma miR-20a was associated with poor differentiation. Further good-designed studies with large sample size are required to validate the potential prevention and clinic value of these miRNAs.
Acknowledgements
This work was supported by The Science and technology Foundation of Education Department of Heilongjiang Province (No. 12511z018).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Lee BI, Hong SP, Kim SE, Kim SH, Kim HS, Hong SN, Yang DH, Shin SJ, Lee SH, Park DI, Kim YH, Kim HJ, Yang SK, Kim HJ, Jeon HJ. Korean guidelines for colorectal cancer screening and polyp detection. Clin Endosc. 2012;45:25–43. doi: 10.5946/ce.2012.45.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yitzhak A, Bayme M, Perry ZH, Mizrahi S. Small bowel perforation after capsule endoscopy in a patient with occult gastrointestinal bleeding and undiagnosed Crohn’s disease. Am Surg. 2012;78:E159–61. [PubMed] [Google Scholar]
- 5.Launoy G, Smith TC, Duffy SW, Bouvier V. Colorectal cancer mass-screening: estimation of faecal occult blood test sensitivity, taking into account cancer mean sojourn time. Int J Cancer. 1997;73:220–224. doi: 10.1002/(sici)1097-0215(19971009)73:2<220::aid-ijc10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Hassan C, Pickhardt PJ, Laghi A, Kim DH, Zullo A, Iafrate F, Di Giulio L, Morini S. Computed tomographic colonography to screen for colorectal cancer, extracolonic cancer, and aortic aneurysm: model simulation with cost-effectiveness analysis. Arch Intern Med. 2008;168:696–705. doi: 10.1001/archinte.168.7.696. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannafon BN, Sebastiani P, de las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miko E, Czimmerer Z, Csanky E, Boros G, Buslig J, Dezso B, Scholtz B. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35:646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 12.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 14.Aslam MI, Taylor K, Pringle JH, Jameson JS. MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg. 2009;96:702–710. doi: 10.1002/bjs.6628. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Hou J, Jin W, Li J, Yue Y, Jin H, Wang X. Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: a meta-analysis. Molecules. 2014;19:6282–93. doi: 10.3390/molecules19056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, Sun Q, Yan F, Yan C, Li H, Ren X. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533:389–97. doi: 10.1016/j.gene.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Yang T, Li X, Yang Q, Liu R, Huang J, Li Y, Yang C, Jiang Y. Alteration of serum miR-206 and miR-133b is associated with lung carcinogenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Toxicol Appl Pharmacol. 2013;267:238–46. doi: 10.1016/j.taap.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Tian Y, Xue Y, Ruan G, Cheng K, Tian J, Qiu Q, Xiao M, Li H, Yang H, Wang L. Interaction of Serum microRNAs and Serum Folate With the Susceptibility to Pancreatic Cancer. Pancreas. 2015;44:23–30. doi: 10.1097/MPA.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J, Yu L. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2011;129:1624–1634. doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Wang Q, Zheng Y, Lv S, Ning S, Sun J, Huang T, Zheng Q, Ren H, Xu J, Wang X, Li Y. Prioritizing human cancer microRNAs based on genes’ functional consistency between microRNA and cancer. Nucleic Acids Res. 2011;39:e153. doi: 10.1093/nar/gkr770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Zhang P, Yang J, Liu Z, Yang Z, Qin H. Candidate microRNA biomarkers in human colorectal cancer: systematic review profiling studies and experimental validation. Int J Cancer. 2012;130:2077–87. doi: 10.1002/ijc.26232. [DOI] [PubMed] [Google Scholar]
- 25.Koga Y, Yamazaki N, Yamamoto Y, Yamamoto S, Saito N, Kakugawa Y, Otake Y, Matsumoto M, Matsumura Y. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013;22:1844–52. doi: 10.1158/1055-9965.EPI-13-0512. [DOI] [PubMed] [Google Scholar]
- 26.Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 28.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL, Shen J, Li T, Yun C, Li H, Shi LH. miR-20a Promotes Prostate Cancer Invasion and Migration Through Targeting ABL2. J Cell Biochem. 2014;115:1269–76. doi: 10.1002/jcb.24778. [DOI] [PubMed] [Google Scholar]
- 30.Chai H, Liu M, Tian R, Li X, Tang H. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin (Shanghai) 2011;43:217–25. doi: 10.1093/abbs/gmq125. [DOI] [PubMed] [Google Scholar]
- 31.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polat E, Duman U, Duman M, Atici AE, Reyhan E, Dalgic T, Bostanci EB, Yol S. Diagnostic value of preoperative serum carcinoembryonic antigen and carbohydrate antigen 19-9 in colorectal cancer. Curr Oncol. 2014;21:e1–7. doi: 10.3747/co.21.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruoquan Y, Wanpin N, Qiangsheng X, Guodong T, Feizhou H. Correlation between plasma miR-122 expression and liver injury induced by hepatectomy. J Int Med Res. 2014;42:77–84. doi: 10.1177/0300060513499093. [DOI] [PubMed] [Google Scholar]
- 36.Song J, Bai Z, Zhang J, Meng H, Cai J, Deng W, Bi J, Ma X, Zhang Z. Serum microRNA-21 levels are related to tumor size in gastric cancer patients but cannot predict prognosis. Oncol Lett. 2013;6:1733–1737. doi: 10.3892/ol.2013.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, Ditzel HJ. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol. 2014;8:874–83. doi: 10.1016/j.molonc.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–7. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–11. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He M, Wang QY, Yin QQ, Tang J, Lu Y, Zhou CX, Duan CW, Hong DL, Tanaka T, Chen GQ, Zhao Q. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ. 2013;20:408–18. doi: 10.1038/cdd.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotta R, Chen L, Costinean S, Josyula S, Mundy-Bosse BL, Ciarlariello D, Mao C, Briercheck EL, McConnell KK, Mishra A, Yu L, Croce CM, Caligiuri MA. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood. 2013;121:3126–3134. doi: 10.1182/blood-2012-12-467597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halappanavar S, Nikota J, Wu D, Williams A, Yauk CL, Stampfli M. IL-1 receptor regulates microRNA-135b expression in a negative feedback mechanism during cigarette smoke-induced inflammation. J Immunol. 2013;190:3679–86. doi: 10.4049/jimmunol.1202456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–13. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, Zhao HW, Chen W, He YL, Wang HY, Xie D, Luo JH. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 45.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 46.Watahiki A, Wang Y, Morris J, Dennis K, O’Dwyer HM, Gleave M, Gout PW, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, Li JY. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91:553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]


