Abstract
Objective: This study aimed to characterize the miR-21 and evaluated its clinical significance. Methods: Total RNA was extracted from 30 pairs of fresh specimens of cervical cancer and normal tissues. The expression levels of the miR-21-3p and miR-21-5p were detected by quantitative reverse transcriptase polymerase chain reaction, with U6 as the internal reference gene. We compared the expression of miR-21-3p and miR-21-5p between study group and control groups, the association between miRNA expression level and clinicopathological factors was investigated. Results: The expression of miR-21-3p and miR-21-5p in HPV positive cervical cancer samples was significantly upregulated compared to that in the paired normal samples (P < 0.05); A multivariate analysis demonstrated that the expression of miR-21 was associated with clinicopathological parameters, including depth of invasion and lymph node metastasis. Conclusions: MiR-21 upregulation is associated with aggressive progression and poor prognosis in cervical cancer, which suggests that miR-21 might be identified as an independent marker for predicting the clinical outcome of cervical cancer patients.
Keywords: miRNA-21, cervical cancer, biomarker
Introduction
Cervical cancer remained one of the leading causes of cancer death in women worldwide. It was an infectious disease that accompanied with gene involved multiple steps and more complex biological processes, which was closely related to persistent infection of high-risk human papilloma virus (HPV) [1], Chromosome mutation and the change of single nucleotide polymorphisms, those were considered the key factors to induce the malignant transformation of cervical epithelial [2]. In recent years, the research of small RNAs (miRNA) regulation of gene expression became a hot spot [3]. Over 1,000 human miRNAs were predicted to regulate approximately 60% of protein-coding genes, indicating their extensive functions in many biological processes. In cervical cancer tissues, microRNA-21 (miR-21) was found to be one of the upregulated miRNAs [4], accumulating evidence for differential expression of miR-21 in cervical cancer suggested that it might play a crucial role in tumor biology.
This study aimed to use real-time fluorescent quantitative PCR method to characterize the miR-21 expression profiles in HPV16 infectious cervical cancers tissues, understanding of miR-21 differential expression with HPV16 infection in cervical cancer, and analyzed the correlation of expression level of miR-21 with clinicopathological factors in patients with cervical cancer, demonstrating how miR-21 behaves as a tumor oncogenic miRNA by upregulating oncogenic components, and thus regulated cell cycle activity in order to provide the clues of miR-21 in regulating malignant transformation of cervical epithelial cells with HPV16 infection and identify the biomarkers for application in early diagnosis of cervical cancer.
Materials and methods
Clinical tissue samples
A total of 30 paired fresh cervical tissue specimens were collected from cervical cancer patients who underwent radical hysterectomy between May 2011 and June 2014 in the Fengxian Hospital, Southern Medical University. Samples (cervical cancer and normal tissues at a distance > 5 cm from the tumor, avoiding obvious necrosis part) were randomly selected in vitro for 0.5 cm × 0.5 cm size each 2 pieces of cervical cancer and normal cervical tissue, rinsed with sterile saline then immediately sent to -80°C cryogenic refrigerator. All of the cases in study group were under new treatments, without any preoperative radiotherapy or chemotherapy before; All cases were preoperative detected by HPV subtype test. All 30 cases of study group were performed extensive hysterectomy and pelvic lymph nodes dissection. The patients’ clinical characteristics are shown in Table 1. All tissue samples were obtained with informed consent and all procedures were performed in accordance with the Human Investigation Ethical Committee of the Fengxian Hospital, Southern Medical University.
Table 1.
Clinical characteristics of patients with cervical cancer
| Patients (n = 30) | |
|---|---|
| Age (years) | |
| < 45 | 18 (60.00%) |
| ≥ 45 | 12 (40.00%) |
| Depth of invasion (T) | |
| T1 | 5 (16.67%) |
| T2 | 7 (23.33%) |
| T3 | 6 (20.00%) |
| T4 | 12 (40.00%) |
| Lymph node metastasis | |
| Negative | 25 (83.33%) |
| Positive | 5 (16.67%) |
| Haematogenous metastasis | |
| Negative | 27 (90.00%) |
| Positive | 3 (10.00%) |
| Stage | |
| Ib1 | 6 (20.00%) |
| Ib2 | 17 (56.67%) |
| IIa | 7 (23.33%) |
Total RNA isolation
MiRcute miRNAs kit, fluorescent quantitative detection kit and total RNA extraction reagent Trizol were provided by Shanghai Biological Engineering Co. LTD. Aanhydrous, miR-21-3p, miR-21-5p detection primer were provided by Guangzhou RuiBo biotechnology company. The total RNA was extracted with Trizol reagent according to manufactures instructions. Its concentration was measured by Thermo 2000D type ultraviolet spectrophotometer, which was determined by absorbance at 260 nm and the ratio of absorbance at 260/280 nm was used as an indicator of RNA purity. Only samples whose 260/280 ratio was P1.8 were used for subsequent procedures. RNA quality was also assessed by electrophoresis on a 1% agarose gel stained with ethidium bromide (EtBr) with evaluation of 28S and 18S bands; only RNA samples with a 28S band twice as thick as the 18S band were used for RT-PCR microRNA amplification.
Total RNA isolation was performed using standard acid: phenol-chloroform extraction methodology. Briefly, 100 mg of tissue was placed in a pre-chilled Falcon tube and homogenized upon the addition of 1 mL TrIzol reagent (Invitrogen, Cat. #15596-018). Samples were incubated for 5 min prior to the addition of 200 lL chloroform. The phenol-chloroform mixture was briefly vortexed for 15 s and allowed to settle. Samples were centrifuged at 10,000 g for 15 min at 4°C. The upper aqueous layer was transferred to a microfuge tube containing 500 lL of isopropanol. RNA was allowed to precipitate over 10 min followed by a second centrifugation as above. The supernatant was removed and the pellet was washed with the addition of 1 mL 70% ethanol. After a final centrifugation, the ethanol was decanted the pellet was air dried for 8 min and resuspended in 50 lL of double-distilled water (ddH2O).
RT and qPCR
Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR) were used to detect miR-21-3p, miR-21-5p expression condition in cervical cancer and normal cervical tissues, adopting HPV subtype check kits (kappe) to defined cervical infection. The reverse transcriptase reactions contained 500 ng of RNA samples respectively put into two groups of cells, each total RNA will be 10 folds diluted, and take 3 µl two kinds of mature miRNAs (miRNA-213p, 5p) for PCR amplification reaction. 5% agarose gel electrophoresis was used to validate the amplification levels of the target genes. LabWorks 4.0 Image Acquisition and Analysis Software were used to quantization and data processing. The qPCR was initiated with an initial denaturation step at 95°C for 30 sec, 40 cycles of 5 sec at 95°C and 30 sec at 60°C. All the reactions were run in triplicate using the 7500 real-time fluorescent quantitative PCR (Applied Biosystems CO, Foster City, CA). The melting curve in 60-95°C was drawn and accuracy of specific fluorescence quantitative was unimodal. Fluorescence dynamic curve of each sample was drawn, U6 small RNA was used as an internal control to normalize the expression levels of the target genes, the experiment was repeated three times. Calculated the ratio of the Ct value of miRNA-21 and U6, the ratio was in inverse proportion to the relative quantitative of micRNAs.
The ΔCt and 2-ΔΔCt methods were used for analysis the value of miR21 expression. The ΔCt value was calculated by the difference between the Ct values of the specific miRNA and U6, ΔCt = CtmiRNA-CtU6. ΔΔCt = ΔCtcancer-ΔCtnormal. In the present study, RQ = 2-ΔCt represented the value of the ratio of miRNA expression in cancer tissue to that in normal tissue. An RQ < 1 indicated that the miRNA expression levels in cancer tissue were lower compared to those in normal tissue. Conversely, an RQ > 1 indicated higher miRNA expression in cancer compared to normal tissues.
Data were analyzed with SPSS software, version 20.0 (SPSS Inc., Chicago, IL, USA) and Excel software (Microsoft, Redmond, USA). The paired samples t-test was used to compare the expression of miRNAs between cancer and normal tissues. One-way ANOVA was used to investigate the association between cancer and normal tissues. And P-values less than 0.05 were considered significant.
Results
The results of qRT-PCR
Total of 30 cases of cervical cancer with 18 were HPV16 positive, all cases of control group were negative. D260/280 values of extraction and purification of micRNAs in two groups cells were within the range of 1.8~2.1, which met the requirement of the miRNA detection.
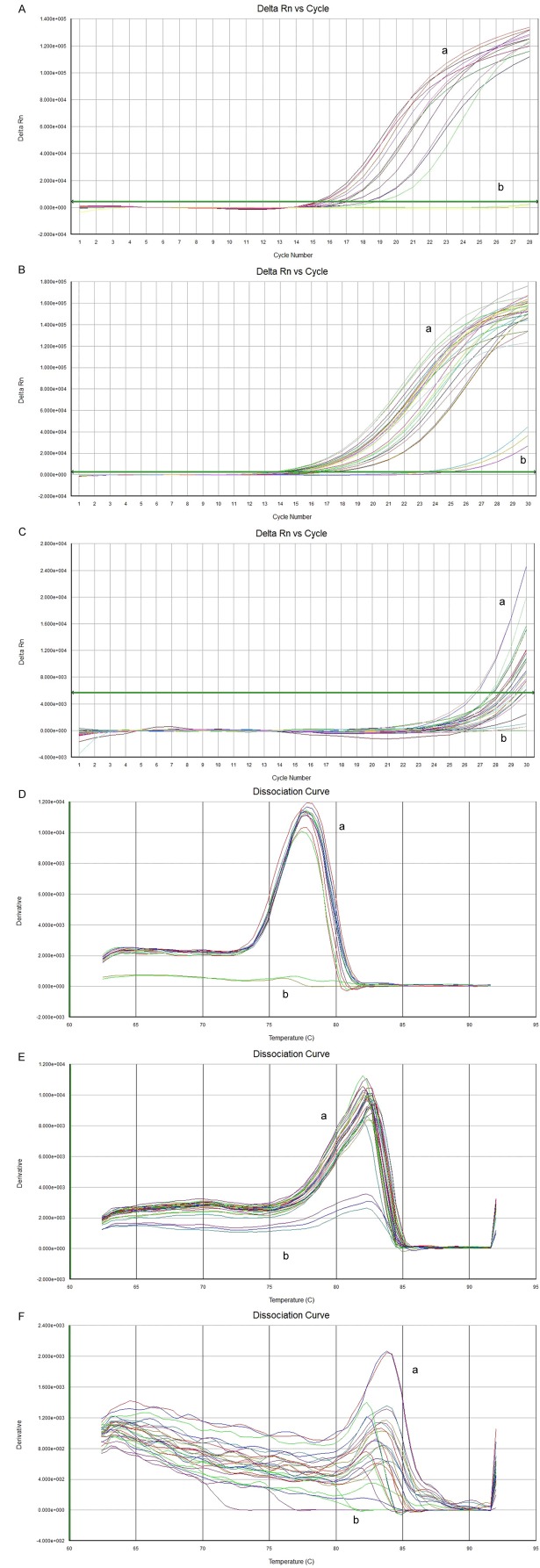
Fluorescent signal of product in each cycle of amplification reaction were detected by real-time fluorescent quantitative PCR for quantitative analysis of the initial template. It can be found that (Figure 1A-C) fluorescence dynamic curve was divided into three stages: phase of fluorescent background signal (the early stage of the logarithmic amplification), stage of index of amplification of fluorescence signal (logarithmic amplification) and stage of platform, which indicated there were enough quantity PCR products and higher sensitivity of primers. In the phase of fluorescence background signal, amplification of fluorescent signal was concealed by fluorescence background signal, the quantity of product cannot be determined. On the plateau, amplification product no longer increased exponentially. Only in the stage of fluorescence signal amplification increased exponentially, there was a linear relationship between the amount of PCR product quantity and initial template’s. The more the quantity of cDNA in template, the lesser of fluorescence cycle number to threshold value, and the smaller the Ct value. In this study, both of the melting curve of miR-21 and internal U6 (Figure 1D-F) were unimodal, which illustrated the specificity of the primers was good, product was pure, no obvious nonspecific product produced and quantitative assay was accuracy.
Figure 1.
MiR-21 fluorescent dynamic curve and melting curve of cervical cancer and normal cervical tissue. A. U6 fluorescent dynamic curve of cervical cancer and normal cervical tissue (a. carcinoma tissue, b. normal tissue); B. MiR-21-5p fluorescent dynamic curve of cervical cancer and normal cervical tissue (a. carcinoma tissue, b. normal tissue); C. MiR-21-3p fluorescent dynamic curve of cervical cancer and normal cervical tissue (a. carcinoma tissue, b. normal tissue); D. Internal U6 melting curve of cervical cancer and normal cervical tissue (a. cancerous tissue, b. normal tissue); E. MiR-21-5p melting curve of cervical cancer and normal cervical tissue (a. cancerous tissue, b. normal tissue); F. MiR-21-3p melting curve of cervical cancer and normal cervical tissue (a. cancerous tissue, b. normal tissue). *note: Cycle Number (cross axis) indicates the cyclic number of fluorescence quantity PCR. Florescence (FI) (vertical axis) indicates the intensity of corresponding PCR.
Among the 30 paired samples, 22 cases were exhibited a higher expression of miR-21 in cervical cancer group, no case express in control group. The average fold change of miR-21 between cervical cancer and normal tissues was 4.0193 ± 0.0023 (P < 0.05) (Table 2; Figure 2).
Table 2.
Expression of miRNA-21 in cervical cancer and normal tissues (x ± s)
| Cervical cancer (n = 30) | Control (n = 30) | P-value | |
|---|---|---|---|
| MiR-21 mean Ct | 26.4231 ± 2.2523 | 29.7435 ± 3.1736 | 0.033 |
| U6 mean Ct | 17.4167 ± 2.1645 | 18.7126 ± 3.2514 | 0.019 |
| ΔCt | 9.0602 ± 2.0015 | 11.0538 ± 3.1047 | 0.028 |
| ΔΔCt | -2.0038 ± 0.0045 | ||
| 2-ΔΔCt (RQ) | 4.0193 ± 0.0023 | ||
| Fold change | 4.0193 ± 0.0023 | ||
Figure 2.
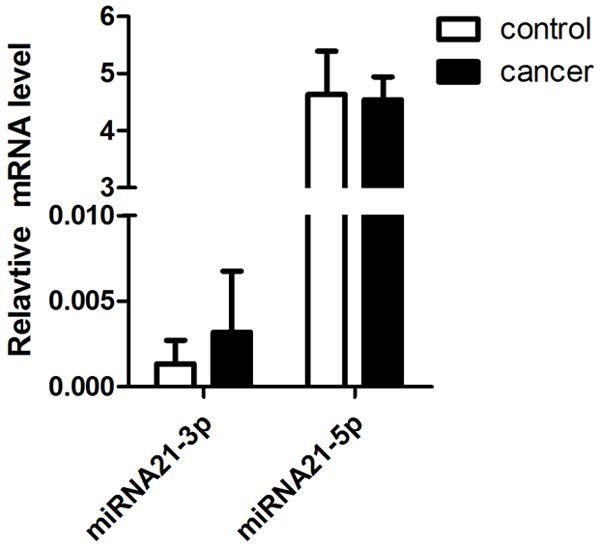
Relative quantity (RQ) of miRNA-21 in cervical cancer and normal tissues. The RQ of miRNA-21-3p was significantly upregulated compared to that in the paired normal samples, but there was no difference between miRNA-21-3p and normal tissues.
18 cases of HPV16 positive demonstrated higher expression of miR-21 RQ in cervical cancer tissues compared to that in HPV16 negative tissues (P < 0.05) (Table 3).
Table 3.
Expression of miRNA-21 in cervical cancer tissues with HPV positive and negative (x ± s)
| HPV16 positive N = 1 | HPV16 negative N = 12 | P-value | |
|---|---|---|---|
| ΔΔCt | -1.7346 ± 0.2554 | -1.5903 ± 0.2872 | 0.043 |
| 2-ΔΔCt (RQ) | 2.3764 ± 0.0341 | 1.9453 ± 0.0290 | 0.016 |
Relationship between MiR-21 expression and cervical cancers
To further investigate the roles of miR-21 expression in cervical cancer progression, the association between the expression of miR-21 level and clinicopathological factors (Tables 4, 5) was analyzed by one-way ANOVA (univariate and multivariate analysis of prognostic parameters in patients with cervical cancers by Cox regression analysis). An increased expression of miR-21 in cervical cancer was not associated with age, tumor size, differentiation and FIGO stage. However, miR-21 overexpression was found to be significantly associated with lymph node metastasis, vascular invasion and depth of invasion (P = 0.015, P = 0.023 and P = 0.031 respectively).
Table 4.
Association of miR-21 expression level with clinical parameters
| cases | Expression Level of miR-21 | P-value | ||
|---|---|---|---|---|
|
| ||||
| High (n, %) | Low (n, %) | |||
| Age (years) | ||||
| < 45 | 18 | 10 (55.56) | 8 (44.44) | NS |
| ≥ 45 | 12 | 6 (50.00) | 6 (50.00) | |
| Tumor size | ||||
| < 4.0 cm | 22 | 12 (54.54) | 10 (45.46) | NS |
| ≥ 4.0 cm | 8 | 4 (50.0) | 4 (50.0) | |
| Histology Type | ||||
| G1 | 18 | 8 (44.4) | 10 (55.6) | 0.065 |
| G2 | 9 | 6 (66.7) | 3 (33.3) | |
| G3 | 3 | 2 (66.7) | 1 (33.3) | |
| FIGO stage | ||||
| Ib1 | 6 | 3 (50.0) | 3 (50.0) | 0.124 |
| Ib2 | 17 | 12 (70.6) | 5 (29.4) | |
| IIa | 7 | 5 (71.4) | 2(28.6) | |
| Lymph node metastasis | ||||
| Absent | 25 | 7 (28.0) | 18 (72.0) | 0.015 |
| Present | 5 | 4 (80.0) | 1 (20.0) | |
| Vascular invasion | ||||
| Absent | 22 | 6 (27.3) | 16 (72.7) | 0.023 |
| Present | 8 | 6 (75.0) | 2 (25.0) | |
| Depth of invasion | ||||
| < 1/2 layer | 22 | 6 (27.3) | 16 (72.7) | 0.031 |
| ≥ 1/2 layer | 8 | 6 (75.0) | 2 (25.0) | |
Table 5.
Association between miRNA level and clinicopathological factors in patients with cervical cancer
| Univariate log-rank test (P) | Cox multivariable analysis (P) | Relative risk (RR) | |
|---|---|---|---|
| Age (years) | |||
| < 45 vs. ≥ 45 | 0.4 | 0.8 | 0.7 |
| Tumor size (cm) | |||
| < 4.0 vs. ≥ 4.0 | 0.6 | 0.7 | 0.5 |
| FIGO stage | |||
| Ia~Ib vs. IIa | 0.001 | 0.02 | 5.8 |
| Histological grades | |||
| G1 vs. G2, 3 | 0.07 | 0.11 | 1.2 |
| Lymph node metastasis | |||
| Absent vs. present | 0.005 | 0.03 | 3.8 |
| Vascular invasion | |||
| Absent vs. present | 0.01 | 0.04 | 2.6 |
| MiR-21 expression | |||
| High vs. Low | < 0.001 | 0.007 | 6.5 |
| Depth of invasion | |||
| Negative vs. positive | 0.06 | 0.1 | 1.1 |
Discussion
Although several miRNAs have been found to function as either tumor metastasis promoters or suppressors [5,6], the exact role that miRNAs play in cervical cancer metastasis is only beginning to be uncovered. Over the past 10 years, accumulating evidence strongly supports the role of miRNAs in crucial cellular processes of cervical cancer development, miRNAs may function as oncogenes or tumor suppressor genes [3,7,8]. This altered miRNA expression pattern provided novel opportunities for the use of biomarkers in cervical cancer diagnosis and built a new way to management of cervical cancer [9].
MiR-21 was found to be one of the upregulated miRNAs in cervical cancer tissues, which accumulated evidence for differential expression of miR-21 in cervical cancer suggesting that it may be play a crucial role in tumor biology [10].
In order to make a comprehensive insight into the clinical value of miR-21, we performed a real-time quantitative RT-PCR assay to explore the expressions profile of miR-21-3p and miR-21-5p in HPV16 positive squamous cell carcinomas in cervix as well as to investigate their association with clinicopathological features of cervical cancer patients. Our data proved that miR-21-3p and miR-21-5p expression significantly increased in cervical cancer compared to paired normal tissues, which suggests that miR-21 may play an oncogenic role in human cervical cancer. Furthermore, we also analyzed the association between miRNA expression level and clinicopathological factors, which demonstrated that the expression of miR-21 was strongly associated with clinicopathological parameters, including depth of invasion and lymph node metastasis. This result is consistent with the report of Deftereos et al and Liu et al [11,12].
In order to know whether the presence of HPV could influences cellular microRNA expression, the analysis was performed in positive HPV and negative HPV cell lines, which found that the expression of miR-21 in HPV16 positive group was higher than those in negative group. These data suggest that the presence of infection of HPV 16 do influence cellular microRNA expression in cervical cancer tissue. The miR-21 gene was located on chromosome 17q23.2, which was inside the common fragile site FRA1 7B, which is one of the HPV16 integration area [13] integration into the host cell genome will genetic alterations (such as deletions, amplifications, or complex rearrangements) and epigenetic alteration, thus intriguing to speculate that the expression of cellular miRNA genes at or near HPV integration sites may contribute to the tumor phenotype [14]. So there was suppose that the HPV16 integration sites near miR-21 genes was likely to become the objective targets which affected the development of cervical cancer, so miRNA-21 may be involved in carcinogenesis and development of cervical cancer. However, In Vanessa’s study didn’t find any differential expression of microRNAs regarding to HPV infection [15]. It suggests the presence of fine molecular events, making to the cervical cancer as more complex disease. More studies are needed to ascertain the results of the present study.
Cox proportional hazards model adjusted for known prognostic variables such as histological grade, FIGO stage, HPV status, lymph node metastasis, and vascular invasion proved that miR-21 was an independent prognostic marker for cervical cancer. Although the precise mechanisms underlying the biological functions of miRNAs have not been fully confirmed, our data demonstrated that miR-21-3p and miR-21-5p was overexpressed in cervical cancer, which may indicate their role as oncogenes in cancerous processes. Thus, miR-21 could be used as a molecular prognostic marker additive to the known prognostic indicator, in order to identify patients who are more likely to have higher risk of death that should require for optimizing treatment selection.
Our founding supplies the microRNA data base associated with cervical cancer. This study of miR-21 in cervical cancer tissues expression and regulation mechanism is still at the initial stage, which still needs to be confirmed by further exploration.
In conclusion, our data indicated that miR-21 upregulation was associated with aggressive progression and poor prognosis in cervical cancer. MiR-21 was identified for the first time as an independent marker for predicting the clinical outcome of cervical cancer patients.
Acknowledgements
The work was supported by a grant (No. 2012-1003 to Y. Han) from the Scientific and Technological Innovation Act Program of Fengxian Science and Technology Commission.
Disclosure of conflict of interest
None.
References
- 1.de Sanjose S, Quint WGV, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GAH, Lombardi LE, Banjo A, Menéndez C, Domingo EJ, Velasco J, Nessa A, Chichareon SCB, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJLM, Al-Jassar WF, Cruz E, Wright TC, Puras A, Llave CL, Tzardi M, Agorastos T, Garcia-Barriola V, Clavel C, Ordi J, Andújar M, Castellsagué X, Sánchez GI, Nowakowski AM, Bornstein J, Muñoz N, Bosch FX. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 2.Sahiner F. Current Problems and Recent Advances in the Molecular Diagnosis of Genital Human Papillomavirus Infections. Mikrobiyol Bul. 2014;48:689–706. doi: 10.5578/mb.7631. [DOI] [PubMed] [Google Scholar]
- 3.Banno K, Iida M, Yanokura M, Kisu I, Iwata T, Tominaga E, Tanaka K, Aoki D. MicroRNA in cervical cancer: OncomiRs and tumor suppressor miRs in diagnosis and treatment. ScientificWorldJournal. 2014;2014:178075. doi: 10.1155/2014/178075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gocze KGK, Kovacs K, Juhasz K, Gocze P, Kiss I. MicroRNA Expressions in HPV-induced Cervical Dysplasia and Cancer. Anticancer Res. 2015;35:523–530. [PubMed] [Google Scholar]
- 5.Buonaguro FM, Pauza D, Tornesello ML, Hainaut P, Franco R, Marincola FM. Cancer diagnostic and predictive biomarkers. Biomed Res Int. 2014;2014:980163. doi: 10.1155/2014/980163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912–1938. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Wang Y, Dong R, Huang X, Ding S, Qiu H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol. 2012;138:671–674. doi: 10.1007/s00432-012-1147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng RQ, Wan HY, Li HF, Liu M, Li X, Tang H. MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7. J Biol Chem. 2012;287:14301–14309. doi: 10.1074/jbc.M111.337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding H, Wu YL, Wang YX, Zhu FF. Characterization of the MicroRNA Expression Profile of Cervical Squamous Cell Carcinoma Metastases. Asian Pac J Cancer Prev. 2014;15:1675–1679. doi: 10.7314/apjcp.2014.15.4.1675. [DOI] [PubMed] [Google Scholar]
- 10.Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–542. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 11.Deftereos G, Corrie SR, Feng Q, Morihara J, Stern J, Hawes SE, Kiviat NB. Expression of mir-21 and mir-143 in cervical specimens ranging from histologically normal through to invasive cervical cancer. PLoS One. 2011;6:e28423. doi: 10.1371/journal.pone.0028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Sun H, Wang X, Yu Q, Li S, Yu X, Gong W. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int J Mol Sci. 2014;15:758–773. doi: 10.3390/ijms15010758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco D, Kivi N, Qian K, Leivonen SK, Auvinen P, Auvinen E. Human papillomavirus 16 E5 modulates the expression of host microRNAs. PLoS One. 2011;6:e21646. doi: 10.1371/journal.pone.0021646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theelen W, Litjens RJ, Vinokurova S, Haesevoets A, Reijans M, Simons G, Smedts F, Herrington CS, Ramaekers FC, von Knebel Doeberitz M, Speel EJ, Hopman AH. Human papillomavirus multiplex ligation-dependent probe amplification assay for the assessment of viral load, integration, and gain of telomerase-related genes in cervical malignancies. Hum Pathol. 2013;44:2410–2418. doi: 10.1016/j.humpath.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Villegas-Ruiz V, Juárez-Méndez S, Pérez-González OA, Arreola H, Paniagua-García L, Parra-Melquiadez M, Peralta-Rodríguez R, López-Romero R, Monroy-García A, Mantilla-Morales A, Gómez-Gutiérrez G, Román-Bassaure E, Salcedo M. Heterogeneity of microRNAs expression in cervical cancer cells: over-expression of miR-196a. Int J Clin Exp Pathol. 2014;7:1389–1401. [PMC free article] [PubMed] [Google Scholar]