Abstract
Lung transplantation has already become the preferred treatment option for a variety of end-stage pulmonary failure. However the long-term results of lung transplantation are still not compelling and the major death reason is commonly due to obliterative bronchiolitis (OB) which is considered as chronic rejection presenting manifests physiologically as a progressive decline in FEV1. Transcription factors (TFs) play a key role in regulating gene expression and in providing an interconnecting regulatory between related pathway elements. Although the transcription factors are required for expression of the proinflammatory cytokines and immune proteins which are involved in obliterative bronchiolitis following lung transplantation, the alterations of the transcription factors in OB have not yet been revealed. Therefore, to investigate the alteration pattern of the transcription factors in OB, we used protein/DNA arrays. Mice orthotopic tracheal transplantation model was used in this studying. In this study, we explored the activity profiles of TFs in Protein/DNA array data of tracheal tissue in 14 and 28 day after transplanted. From a total of 345 screened TFs, we identified 42 TFs that showed associated with OB progression. Our data indicate that TFs may be potentially involved in the pathogenesis of OB, and can prevent, diagnose and treat OB after lung transplantation. In development of OB, some of the TFs may have ability to modulate the transcription of inflammatory proteins such cytokines, inflammatory enzymes and so on.
Keywords: Protein-DNA array, transcription factor, obliterative bronchiolitis
Introduction
Lung transplantation has already become the preferred treatment option for a variety of end-stage pulmonary failure [1]. Compared with other solid organ transplantations, the operation and preservation of donor techniques have been developed and the result is satisfactory in short-time [1,2]. However long-term results are still not compelling and the major death reason is commonly due to obliterative bronchiolitis (OB) which is considered as chronic rejection presenting manifests physiologically as a progressive decline in FEV1 [3]. The criterion for diagnosis of OB is the loss of allograft function. The pathogenesis of OB is initiated by a peribronchiolar leukocyte infiltration that disrupts the underlying submucosa and epithelial layers of the transplanted airway resulting in a progressive loss of airway structure. And the pathological feature of OB shows luminal obliteration, bronchiolar epithelial cell injury and cicatrix formation which leads to airway fibrosis and disappearance of small airways. The airway changes of OB are perceived as alloimmune or autoimmune disorders respectively, it shows similarities to the OB that can occur in allogengeic bone marrow or stem cell transplant recipients as well as constrictive bronchiolitis in patients with connective tissue diseases. Many observations indicated that OB is the effects of an alloimmune response to foreign tissue associated with acute cellular rejection [4] and greater degrees of HLA mismatch [5]. The acute rejection and late lymphocytic bronchiolitis (LB) are prime risk factors for developing OB [6-13] and humoral rejection has also been associated with the development of OB [14,15]. More recent investigations have linked alloimmune responses to OB, which may involve Th17 cells and IL-17 [16]. Interleukin-17-producing cells (Th17 cell which produce interleukin-17 may be the prime stimulus for upregulation of IL-8, which is a potent chemoattractant of neutrophils. And some researches shows the constrictive bronchiolitis of OB may associated with these chemoattractant of neutrophils [17,18]. At present, OB is be reliably diagnosed by either spirometric or histopathological biopsy findings [19], since OB is easily overlooked. Early diagnosis of OB is important to immunosuppressive treatment which could lead to increased survival rate. For this purpose, many researchers have aspired to seek better specific biomarkers that could be able to predict and monitor development of OB in lung transplant recipients. And clinical management of lung transplant patients would be improved if some novel biomarker for rejection as OB could be found. To search these biomarkers, such as bronchoalveolar lavage fluid granulocyte and measurements of circulating fibrocytes have been attempted. Increased circulating fibrocyte levels correlate with the development of OB following lung transplantation. Increasing fibrocyte levels positively correlates with advancing OB stage [20]. Elevated bronchoalveolar lavage fluid neutrophil percentage as well as levels of the granulocyte activation markers myeloperoxidase and eosinophil cationic protein appears to be early signs of development of OB in lung transplant recipients [21]. Plasma MBL which localizes within the lung during graft ischemia and OB, its levels increase after lung transplantation are associated with poor long-term outcomes, it may serve as a biomarker for poorer outcome post-lung transplantation [22,23]. OB induction is in part complement dependent due to IL-17-mediated downregulation of CRPs on airway epithelium. C3a and IL-17 are part of a feed-forward loop that may enhance CRP downregulation [24]. Despite all the work done, we are still far away from success. Novel methods should be looked into in this area.
As mentioned above, an increased expression of many proteins is involved in the complex immune response in OB. The expression of these immune proteins such as cytokines, inflammatory enzymes, receptors and adhesion molecules, is mainly regulated by gene transcription by an activation or inactivation of certain transcription factors (TFs), the DNA-binding proteins. As a group of crucial components in the regulatory net works, transcription factors serve as a major link between the activation of various signal transduction pathways and nuclear gene expression [25]. Therefore, TFs for these proteins may play a key role in the pathophysiology of OB. In the present study, protein/DNA arrays for 345 TFs were performed in nuclear proteins of orthotopic tracheal transplanted mice.
Materials and methods
Animals
Specific pathogen-free, female C57BL/6 (H-2b) mice and Balb/C (H-2d) mice (Tianjin Medical University Laboratory) weighing 20-24 g were used. All animals were housed in the specific pathogen-free facility (Tianjin Medical University, Tianjin, China) and had access to water and food ad libitum. All the studies were performed in compliance with the Principles of Laboratory animal care (NIH publication Vol 25, No. 28 revised 1996) and the Tianjin Medical University Animal Care and Use Committee Guidelines. Tracheas from C57BL/6 mice were implanted into Balb/c mice (allogeneic) or C57BL/6 mice (syngeneic). Each donor tracheal graft was simultaneously implanted into orthotopic [26]. Grafts were harvested on Days 14, 28 after transplantation for histological analyses.
Histology
The grafts were harvested from CO2 euthanized recipient mice on Day 14, 28 after transplantation for histological analyses. Tracheal grafts were cut into longitudinal sections for hematoxylin and eosin (H&E) staining. Each graft segment for H&E staining was fixed in 4% formalin at room temperature for 24 h. The formalin-fixed tissues were embedded in paraffin, later cut into 4-μm sections and then stained with H&E.
Extraction of nuclear proteins
Nuclear protein of bronchial tissue was purified by using Nuclear Extract Kit (Panomics Nuclear Extraction Kit) as per the manufacturer’s instructions. The concentration of nuclear protein was determined with the bicincho-ninic acid (BCA) protein assay reagent kit (Pierce, Rock -ford, IL, USA) to normalize for the amounts of protein within each experiment.
Transcription factor array method (Transignal™ protein/DNA array)
Nuclear extracts were prepared using Nuclear Extract Kit (Panomics Nuclear Extraction Kit, Cat.) as per the manufacturer’s instructions. The concentration of nuclear protein was determined with the bicinchoninic acid protein assay reagent kit (Pierce, Rockford, IL) to normalize for the amounts of protein within each experiment. Transcription factor array analysis (Panomics Transcription factor pathfinder array Tran Signal kit, Redwood City, CA) was used to profile activities of 345 transcription factors. To highlight TFs that characterize each group, a per-gene on median normalization was performed, which normalizes the expression of every TF on its median among samples using saved data in an Excel spreadsheet and calculating the ratio of the data collected from the images. Any spots with a two-fold increase or decrease are considered significant.
Reuslts
Gross pathology and histology
At 14, 28 day postransplant periods, random sections from the mid-portion of the tracheal grafts were evaluated. The syngeneic group show normal luminal ciliated mucosa. And there was no evidence of cellular infiltrate or edema. The allografts group show clear evidence of rejection manifest by edema and lymphocyte infiltration in 14 and 28 day, the luminal obliteration of > 30% after 28 days. Lymphocytic tracheitis and epithelial ulcerations are noted in the allografts soon after the transplant, indicating alloimmune injury. Accompanying mononuclear cell infiltration is also seen within the host tracheal segments adjacent to the graft. Day 28 allografts demonstrate progressive obliterative airway disease, consisting of granulation tissue within the airway lumen formed by spindled cells, matrix, and inflammatory cells (Figure 1). And there was a relatively steady progressing in the feature of pathology and time progress after transplanted.
Figure 1.
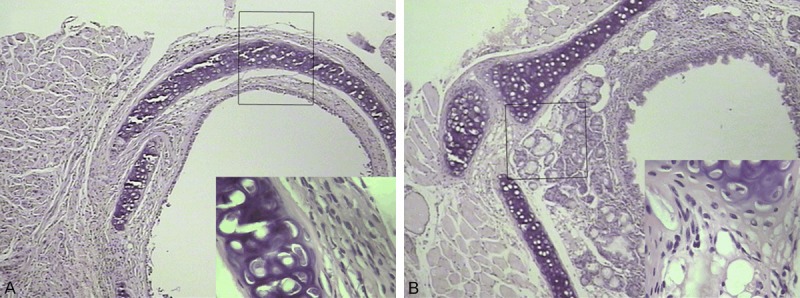
Histological features of orthotopic allografts grafts on D 14 (A) and 28 (B) post-transplant. peribronchiolar leukocyte infiltration that disrupts the underlying submucosa and epithelial layers of the transplanted airway (H&E; original magnification: × 100; inset magnification: × 400.
Protein/DNA array study
Relative TFs activation or inactivation in bronchus was compared with such profiles in syngeneic control mice. Changes in 42 TFs were followed by protein/DNA array. Array membranes were treated with nuclear extracts isolated from control, D14 and 28 groups. After normalization of the raw data, 345TFs were detected in three groups; with 42 of them differentially expressed by TF array and all of were up-regulated in both D14 and 28 groups compared with the syngeneic control group (Table 1). These data provide the first evidence that the activities of various transcription factors are differentially regulated in OB of allogeneic mice orthotopic tracheal transplantation and syngeneic control group. The expression level of these TFs increases with the progress of OB (Figure 2).
Table 1.
Transcription factors (TFs) with altered expression by orthotopictracheal transplantation in mice
| Transcription Factor/Cis-element | Description | D14 | D28 |
|---|---|---|---|
| CdxA/NKX2 | Caudal-type homeodomain protein/cardiac-specific homeo box | 7.2284311 | 19.108173 |
| Freac-2 (1) | forkhead box F2 (mouse) | 5.8721234 | 13.101664 |
| TGT3 | TTF-1 (Thyroid Transcription Factor 1) binding element | 3.1763948 | 6.9544256 |
| Pur-1 | MYC-associated zinc finger protein (purine-binding transcription factor) | 3.5535206 | 8.5973831 |
| AP3 | activator protein 3 | 2.1459901 | 6.3927236 |
| GATA1 (2) | 3.7750988 | 11.853023 | |
| Freac-7 | forkhead box L1 | 5.1269967 | 12.819287 |
| HNF-3 (a, b, g) | hepatocyte nuclear factor 3 (a, b, g) | 2.8391372 | 11.045721 |
| GBF1/2/3/HY5 | G-box binding factor 1 | 2.323852 | 5.0370897 |
| LF-A1 (2) | Liver-specific transcription factor | 3.7001827 | 13.529127 |
| myc-CF1 | common factor 1; CF1 | 2.6879086 | 10.570438 |
| GKLF | gut-enriched Krueppel-like factor; EZF; epithelial zinc-finger | 3.3945985 | 8.2673506 |
| myc-PRF | A transcriptional repressor of c-myc | 3.6417267 | 20.370261 |
| PUR | Pur factor | 3.0497692 | 6.2351248 |
| LF-B2 | liver-specific factors | 4.9896092 | 19.762222 |
| Myc/Max | myc-associated factor X | 64.628873 | 866.443 |
| Elf-1 | E74-like factor 1, a novel Ets family member | 3.5593017 | 20.000568 |
| RORE | RAR-related orphan receptor | 2.6153743 | 7.3188295 |
| Smad SBE | MADH: MAD, mothers against decapentaplegic homolog | 480.48037 | 1234.3028 |
| Ikaros | Ikaros protein (zinc-finger protein) | 7.3938934 | 15.171127 |
| SIF1 | Sucrase-isomaltase (SI) is an enterocyte-specific gene which exhibits a complex pattern of expression during intestinal development and in the adult intestinal mucosa. SIF1SI promoter 1 | 3.279225 | 8.9836184 |
| C/EBPa/g | CCAAT/enhancer binding protein alpha, gamma | 7.9115469 | 16.866061 |
| DE I | Rat DE1 element from albumin gene | 17.987428 | 38.570058 |
| IRF-1, IRF-2 | interferon regulatory factor 1/2 binding element | 13.776668 | 27.660436 |
| XBP1, X2BP | X-box-binding protein 1 | 18.920963 | 46.028576 |
| CD28RC, NF-IL2B | T-cell accessory molecule CD28 | 23.549489 | 92.518538 |
| LR1 | LR1 is a 106-kDa sequence-specific DNA-binding protein first identified as a potential regulator of immunoglobulin class switch recombination in B lymphocytes | 14.937349 | 32.436226 |
| SIF3 | SI promoter 3 | 5.5794092 | 18.138106 |
| ZNF174 | zinc-finger protein 174 | 10.194376 | 20.948212 |
| SPERM1 | A Pou domain gene transiently expressed immediately prior to meiosis I in the male germ cell | 2.620666 | 6.0012397 |
| Pax2 | paired box gene 2 (gene5/gene8) | 4.9965179 | 14.07995 |
| HLF | hepatic leukemia factor | 3.0952976 | 13.906064 |
| NRF-1 | nuclear respiratory factor 1 | 8.9915515 | 23.811503 |
| AAF | an IFN-gamma-regulated DNA-binding factor | 17.684316 | 46.073765 |
| CBFB | CCAAT-binding factor | 21.862635 | 44.059122 |
| FKHR | forkhead box O1A (rhabdomyosarcoma) human | 174.26371 | 460.03887 |
| CBF | mouse CCAAT-binding factor, CP1 (human, rat); NF-Y | 2.5223519 | 10.014016 |
| PO-B | Stimulatory factor, binds at -15 in proopiomelanocortin (POMC) gene | 5.8450886 | 11.857744 |
| WAP BP | whey acidic protein | 2.9996083 | 6.523301 |
| AFP1 | alpha-fetoprotein | 4.7012304 | 9.7924955 |
| HOXA4 | Chox-1.4; Chox1.4; Hox-1.4 (mouse) | 8.4866902 | 39.80197 |
| IL-6 RE-BP | interleukin-6 response element binding protein | 3.2893583 | 12.353047 |
D14 and D28 were Protein/DNA array expression folds compared with syngeneic control group.
Figure 2.

The expression level of these TFs increases with the progress of OB.
Discussion
In the present study, we have identified a number of transcription factors that are highly activated in the OB mice. The activation of 42 TFs after orthotopic tracheal transplantation, all of them were up-regulated and the expression level of these TFs increases with the progress of OB. Transcription factors represent one of the most powerful drug development targets [27]. Modulation of the activation of these signaling pathways and/or their downstream target proteins may serve as potential therapeutics to protect lung function and to improve quality and length of life for patients after lung transplantation. To data, the exact mechanisms involved in OB after lung transplantation are not completely understood. TFs play an important role in the development and function of the immune system [28-30]. It’s proven that, a lot of human disease was linked with TFs. Therefore, we apply TF array to analyze the relationship between TFs and immune rejection, which has not been studied before. The aim of our study was to reveal the relationship between TFs and OB to the public, function research on TFs in OB is our next plan. FKHR (FOXO1) is a member of forkhead transcription factors family which is characterized by the presence of a highly conserved, monomeric DNA-binding domain. FOXO members could play a decisive role in the life and death of cells of the immune system, as their activity appears to be under tight control of a variety of cytokines, and they appear to play an active role in programmed cell death. Stress stimuli also trigger the relocalization of forkhead transcription factors into the nucleus, thus allowing an adaptive response to stress stimuli. Forkhead transcription factors translocate to the nucleus and upregulate a series of target genes, thereby promoting cell cycle arrest, stress resistance, or apoptosis [31]. FOXO proteins translate environmental stimuli into changes in gene expression programs that may induce the progression of OB. Forkhead Box M1 (foxm1) is transcription factor, which orchestrates complex temporal and spatial gene expression throughout embryonic and foetal development as well as during adult tissue homeostasis and repair. Mouse genetic deletion studies have revealed that foxm1 plays a critical role in the formation of respiratory epithelium during initial stages of lung development, whereby deletion of foxm1 gene impairs lung maturation and results in respiratory failure after birth. Conversely, expression of a constitutively active form of foxm1 in the lung epithelium causes epithelial hyperplasia, and inhibits both lung sacculation and the expression of the type II epithelial marker [32]. Epithelial to mesenchymal transition (EMT) is a crucial developmental phenomenon during which epithelial cells lose the characteristic epithelial features and their polarity to attain a mesenchymal phenotype, which will bestow cells increased migratory capabilities. In adults, EMT is engaged in wound healing, organ fibrosis and metastasis during cancer progression. The acquisition of fibroblast-like spindle shaped morphology with cytoskeleton re-organization, increased motility, invasiveness and metastatic capabilities, the accumulation of β-catenin as well as the E-cadherin switching (down-regulation of E-cadherin and up-regulation of N-cadherin and cadherin-11). EMT is mediated by a series of transcription and there is strong evidence that FOXM1 is a key regulator of EMT [33-35]. Foxm1 overexpression also drives EMT and the expression of EMT-regulatory proteins, such as ZEB1 (E-cadherin inhibitor), Vimentin, Snail-2 [35]. Controlled Foxm1 expression and activity deliver a balanced transcriptional programme to homeostasis and repair of adult tissues. However, targeting Foxm1 can be a double-edged sword as its role in adult tissue repair and homeostasis. The abnormal expression of foxm1 may be caused by stimuli of transplantation and lead to OB through interference the reparation and homeostasis of lung. Freac-7 (Foxl1) proteins belong to the forkhead box (Fox) family of transcription factors which has been implicated in the regulation of epithelial cell proliferation in gastrointestinal tracts. Loss of Foxl1led to a marked increase in cellular proliferation of intestinal epithelia in mice, leading to the distortion in the tissue architecture of the stomach and small intestine [36]. A recent study revealed that FOXL1 could bind to the promoter of ZEB1 and suppresses ZEB1 transcription [37] and overexpression of FOXL1 suppressed ZEB1 expression, which hence induced the expression of E-cadherin. In addition, the role of E-cadherin in apoptosis has been investigated in several studies. E-cadherin overexpression could induce apoptosis, increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines [38]. The induction of apoptosis by E-cadherin expression may be mediated by its suppressive activities on Bcl-2 expression [39]. Based on the fact that FOXL1 overexpresion decreased the expression of Bcl-2 and induced mitochondrial dysfunction, it is suggested that E-cadherin may play a role in induction of apoptosis by FOXL1 overexpression. At the transcriptional level the transcription factor Myc, which heterodimerizes with Max (Myc/Max) regulates a large number of genes including genes involved in ribosome biogenesis, and control the progression through the cell cycle and cell growth in somatic cells [40]. The Myc/Max heterodimer binds the DNA sequence CACGTG (E box element) and activates transcription through the transactivation domain of Myc. Thus, Myc/Max induces proliferation and apoptosis in cells while are involved in growth arrest, differentiation, and cell survival [41]. Smad-SBEs (Smad-binding elements) located within promoters, and bind with transcriptional coactivators and corepressors to modulate gene expression. The classic pathway is TGF-β pathway. TGF-β is a cytokine secreted by numerous cell types to regulate cellular processes such as proliferation, differentiation, apoptosis, migration and immune responses. Signaling via the TGF-β pathway alters the expression of more than 500 genes to exert a variety of cellular responses. The signal is propagated by TGF-β type Ι receptor (TβRΙ) phosphorylating the intracellular proteins Smad2 and Smad3, which then complex with Smad4. This activated complex can shuttle to the nucleus phosphor, where it modulates gene expression by binding along with transcriptional coactivators to Smad-SBEs [42]. The expression of Smad-SBEs may suggest the activation of TGF-β pathway, and related with OB.
Until now, the diagnosis of OB can only be made by bronchoscope, which is costly, inconvenient, and carries a small risk of complications. Clinical management of lung transplant patients would be improved if some novel biomarker for BOS could be found. As a group of crucial components in the regulatory net works, transcription factors (TFs) regulate the initial step of gene expression and transcription.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (863 Program: 2012AA021003) and Science and technology foundation of Tianjin Health Bureau (2014KZ127).
Disclosure of conflict of interest
None.
Abbreviations
- OB
obliterative bronchiolitis
- TFs
transcription factors
References
- 1.Trulock E, Christie JD, Edwards LB, Boucek MM, Aurora P, Taylor DO, Dobbels F, Rahmel AO, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: twenty-fourth official adult lung and heartlungtransplantation report-2007. J Heart Lung Transplant. 2007;26:782–795. doi: 10.1016/j.healun.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Hachem RR, Trulock EP. Bronchiolitis obliterans syndrome: pathogenesis and management. Semin Thorac Cardiovasc Surg. 2004;16:350–355. doi: 10.1053/j.semtcvs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Goves S, Galazka M, Johnson B, Corcoran T, Verceles A, Britt E, Todd N, Griffith B, Smaldone GC, Iacono A. Inhaled cyclosporine and pulmonary function in lung transplant recipients. J Aerosol Med Pulm Drug Deliv. 2010;23:31–39. doi: 10.1089/jamp.2009.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton CM, Iversen M, Carlsen J, Mortensen J, Andersen CB, Steinbrüchel D, Scheike T. Acute cellular rejection is a risk factor for bronchiolitis obliterans syndrome independent of post-transplant baseline FEV1. J Heart Lung Transplant. 2009;28:888–93. doi: 10.1016/j.healun.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Schulman LL, Weinberg AD, McGregor C, Galantowicz ME, Suciu-Foca NM, Itescu S. Mismatches at the HLA-DR and HLA-B loci are risk factors for acute rejection after lung transplantation. Am J Respir Crit Care Med. 1998;157:1833–7. doi: 10.1164/ajrccm.157.6.9707007. [DOI] [PubMed] [Google Scholar]
- 6.Girgis RE, Tu I, Berry GJ, Reichenspurner H, Valentine VG, Conte JV, Ting A, Johnstone I, Miller J, Robbins RC, Reitz BA, Theodore J. Risk factors for the development of obliterative bronchiolitis after lung transplantation. J Heart Lung Transplant. 1996;15:1200–8. [PubMed] [Google Scholar]
- 7.Kroshus TJ, Kshettry VR, Savik K, John R, Hertz MI, Bolman RM 3rd. Risk faactors for the development of bronchiolitis obliterans syndrome after lung transplantation. J Thorac Cardiovasc Surg. 1997;114:195–202. doi: 10.1016/S0022-5223(97)70144-2. [DOI] [PubMed] [Google Scholar]
- 8.Pu Q, Ma L, Mei J, Zhu Y, Che G, Lin Y, Wu Z, Wang Y, Kou Y, Liu L. Video-assisted thoracoscopic surgery versus posterolateral thoracotomy lobectomy: A more patient-friendly approach on postoperative pain, pulmonary function and shoulder function. Thoracic Cancer. 2013;4:84–89. doi: 10.1111/j.1759-7714.2012.00153.x. [DOI] [PubMed] [Google Scholar]
- 9.Heng D, Sharples LD, McNeil K, Stewart S, Wreghitt T, Wallwork J. Bronchiolitis obliterans syndrome: incidence, natural history, prognosis, and risk factors. J Heart Lung Transplant. 1998;17:1255–63. [PubMed] [Google Scholar]
- 10.Burton CM, Iversen M, Scheike T, Carlsen J, Andersen CB. Is lymphocytic bronchiolitis a marker of acute rejection? An analysis of 2,697 transbronchial biopsies after lung transplantation. J Heart Lung Transplant. 2008;27:1128–34. doi: 10.1016/j.healun.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Ross DJ, Marchevsky A, Kramer M, Kass RM. “Refractoriness” of airflow obstruction associated with isolated lymphocytic bronchiolitis/bronchitis in pulmonary allografts. J Heart Lung Transplant. 1997;16:832–8. [PubMed] [Google Scholar]
- 12.Kapucu S, Karacan Y. Physiological problems in patients undergoing autologous and allogeneic hematopoietic stem cell transplantation. Asia-Pacific J Oncol Nurs. 2014;1:50–54. doi: 10.4103/2347-5625.135821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos R, Vanaudenaerde BM, Verleden SE, De Vleeschauwer SI, Willems-Widyastuti A, Van Raemdonck DE, Dupont LJ, Nawrot TS, Verbeken EK, Verleden GM. Bronchoalveolar lavage neutrophilia in acute lung allograft rejection and lymphocytic bronchiolitis. J Heart Lung Transplant. 2010;29:1259–69. doi: 10.1016/j.healun.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 14.Palmer SM, Davis RD, Hadjiliadis D, Hertz MI, Howell DN, Ward FE, Savik K, Reinsmoen NL. Development of an antibody specific to major histocompatibility antigens detectable by flow cytometry after lung transplant is associated with bronchiolitis obliterans syndrome. Transplantation. 2002;74:799–804. doi: 10.1097/00007890-200209270-00011. [DOI] [PubMed] [Google Scholar]
- 15.Girnita AL, Duquesnoy R, Yousem SA, Iacono AT, Corcoran TE, Buzoianu M, Johnson B, Spichty KJ, Dauber JH, Burckart G, Griffith BP, McCurry KR, Zeevi A. HLA-specific antibodies are risk factors for lymphocytic bronchiolitis and chronic lung allograft dysfunction. Am J Transplant. 2005;5:131–8. doi: 10.1111/j.1600-6143.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- 16.Saini D, Weber J, Ramachandran S. Alloimmunity-induced autoimmunity as a potential mechanism in the pathogenesis of chronic rejection of human lung allografts. J Heart Lung Transplant. 2011;30:624–31. doi: 10.1016/j.healun.2011.01.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanaudenaerde BM, De Vleeschauwer SI, Vos R, Meyts I, Bullens DM, Reynders V, Wuyts WA, Van Raemdonck DE, Dupont LJ, Verleden GM. The role of the IL23/IL17 axis in bronchiolitis obliterans syndrome after lung transplantation. Am J Transplant. 2008;8:1911–20. doi: 10.1111/j.1600-6143.2008.02321.x. [DOI] [PubMed] [Google Scholar]
- 18.DiGiovine B, Lynch JP 3rd, Martinez FJ, Flint A, Whyte RI, Iannettoni MD, Arenberg DA, Burdick MD, Glass MC, Wilke CA, Morris SB, Kunkel SL, Strieter RM. Bronchoalveolar lavage neutrophilia is associated with obliterative bronchioliltis after lung transplantation: role of IL-8. J Immunol. 1996;157:4194–202. [PubMed] [Google Scholar]
- 19.Cooper JD, Billingham M, Egan T, Hertz MI, Higenbottam T, Lynch J, Mauer J, Paradis I, Patterson GA, Smith C, et al. A working for mulation for the standardization of nomenclature and for clinical staging of chronic dysfunction in lung allografts. J Heart Lung Transplant. 1993;12:713–716. [PubMed] [Google Scholar]
- 20.LaPar DJ, Burdick MD, Emaminia A, Harris DA, Strieter BA, Liu L, Robbins M, Kron IL, Strieter RM, Lau CL. Circulating fibrocytes correlate with bronchiolitis obliterans syndrome development after lung transplantation: a novel clinical biomarker. Ann Thorac Surg. 2011;92:470–7. doi: 10.1016/j.athoracsur.2011.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riise GC, Andersson BA, Kjellström C, Martensson G, Nilsson FN, Ryd W, Scherstén H. Persistent high BAL fluid granulocyte activation marker levels asearly indicators of bronchiolitis obliterans after lung transplant. Eur Respir J. 1999;14:1123–30. doi: 10.1183/09031936.99.14511239. [DOI] [PubMed] [Google Scholar]
- 22.Carroll KE, Dean MM, Heatley SL, Meehan AC, Mifsud NA, Kotsimbos TC, Snell GI, Westall GP. High levels of mannose-binding lectin are associated with poor outcomes after lung transplantation. Transplantation. 2011;91:1044–9. doi: 10.1097/TP.0b013e318212c7d6. [DOI] [PubMed] [Google Scholar]
- 23.Budd SJ, Aris RM, Medaiyese AA, Tilley SL, Neuringer IP. Increased plasma mannose binding lectin levels are associated with bronchiolitis obliterans after lung transplantation. Respir Res. 2012;13:56. doi: 10.1186/1465-9921-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Lasbury ME, Fan L, Vittal R, Mickler EA, Benson HL, Shilling R, Wu Q, Weber DJ, Wagner SR, Lasaro M, Devore D, Wang Y, Sandusky GE, Lipking K, Pandya P, Reynolds J, Love R, Wozniak T, Gu H, Brown KM, Wilkes DS. Role of Complement Activation in Obliterative Bronchiolitis Post-Lung Transplantation. J Immunol. 2013;191:4431–9. doi: 10.4049/jimmunol.1202242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitmarsh AJ, Davis RJ. Regulation of transcription factorfunction by phosphorylation. Cell Mol Life Sci. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrepfer S, Deuse T, Hoyt G, Sheikh AY, Hoffmann J, Reichenspurner H, Robbins RC, Pelletier MP. Experimental orthotopic tracheal transplantation: The Stanford technique. Microsurgery. 2007;27:187–9. doi: 10.1002/micr.20329. [DOI] [PubMed] [Google Scholar]
- 27.Latchman DS. Transcription factors as potential targets for therapeutic drugs. Curr Pharm Biotechnol. 2000;1:57–61. doi: 10.2174/1389201003379022. [DOI] [PubMed] [Google Scholar]
- 28.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–84. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 29.Liu CH, Wang HH, Perng CL, Peng CK, Chian CF, Shen CH. Primary extranodal NK/T-cell lymphoma of the lung: Mimicking bronchogenic carcinoma. Thoracic Cancer. 2014;5:93–96. doi: 10.1111/1759-7714.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Lu Y, Wang J, Liu X, Keller ET, Liu Q, Zhou Q, Zhang J. Targeting the Notch signaling pathway in cancer therapeutics. Thoracic Cancer. 2014;5:473–486. doi: 10.1111/1759-7714.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–25. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 32.Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, Park HJ, Whitsett JA, Kalinichenko VV. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev Biol. 2010;347:301–14. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C, Xie D, Cui J, Li Q, Gao Y, Xie K. FOXM1c promotes pancreatic cancer epithelial-to-mesenchymal transition and metastasis via upregulation of expression of the urokinase plasminogen activator system. Clin Cancer Res. 2014;20:1477–88. doi: 10.1158/1078-0432.CCR-13-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu N, Jia D, Chen W, Wang H, Liu F, Ge H, Zhu X, Song Y, Zhang X, Zhang D, Ge D, Bai C. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One. 2013;8:e59412. doi: 10.1371/journal.pone.0059412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, Li Y, Azmi AS, Miele L, Sarkar FH. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Kaestner KH, Silberg DG, Traber PG, Schutz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–95. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G, He P, Gaedcke J, Ghadimi BM, Ried T, Yfantis HG, Lee DH, Hanna N, Alexander HR, Hussain SP. FOXL1, a novel candidate tumor suppressor, inhibits tumor aggressiveness and predicts outcome in human pancreatic cancer. Cancer Res. 2013;73:5416–5425. doi: 10.1158/0008-5472.CAN-13-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H, Zhang X, Cho BC, Park HJ, Kim JH. Tumor MET expression profile predicts the outcome of non-small cell lung cancer patients receiving epidermal growth factor receptor tyrosine kinase inhibitors. Thoracic Cancer. 2014;5:517–524. doi: 10.1111/1759-7714.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki CY, Lin Hc, Passaniti A. Expression of E-cadherin reduces bcl-2 expression and increases sensitivity to etoposide-induced apoptosis. Int J Cancer. 2000;86:660–666. doi: 10.1002/(sici)1097-0215(20000601)86:5<660::aid-ijc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 40.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–9. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 41.Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg Med Chem Lett. 2009;19:6038–41. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iempridee T, Das S, Xu I, Mertz JE. Transforming growth factor beta-induced reactivation of Epstein-Barr virus involves multiple Smad-binding elements cooperatively activating expression of the latent-lytic switch BZLF1 gene. J Virol. 2011;85:7836–48. doi: 10.1128/JVI.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


