Abstract
In this study, we examined the subcellular distribution and functions of LIMK1 in developing neurons. Confocal microscopy, subcellular fractionation, and expression of several epitope-tagged LIMK1 constructs revealed that LIMK1 is enriched in the Golgi apparatus and growth cones, with the LIM domain required for Golgi localization and the PDZ domain for its presence at neuritic tips. Overexpression of wild-type LIMK1 suppresses the formation of trans-Golgi derived tubules, and prevents cytochalasin D-induced Golgi fragmentation, whereas that of a kinase-defective mutant has the opposite effect. Transfection of wild-type LIMK1 accelerates axon formation and enhances the accumulation of Par3/Par6, insulin-like growth factor (IGF)1 receptors, and neural cell adhesion molecule (NCAM) at growth cones, while inhibiting the Golgi export of synaptophysin-containing vesicles. These effects were dependent on the Golgi localization of LIMK1, paralleled by an increase in cofilin phosphorylation and phalloidin staining in the region of the Golgi apparatus, and prevented by coexpression of constitutive active cofilin. The long-term overexpression of LIMK1 produces growth cone collapse and axon retraction, an effect that is dependent on its growth cone localization. Together, our results suggest an important role for LIMK1 in axon formation that is related with its ability to regulate Golgi dynamics, membrane traffic, and actin cytoskeletal organization.
INTRODUCTION
During recent years, a growing body of evidence has accumulated suggesting that the actin cytoskeleton has a crucial role in regulating axon formation (Bradke and Dotti, 1999). In particular, signaling pathways that include the small GTPases of the Rho/Rac/Cdc42 superfamily (Bishop and Hall, 2000) have been shown to be important for actin reorganization and dynamics during neuronal polarization (Bradke and Dotti, 1999; Kunda et al., 2001). Among the downstream effectors of these GTPases are proteins that directly regulate actin turnover, such as the actin-depolymerizing factor (ADF) and cofilin (Bamburg, 1999; Sarmiere and Bamburg, 2004). They are abundant in neuronal growth cones (Bamburg and Bray, 1987; Khun et al., 2000) where the regulation of actin dynamics is essential for axon outgrowth (Bradke and Dotti, 1999; Kunda et al., 2001); not surprisingly, overexpression of ADF-cofilin enhances neurite outgrowth and growth cone motility in developing neurons (Meberg et al., 1998; Meberg and Bamburg, 2000).
Recently, in vivo and in vitro studies have demonstrated that ADF-cofilin is a substrate of a novel family of kinases, designated as LIMKs. Phosphorylation of ADF-cofilin at a single site (ser 3) by LIMKs inhibits its binding to actin monomers and its actin-depolymerizing activity (Arber et al., 1998; Yang et al., 1998; Sumi et al., 1999). The LIMK family of serine-threonine kinases is composed of two major members, one of which, LIMK1 is predominantly expressed in the nervous system (Proschel et al., 1995). Because deletion of the region coding for the LIMK1 gene induces nervous system impairment, resulting in Williams syndrome (Belluji et al., 1999), this protein may be critically involved in brain function. In favor of this, it has recently been showed that LIMK1 knockout mice exhibit a significant reduction in the amount of phosphorylated ADF-cofilin in brain slices, as well as alterations in spine morphology, and in synaptic function, including enhanced long-term potentiation (Meng et al., 2002). Unfortunately, up to present a clear picture regarding the role of LIMK1 in neuronal development has not yet emerged. For example, although the complexity of dendritic branches from cultured hippocampal pyramidal neurons obtained from LIMK1 -/- mice was similar to that of wild-type neurons, the LIMK1-deficient neurons display a dramatic reduction in growth cone size and significant alterations in actin organization. Also, although phosphorylation of cofilin by LIMK1 is necessary for semaphorin 3A-induced growth cone collapse in dorsal root ganglion (DRG) cells, overexpression of constitutively active LIMK1 or dominant-negative LIM-kinase has been reported to have either little effect on neurite outgrowth (Aizawa et al., 2001) or to reduce growth cone size or repress growth cone motility and neurite extension (Endo et al., 2003). Inhibition of neurite extension by overexpression of individual domains of LIMK1 also has been reported in PC12 cells (Birkenfeld et al., 2001).
Therefore, to obtain additional information about the role of LIMK1 during neuronal development, and particularly during axon formation, we have now used a series of recombinant constructs to overexpress LIMK1 or its mutants in cultured hippocampal pyramidal neurons and analyze effects on its subcellular distribution, process formation, and cytoskeletal organization. The results obtained provide evidence about a novel subcellular localization of LIMK1, namely, the Golgi apparatus. More importantly, we show that in developing neurons LIMK1 regulates Golgi dynamics and that its presence within the Golgi apparatus is important for promoting axon outgrowth and the delivery to growth cones of proteins (e.g., Par3/Par6; Shi et al., 2003) critically involved in the development of neuronal polarity.
MATERIALS AND METHODS
Cell Culture
Dissociated cultures of hippocampal pyramidal cells from embryonic rat brain tissue were prepared as described previously (Kunda et al., 2001). All cultures were maintained in a humidified 37°C incubator with 5% CO2. For some experiments, Jasplakinolide (Jasp; Molecular Probes, Eugene, OR) was dissolved in dimethyl sulfoxide and added to the cultures at concentration ranging from 5 to 50 nM. In other cases cytochalasin D was added to the cultures at a final concentration of 1 μM.
Expression Plasmids and Transfection
Expression plasmids for hemagglutinin (HA)-tagged LIMK1 and their mutants were constructed as described previously (Sumi et al., 1999). To generate plasmids encoding ΔLIM (147-547 amino acids) and ΔN (338-647 amino acids) mutants of wild-type or kinase dead LIMK1, cDNA fragments were amplified by polymerase chain reaction, with the following primers: ΔLIM: forward, 5′GCAAGCTTGCCATGATCGAACAGATCCTC3′, and the reverse primer designated as (J)5′GCTCTAGACTCAGCGTAATCTGGAAC3′; ΔN: forward, 5′CCATGGACCTCAT-CCATGG3′, and the reverse primer (J). PCR products were digested with HindIII and XbaI and ligated to HindIII- and XbaI-digested pcDNA3 o pRo/RsV vector (Invitrogen, Carlsbad, CA). To generate plasmids encoding: ΔPK (1-255 amino acids), HA-tagged LIMK1 cDNA was digested with HindIII and XhoI, and ligated to HindIII and XhoI-digested pcDNA3/LIMK2-HA to replace the original HindIII-XhoI fragment of LIMK2 cDNA with LIMK1 cDNA; or ΔPDZ-LIMK1, HA-tagged LIMK1 cDNA was digested with XhoI and SalI and ligated to XhoI and SalI digested pcDNA3. These expression plasmids also were inserted into pEF-BOS vector by using an XbaI linker. We also used 1) a plasmid (pEYFP-N1; BD Biosciences Clontech, Palo Alto, CA) containing the cDNA coding for the N-terminal domains (cytosolic tail, transmembrane domain, and a few amino acids of the stem region) of galactosyl-transferaseT2 fused to the N terminus of the enhanced yellow fluorescent protein (Giraudo et al., 2001); 2) a plasmid (p38-EGFP) containing the cDNA coding for p38 synaptophysin fused to the N terminus of the enhanced green fluorescent protein (Kaether et al., 2000); 3) a plasmid (NCAM-GFP) containing the cDNA coding for the chicken neuronal cell adhesion molecule (NCAM-180) fused to the N terminus of enhanced green fluorescent protein (Musch et al., 2001); 4) a plasmid containing the cDNA for rat Par3 (a generous gift of Dr. T. Pawson, Samuel Lunenfeld Research Institute, Mt. Sinai Hospital, Toronto, ON, Canada); and 5) a plasmid containing a cDNA coding for constitutively active mutant cofilin (S3A mutant) in which Ser 3 was substituted by Ala by using site-directed mutagenesis (Sumi et al., 1999). Transient transfection of cultured neurons was performed using either the modified calcium phosphate precipitation method described by Kunda et al., (2001) or Lipofectamine 2000 (Invitrogen, Carlsbad). Cells were then analyzed at different posttransfection intervals ranging from 12 to 36 h.
Primary Antibodies
The following primary antibodies were used in this study: a monoclonal antibody (mAb) against tyrosinated α-tubulin (clone TUB-1A2, mouse IgG; Sigma-Aldrich, Louis, MO, or clone YL1/2, rat IgG, provided by Dr. Geri Kreitzer, Cornell Medical College, Ithaca, NY) diluted 1:1000; a mAb against the class III neuron-specific β-tubulin isotype diluted 1:1000; a mAb against tau protein (clone Tau-1; Kunda et al., 2001) diluted 1/100; affinity-purified rabbit polyclonal antibodies raised against peptides corresponding to amino acid sequences mapping at either the amino or carboxy termini of LIMK1 of human origin (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:100 or 1:500; a rabbit polyclonal antibody against phosphorylated (Thr 501) LIMK1 (Cell Signaling Technology, Beverly, MA) diluted 1:50; an affinity-purified rabbit polyclonal antibody against phosphorylated cofilin (SER-3; Santa Cruz Biotechnology) diluted 1:100; an affinity-purified rabbit polyclonal antibody against nonphosphorylated cofilin (Santa Cruz Biotechnology) diluted 1:100; a rabbit polyclonal antibody against phosphorylated cofilin (provided by Dr. James Bamburg, Colorado State University, Fort Collins, CO; Meng and Bamburg, 2000) diluted 1:100 or 1:1000; a rabbit polyclonal antibody against Par3 (Cell Signaling Technology) diluted 1:50; a goat polyclonal antibody against Par6 (Santa Cruz Biotechnology); a rabbit polyclonal antibody against phospho-AKT (p-AKT; Cell Signaling Technology) diluted 1:50; a rabbit polyclonal antibody against a β-subunit of the IGF1 receptor (β-gc, Mascotti et al., 1997); a rabbit polyclonal antibody against TrkB (Santa Cruz Biotechnology) diluted 1:100; a mAb against mannosidase II (provided by Dr. Carlos Dotti, European Molecular Biology Laboratory, Germany); a rabbit polyclonal antibody against the influenza protein hemagglutinin (HA, Y-11; Santa Cruz Biotechnology) diluted 1:1000; and a mAb against HA (clone F-7; Santa Cruz Biotechnology) diluted 1:1000.
Immunofluorescence
Cells were fixed before or after detergent extraction under microtubule-stabilizing conditions and processed for immunofluorescence as described previously (Kunda et al., 2001). For some experiments, and to visualize the rhodamine-phalloidin staining of the Golgi apparatus and the perinuclear region, cells were fixed according to the procedure described by Nakata and Hirokawa, (1987; also see Paglini et al., 1998). The antibody staining protocol was as described previously (Kunda et al., 2001), except that fixation and staining for Par3/Par6 or p-AKT were performed as described by Shi et al. (2003) and that cultures stained for phosphorylated LIMK1 were fixed with 4% paraformaldehyde-4% sucrose in Tris-saline (pH 7.2). The cells were analyzed with a Zeiss or Leica confocal scanning microscope and with a Zeiss inverted microscope equipped with epifluorescence and differential interference contrast optics. The relative intensities of HA-tagged LIMK1, or its mutants, as well as of tubulin, cofilin, phospho-cofilin, Par3, and F-actin staining, were evaluated in fixed unextracted cells or in detergent-extracted cytoskeletons by using quantitative fluorescence techniques (Paglini et al., 1998). Synaptophysin-GFP, NCAM-GFP, or Par3 fluorescence intensities were quantified after fixation in a defined area (20-30 μm in length) at the proximal and distal ends of individual neurites. Images were collected using a charge-coupled device camera (Orca 1000; Hamamatsu, Middlesex, NJ) and Meta-Morph software (Universal Imaging, West Chester, PA).
Time-Lapse Videomicroscopy
For time-lapse fluorescence microscopy, transfected hippocampal pyramidal cells were cultured in special Petri dishes prepared according to the procedures described by Paglini et al. (1998). In all cases, cells were observed using a 63× immersion objective. Images were collected using the charge-couple device camera and MetaMorph software.
Subcellular Fractionation Techniques
Fetal rat brain (E18) was fractionated according to Pfenninger et al., (1983) (also see Paglini et al., 1998) to obtain growth cone particles (GCPs) and Golgi membranes were prepared as described by Paglini et al. (2001). Briefly, total embryonic (E19) microsomal membranes from rat cerebral cortex were adjusted to 1.24 M sucrose and loaded at the bottom of a 32-ml discontinuous sucrose gradient and centrifuged at 82,000 × g for 3 h. Bands at the interface between 0.25 M/0.86 M and 0.86 M/1.14 M sucrose, which are enriched in Golgi elements, were collected and designated as Golgi light (F1) and Golgi heavy (F2) fractions. Fraction 1.24 was defined as the residual microsomal fraction (F3). Sucrose density gradient centrifugation of microsomal fractions and immunoisolation of vesicles was as described by Peretti et al. (2000).
Western Blot Analysis of LIMK1 Protein Expression
Changes in the levels of LIMK1 during neuronal development as well as subcellular fractions were analyzed by Western blotting as described previously (Kunda et al., 2001). Briefly, equal amounts of samples were separated on SDS-PAGE and transferred to polyvinylidene difluoride membranes in a Tris-glycine buffer, 20% methanol. The filters were dried, washed several times with Tris-buffered saline) and blocked for 1 h in TBS containing 5% bovine serum albumin. The filters were incubated for 1 h at room temperature with the primary antibodies in TBS containing 1% bovine serum albumin, and then washed three times in TBS containing 0.05% Tween 20. They were then incubated with a secondary horseradish peroxidase-conjugated antibody for 1 h. After five washes with TBS and 0.05% Tween 20, the blots were developed using a chemiluminescence detection kit (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom).
Morphometric Analysis of Neuronal Shape Parameters
Images were digitized on a video monitor using MetaMorph software. To measure neurite length or growth cone shape parameters, antibody-labeled cells were randomly selected and traced from a video screen by using the morphometric menu of the MetaMorph (Paglini et al., 1998; Kunda et al., 2001). Differences among groups were analyzed by the use of analysis of variance and Student-Newman-Keuls test.
RESULTS
LIMK1 Localizes to the Golgi Apparatus and Growth Cones in Developing Neurons
To begin analyzing the possible involvement of LIMK1 in the regulation of neuronal morphology, we first examined its time course of expression in the developing brain and in cultured neurons. In the cerebral cortex and hippocampus, the expression of LIMK1 begins at late embryonic days, having a peak at the end of the second postnatal week (Figure 1A, lanes 1-3), which is then followed by a progressive decline until adulthood. A similar analysis performed with cell extracts obtained from cultured neurons revealed an increase in LIMK1 protein levels 24 h after plating, just at the time in which cells are beginning to extend axons; after that time point, LIMK1 protein levels remain unchanged until the end of the second week in vitro when an additional and significant increase is detected (Fig. 1 A, lanes 4-8).
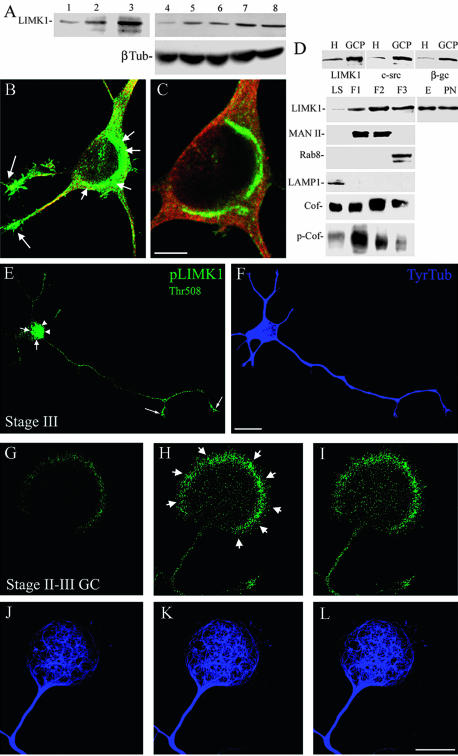
Figure 1.
Endogenous LIMK1 localizes to the Golgi apparatus and growth cones. (A) Western blots showing the time course of expression of LIMK1 in the cerebral cortex of developing rats (lanes 1-3) and in cultured hippocampal pyramidal neurons (lanes 4-8); β-tubulin (β-Tub) protein levels also are shown in samples obtained from cultured cells. Whole tissue extracts were obtained at embryonic day 18 (E18, lane 1), postnatal day 5 (P5, lane 2), and postnatal day 15 (P15, lane 3). Cells tissue extracts were obtained 4 h (lane 4), 1 d (lane 5), 7 d (lane 6), 14 d (lane 7), and 21 d (lane 8) after plating. 10 μg of protein was loaded in each lane. (B) Confocal image showing the distribution of LIMK1 (green) and tubulin (red) in a cultured hippocampal pyramidal neuron. Note the intense labeling for LIMK1 in the perinuclear region (short arrows) and at growth cones (long arrows); staining is also detected in the nuclear region. (C) A high-power view of an adjacent confocal plane from the cell body region of the neuron shown in B. For this experiment, cultures were fixed 3 d after plating and stained with a rabbit polyclonal antibody against LIMK1 diluted 1/100 (1 μg/ml). Bar, 10 μm (A) and 5 μm (B). (D) Western blots showing the enrichment of LIMK1, c-src, and β-gc in GCPs. H, whole tissue homogenate. Ten micrograms of protein was loaded in each lane. (D) Western blots showing the distribution of LIMK1 and several other proteins in the low-speed supernatant (LS) and Golgi fractions (F1-F3). Note that Golgi fractions are enriched in LIMK1, cofilin (Cof), and phospho-cofilin (p-Cof). Note that there are no significant increases in the levels of LIMK1 between Golgi fractions obtained from embryonic (E) or postnatal (PN) brains. Twenty micrograms of protein was loaded in each lane, except for the E and PN lanes that were loaded with 10 μg. For these experiments, the LIMK1 antibody was used at a concentration of 0.5 μg/ml. (E and F). Confocal micrographs showing the distribution of p-LIMK1 (green) and tubulin (blue) in a hippocampal pyramidal neuron maintained in culture for 1 d. Note the intense staining for p-LIMK1 in the Golgi region (short arrows), the cell nucleus (arrowheads), and the axonal growth cone (long arrows). Punctate p-LIMK1 immunolabeling is also detected along neuritic shafts. Bar, 10 μm. (G-L) Series of confocal images showing the distribution of p-LIMK1 (green) and tubulin (blue) in a large growth cone of a stage II-III cultured hippocampal pyramidal neuron. Note that p-LIMK1 immunolabeling is highly enriched at the peripheral rim (short arrows) of the growth cone. Bar, 10 μm.
Next, we examined LIMK1 subcellular distribution by confocal fluorescence microscopy in cultured hippocampal pyramidal neurons. In unpolarized cells, light LIMK1 immunofluorescence was found in the cell body, while in neurite-bearing cells a strong immunofluorescence signal was detected in growth cones and in a region of the cell body localized in close apposition to the cell nucleus that resembled the Golgi apparatus (Figure 1, B and C). Confocal microscopy revealed extensive colocalization of LIMK1 and mannosidase II in the region corresponding to the Golgi apparatus (see below); LIMK1 immunofluorescence was also detected in the cell nucleus, and in vesicle-like structures located along neurites.
To confirm biochemically these observations, growth cone particles (GCPs) and Golgi membranes were isolated from fetal brains and probed with the antibody against LIMK1 used at a concentration of 1 μg/ml. The purity of GCPs was confirmed by its enrichment in c-src and β-gc, a novel subunit of the IGF1 receptor highly enriched in growth cones (Mascotti et al., 1997; Paglini et al., 1998). On the other hand, the purity of Golgi fractions was evaluated by the presence of mannosidase II, a well established Golgi marker, and rab 8, a marker of the trans-Golgi network, as well as by the absence of rab5, a marker of early endosomes, or LAMP1, a marker of lysosomes (Paglini et al., 2001). The results obtained (Figure 1D) clearly revealed that LIMK1 was not only present but also enriched in GCPs and Golgi membranes obtained from the embryonic rat cerebral cortex. Because the presence of LIMK1 in the Golgi apparatus represents a novel subcellular localization with potential great functional significance, we also examined whether cofilin and/or phosphorylated cofilin were present in this fraction. The results obtained clearly revealed the presence of cofilin in Golgi membranes and that all Golgi subfractions (F1-F3 fractions) contain phosphorylated cofilin (Figure 1D). Similar LIMK1 (Figure 1D) and p-cofilin (our unpublished data) protein levels were detected between Golgi fractions obtained from fetal and 10-d postnatal brain.
A downstream target of Rac and cdc42 is PAK1 (Sarmiere and Bamburg, 2004), which has been shown to promote neurite outgrowth in PC12 cells (Daniels et al., 1998), and is a direct activator of LIMK1 through phosphorylation of Thr508. Therefore, it became of interest to analyze the subcellular distribution of activated LIMK1 (p-LIMK1) by using a rabbit polyclonal antibody against the phosphorylated Thr 508 epitope of LIMK1. The results obtained show a strong immunofluorescence signal for p-LIMK1 in the cell body of neurons bearing immature neurites (stage II) and also in those that have develop an axon-like neurite (stage III); the cell body staining comprised the region of the Golgi apparatus (Figure 1, E and F, arrows) and the nucleus (Figure 1, E and F, arrowheads). In addition, punctate p-LIMK1 immunofluorescence was detected along neuritic processes; this staining was much more intense in the axon and its growth cone than in minor processes (Figure 1, E and F). The conversion of one minor process into an axon (stage II-III transition) requires a transient engorgement of the growth cone and penetration of microtubules to form a central domain (c-domain) in which actin filaments have been eliminated (Paglini et al., 1998; Kunda et al., 2001). Interestingly, prominent p-LIMK1 labeling was detected in the shaft and peripheral F-actin rich domain (p-domain) of the large growth cone of the prospective axon (Figure 1, G-L).
Different Protein Domains Determine LIMK1 Subcellular Localization
In a complementary series of experiments, we sought to determine protein domains responsible for LIMK1 subcellular localization. For such a purpose 1- to 3-d-old hippocampal pyramidal cultures were transiently transfected with full-length LIMK1 (wt-LIMK1) or with LIMK1 mutants carrying deletions of 1) the NH2 terminus (ΔN), 2) the LIM domain (ΔLIM), 3) the PDZ domain (ΔPDZ), or 4) the entire kinase domain (ΔPK); all these constructs were tagged at the COOH terminus with HA. Cells were transfected using a modified calcium phosphate precipitation protocol (Kunda et al., 2001) and the constructs used at a concentration of 6 μg/ml, unless otherwise stated. Several hours later, the cultures were fixed and processed for immunofluorescence with antibodies against HA or LIMK1. Staining with the LIMK1 antibody used at a dilution of 1:1000 (0.05 μg/ml) allows for a rapid and reliable identification of transfected neurons, because cells expressing normal levels of LIMK1 display very light immunofluorescence, whereas in those overexpressing LIMK1 a high signal was easily detected (Figure 2); by using this criterion, we estimated a transfection efficiency of ∼15-20%. In addition, levels of expression of HA-tagged wt-LIMK1 and its mutants were evaluated at different time points after transfection by quantitative fluorescence. This analysis revealed a two- to threefold increase in LIMK1 fluorescence intensity 12-15 h after transfection; these values increase considerably (more than fourfold) after a 24-h posttransfection interval. Therefore, most of our experiments were performed in cells that survived transfection no more than a day.
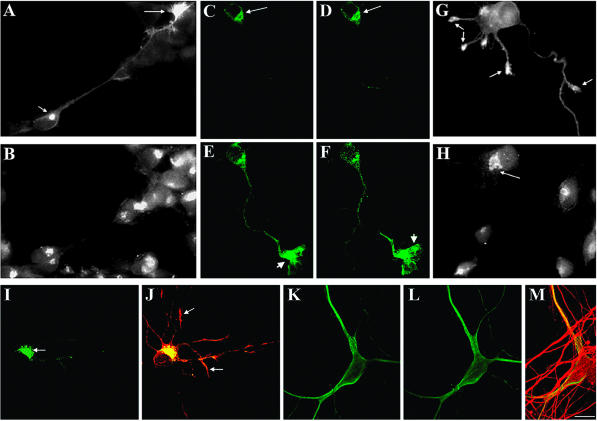
Figure 2.
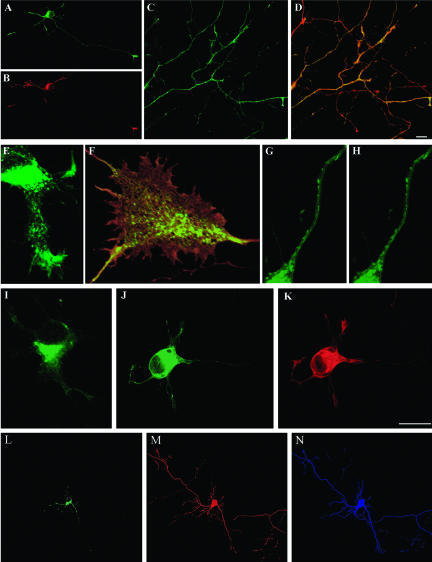
Subcellular distribution of HA-tagged wt-LIMK1 and its mutants. (A and B) Double immunofluorescence micrographs showing the distribution of HA-tagged wt-LIMK1 (A) and mannosidase II (B) in cultured hippocampal pyramidal neurons. Note that HA-tagged wt-LIMK1 localizes to the Golgi apparatus (short arrow) and the growth cone (long arrow). (C-F) Series of confocal images showing another example of the distribution of HA-tagged wt-LIMK1 in the Golgi apparatus (long arrows) and the axonal growth cone (short arrows) of a cultured hippocampal pyramidal neuron. (G and H) Double immunofluorescence micrographs showing the subcellular distribution of HA-tagged Δ-LIM (G) and mannosidase II (H) 12 h after transfection. Note the absence of HA-tagged Δ-LIM immunofluorescence in the region of the Golgi apparatus, but its presence within growth cones (arrows in G). (I) Immunofluorescence micrograph showing the distribution of HA-tagged ΔPDZ-LIMK1 in a cultured hippocampal pyramidal neuron. Note that most of the labeling is found concentrated in the region of the Golgi apparatus (arrows). (J) Red-green overlay showing the distribution of HA-tagged ΔPDZ-LIMK1 (green) and endogenous LIMK1 (red). Note that HA-tagged ΔPDZ-LIMK1 is absent from neuritic tips (arrows). (K and L) Series of confocal images showing the distribution of HA-tagged Δ-LIM. Note the diffuse distribution of the labeling. (M) Red-green overlay showing the distribution of HA-tagged Δ-LIM (green) and of tyrosinated α-tubulin (red). The cell shown in C-F was stained with a rabbit polyclonal antibody against LIMK1 diluted 1/1000. All other transfected cells were stained with a rabbit antibody against HA. Bar, 10 μm.
As shown in Figure 2, HA-tagged full-length LIMK1 (wt-LIMK1) localized in a pattern similar to that of endogenous LIMK1 (Figure 2, A-F). Colocalization with mannosidase II (Figure 2, A and B), or β-COP (our unpublished data), and sensitivity to brefeldin A (our unpublished data) confirmed that the structure labeled within the cell body was the Golgi apparatus. By contrast, deletion of the entire LIM domain (ΔLIM) or of any one of the two tandemly arranged LIM domains (either LIM1 or LIM2) results in the loss of Golgi localization (Figure 2, G and H) and in its association with vesicle-like structures; interestingly, HA-tagged ΔLIM, ΔLIM1, or ΔLIM2 localize to growth cones (Figure 2, G and H). By contrast, deletion of the PDZ domain (ΔPDZ) prevents LIMK1 localization to growth cones without altering its association with the Golgi apparatus (Figure 2, I and J) or vesicles. Deletion of the entire NH2-terminal domain also results in a striking alteration in LIMK1 subcellular distribution; thus, HA-tagged ΔN initially (12-18 h posttransfection) displays a diffuse cytoplasmic distribution, with no evident enrichment in the perinuclear region, the Golgi apparatus, or growth cones (Figure 2, K-M). However, at later posttransfection intervals (20-30 h), most of the labeling was found in the cell nucleus (our unpublished data). Finally, deletion of the entire PK domain does not result in significant alterations of HA-tagged LIMK1 subcellular distribution (our unpublished data), with the exception that it completely abolished its nuclear localization. Together, these observations suggests that LIM domains are required for LIMK1 localization to the Golgi apparatus, whereas a region containing the PDZ domain is required for its localization at growth cones, and its exclusion from the cell nucleus.
LIMK1 Regulates Golgi Morphology and Dynamics
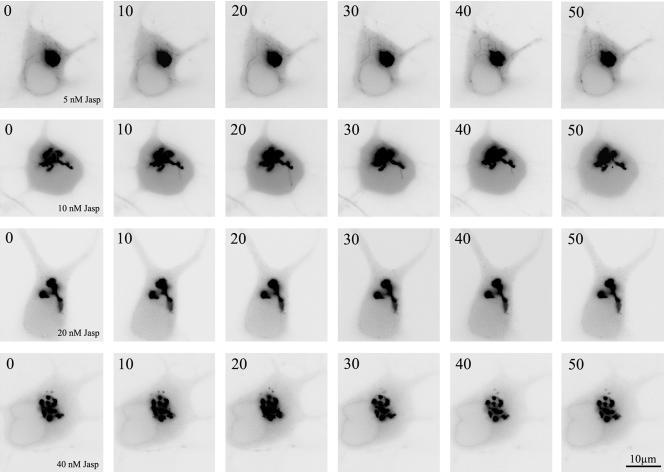
Because a growing body of evidence has recently emerged showing that small Rho-GTPases as well as some of their effector proteins associate with Golgi membranes (Ridley, 2001) it became of considerable interest to analyze the morphology and dynamics of this organelle after expression of wt-LIMK1 or its mutants. To visualize the Golgi apparatus neurons were cotransfected with a cDNA coding for the galactosyl-transferase T2 (Gal-T2, a resident enzyme of the trans-Golgi network; see Giraudo et al., 2001) fused to the NH2-terminal domain of enhanced yellow fluorescent protein (EYFP), and either wt-LIMK1 or full-length LIMK1 carrying a point mutation at the C terminus that abolishes its kinase activity (LIMK1-kd; Sumi et al., 1999), or a mutated form of cofilin that renders it constitutive active (S3A cofilin). Immunofluorescence examination of >20 cultures revealed a high degree of cotransfection (>80%). This analysis showed that while the Golgi apparatus seems to be slightly smaller and more compact in neurons overexpressing wt-LIMK1 than in control ones, its overall morphology does not change considerably localizing either in the perinuclear region or in the initial segment of one primary dendrite (Figure 3, A-C; see also Figure 1, Supplementary Material). By contrast, the Golgi apparatus of neurons overexpressing LIMK1-kd or S3A cofilin display a less compact appearance with long cisternae extending into dendritic processes (Figure 1, Supplementary Material). Time-lapse recordings of living neurons confirmed these observations and revealed additional features. Thus, hippocampal pyramidal neurons cotransfected with GalT2-EYFP (2 μg of DNA) alone or GalT2-EYFP (2 μg of DNA) plus Δ-LIM (4 μg of DNA) revealed that the Golgi apparatus of hippocampal pyramidal neurons is a dynamic structure displaying several long (3-5 μm in length) tubulo-vesicular processes emerging from the main Golgi stacks that extend and retract over short periods of time (Figure 3, D-F). Interestingly, this dynamic behavior was dramatically reduced in neurons cotransfected with GalT2-EYFP (2 μg of DNA) and wt-LIMK1 (4 μg of DNA). Under this condition very few, if any, tubulo-vesicular processes were observed emerging from the Golgi stacks; a small number of vesicle-like structures containing GalT2-EYFP were detected in the vicinity of the Golgi apparatus (Figure 3, G-I). By contrast, overexpression of LIMK1-kd produces a different effect. Thus, time-lapse recordings of neurons overexpressing GalT2-EYFP and LIMK1-kd revealed a dynamic Golgi apparatus displaying long tubulo-vesicular processes (10-15 μm) that persist for longer periods of time than the ones of control cells (Figure 3, J-O). Quantitative analysis of the number, length, and persistence of these Golgi-derived tubulo-vesicular structures confirmed these observations and clearly revealed that overexpression of wt-LIMK1 suppresses this dynamic behavior, whereas that of the kinase-defective form tends to enhance it (Table 1). A similar phenomenon was observed after transfection of S3A cofilin; these neurons display an extended Golgi apparatus with very long tubules (>30 μm), some of which persist for more than a minute (Figure 4). Interestingly, this behavior was suppressed in a dose-dependent manner by Jasplakinolide (Figure 5), a cell-permeable macro-cyclic peptide that inhibits actin turnover (Gallo et al., 2002).
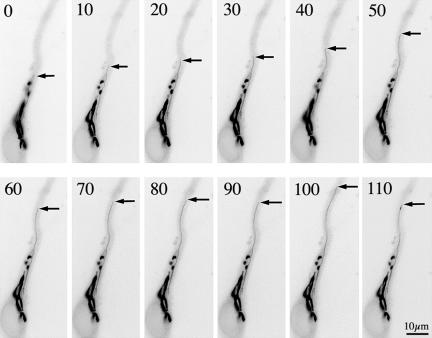
Figure 3.
LIMK1 regulation of Golgi dynamics. (A-C) Fluorescence micrographs showing the distribution of galactosyl-transferase T2-EYFP fusion protein (A) and HA-tagged wt-LIMK1 (B) in a cultured hippocampal pyramidal neuron; the image in C shows the red-green overlay of the images shown in A and B. Note the strong colocalization of both proteins at the Golgi apparatus. (D-F) Sequence of fluorescence images showing the dynamics of the Golgi apparatus from a neuron transfected with galactosyl-transferaseT2-EYFP; the EYFP signal is shown in black. Note the presence of tubulo-vesicular processes emerging from the Golgi stacks. (G-I) A similar sequence to that described previously, but from a cell cotransfected with galactosyl-transferase T2-EYFP and HA-tagged wt-LIMK1. Note the absence of tubulo-vesicular processes emerging from the Golgi stacks and the presence of small vesicles within the cell cytoplasm. (J-O) A sequence of fluorescence images showing the dynamics of the Golgi apparatus from a neuron cotransfected with galactosyl-transferase T2-EYFP and HA-tagged LIMK1-kd. Note the presence of several tubulo-vesicular processes (arrows) emerging from the Golgi stacks that persist for longer periods of time than the ones observed in cells transfected with galactosyl-transferaseT2-EYFP alone; parts of the labeled tubules are out of the plane of focus. For all these experiments cells were visualized 12 h after transfection and each image was taken at 30-s intervals. Bars, 10 μm (A-C) and 2.5 μm (D-O).
Table 1.
Quantification of Golgi-derived tubule formation in neurons overexpressing wt-LIMK1, LIMK1-kd, and Δ-LIM
| Groups | Numbera | Length (μm) | Duration (sec)b |
|---|---|---|---|
| Control | 2.6 ± 0.2 | 4.2 ± 0.4 | 18 ± 6 |
| wt-LIMK1 | 0.02 ± 0.01c | — | — |
| LIMK1-kd | 2.2 ± 0.2 | 8.5 ± 0.6* | 45 ± 8 |
| Δ-LIM | 2.5 ± 0.2 | 3.8 ± 0.6 | 21 ± 4 |
Number of tubules emerging from the Golgi stacks per minute.
Duration represents the period of time elapsed between the formation of a new tubule and its retraction and disappearance into the main Golgi stacks.
Values significantly different from those of control cells or cells contransfected with Gal-T2-EYFP (2 μg DNA) plus Δ-LIM (4 μg DNA). Each value represents the mean ± SEM. Seventy transfected cells were analyzed for each experimental condition. Each value is the mean ± S.E.M.
Figure 4.
Sequence of fluorescence images showing the dynamics of the Golgi apparatus from a neuron cotransfected with galactosyl-transferase T2-EYFP and FLAG-tagged S3A-cofilin. Note the extended morphology of the Golgi apparatus and the presence of a long tubulo-vesicular process (arrows) emerging from the Golgi stack that elongates and persists for more than a minute; other labeled tubules are out of the plane of focus. Images were taken every 10 s. For this experiment, cells were visualized 12 h after transfection.
Figure 5.
Jasp reduces Golgi tubule formation induced by S3A-cofilin expression. A sequence of fluorescence images showing the dynamics of the Golgi apparatus from neurons cotransfected with galactosyl-transferase T2-EYFP plus FLAG-tagged S3A-cofilin and treated for 30-45 min before the beginning of the time-lapse recording with different doses of Jasp. Images were taken every 10 s. Note the disappearance of membrane tubules with increasing doses of Jasp.
Finally, we noticed that the long-term overexpression (>20 h posttransfection) of LIMK1-kd of S3A cofilin produces a fragmentation of the Golgi apparatus (Figure 2, Supplementary Material), suggesting that regulation of actin turnover is also important for maintaining Golgi structure. One prediction of this observation is that overexpression of wt-LIMK1 should delay the occurrence of cytochalasin D-induced Golgi disruption, whereas that of LIMK1-kd or S3a cofilin should accelerate this effect. The results obtained, which are shown in Figure 2 (Supplementary Material) fully support the proposed scenario.
Effects of the Overexpression of LIMK1 or Its Mutants on Neuronal Morphology and Axon Formation
We next examined the consequences of LIMK1 overexpression on neurite extension and axon formation, being particularly interested in determining whether the Golgi localization of LIMK1 was required for any neuritogenic function it may have. First, we analyzed the consequences of LIMK1 overexpression in neurons that have already developed morphological polarity. Analysis of neurons (2-3 days in vitro [DIV]) overexpressing wt-LIMK1 revealed that 12-18 h after transfection >95% of these cells exhibited a high level of morphological differentiation with neurites frequently taking complex pathways. Thus, neurons overexpressing wt-LIMK1 display several neurites of >75 μm in length; usually, one of these neurites, presumably the axon, extended for >400 μm, displaying multiple collateral branches (Figure 6, A and B). High-resolution fluorescence microscopy or confocal microscopy also revealed that axonal shafts display many ectopic cytoplasmic expansions with prominent lamellipodial veils that resemble growth cones (“waves;” Ruthel and Banker, 1998, 1999) and contain accumulations of LIMK1 and F-actin (Figure 6, B-D). An additional and distinctive feature of neurons overexpressing wt-LIMK1 was an increase in F-actin and phospho-cofilin staining in the region of the Golgi apparatus (Figure 6, E-I), as well as the presence of large growth cones containing abundant F-actin in both its central and peripheral regions (Figure 6, J and K), and a significant increase (2- to 3-fold) in phospho-cofilin immunolabeling (Table 2). Neurons overexpressing wt-LIMK1 also were analyzed at longer posttransfection intervals. Thus, 24-30 h after transfection, we detected a significant reduction in the number of collateral branches, and a retraction of growth cones and neuritic processes (Figure 6L); this phenomenon was accompanied by a dramatic increase (more than sixfold) in the phospho-cofilin immunolabeling of neuritic tips. Similar alterations were detected after overexpression of Δ-LIM, which is known for having a three- to fourfold increase in kinase activity (Sumi et al., 1999; Rosso and Caceres, unpublished observations); in these neurons, we initially (10-15 h posttransfection) detected an increase in the number of waves, and the size of growth cones, as well as in the staining for F-actin and phospho-cofilin, which was later (18-24 h posttransfection; Table 2 and Figure 3, Supplementary Material) followed by growth cone collapse and axon retraction. A different picture was observed after transfection with LIMK1-kd (Figure 4, Supplementary Material) or S3A cofilin (our unpublished data). Thus, at all posttransfection intervals (12, 15, and 24 h) the transfected neurons display a single long axon with many collateral branches (Figure 4, Supplementary Material), and several (4-5) much shorter neurites; even though axonal length seems to increase in the transfected cells, their overall morphologies were similar to those of nontransfected neurons. Neurons overexpressing LIMK1-kd or S3A cofilin display small growth cones that stain faintly with rhodamine-phalloidin (Table 2; Figure 4, Supplementary Material) or the phospho-cofilin antibody (Table 2). Interestingly, deletion of the LIM domain in LIMK1-kd seems to enhance its neuritogenic effect, whereas deletion of the PDZ domain has the opposite effect (see below). In addition, expression of the isolated C-terminal domain of LIMK1, either of the wt-form or kd-mutant, had no effect on axon morphology, growth cone size, or actin cytoskeletal organization. Together, these data indicate that the kinase domain requires the N terminus to exert its specific functions in vivo, which involve the targeting of LIMK1 to either the Golgi apparatus or the axonal growth cone.
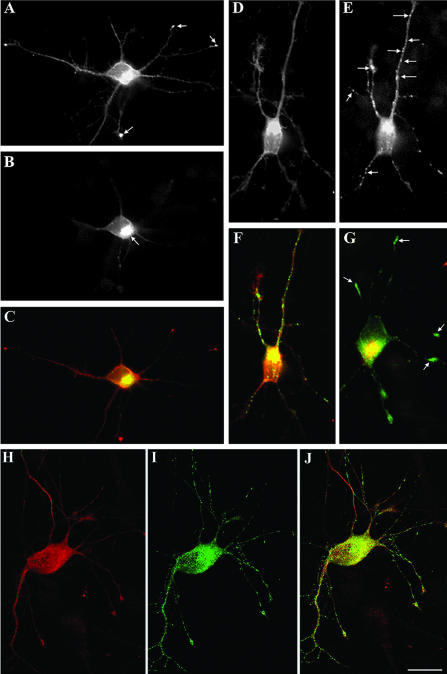
Figure 6.
Effect of the expression of HA-tagged wt-LIMK1 on neuronal morphology. For this experiment, the cells were transfected with 6 μg of HA-tagged wt-LIMK1 and fixed 12 (A-I), 20 (J and K), and 24 (L) h later. The cells were stained with a rabbit polyclonal antibody against HA (green) and either tyrosinated α-tubulin (red in A, C, and D) or rhodamine-phalloidin (red in B, I and L), or p-cofilin (F). (A-D) Image in A shows the general morphology of a neuron overexpressing wt-LIMK1; note the presence of growth cone-like structures along the axon (arrows). The image in B shows accumulations of F-actin at the tip of neuritic branches of the transfected neuron. The image in C shows a high power view of a growth cone located along an axonal process; note the accumulation of LIMK1 (arrow) at the periphery of the growth cone. These accumulations also are observed in growth cones located at the tip of neuritic processes (arrow in D). (E-G) Double immunofluorescence micrographs showing the distribution of HA-tagged wt-LIMK1 (E) and phospho-cofilin (F) in a cultured hippocampal pyramidal neuron 12 h after transfection. Note the dramatic increase in phospho-cofilin fluorescence intensity in the Golgi region of the transfected cell compared with a nontransfected one (arrow in F). (G) Red-green overlay of the images shown in E and F. (H and I) Confocal images showing the distribution of HA-tagged LIMK1 (H) and rhodamine-phalloidin (I) in cultured neurons; note the intense phalloidin staining of the Golgi region (arrow) in the transfected neuron. (J and K) Confocal images showing examples of growth cones 20 h after transfection with wt-LIMK1. These growth cones have abnormal shapes, are several times larger than the ones of nontransfected cells (arrow in L) and stained strongly for F-actin. (L) This image shows an example of a neuritic tip (green-yellow) of a neuron overexpressing wt-LIMK1 24 h after transfection. Note the disappearance of the growth cone and the presence of patches of F-actin along the distal end of the neuritic process. Bars, 15 μm (A and B) and 10 μm (C-L).
Table 2.
Quantification of cofilin, phospho-cofilin, and phalloidin fluorescence intensity in neurons overexpressing wt-LIMK1, LIMK1-kd, and Δ-LIM
| Anti-cofilin (1 μg/ml) | Golgi region | Growth cones |
|---|---|---|
| Non transfected | 1250 ± 65 | 1640 ± 20 |
| wt-LIMK1 | 1225 ± 58 | 1750 ± 56 |
| LIMK1-kd | 1356 ± 46 | 1630 ± 48 |
| Δ-LIM | 1278 ± 84 | 1670 ± 38 |
| Anti-phospho cofilin (0.5 μg/ml) | Golgi region | Growth cones |
| Nontransfected | 458 ± 72 | 680 ± 26 |
| wt-LIMK1 | 1852 ± 64a | 2240 ± 44a |
| Δ-LIM | 450 ± 24 | 2720 ± 28a |
| Anti-phospho cofilin (1 μg/ml) | Golgi region | Growth cones |
| Nontransfected | 932 ± 68 | 1125 ± 55 |
| LIMK1-kd | 184 ± 15a | 272 ± 14a |
| Rhodamine-phalloidin (0.2 μg/ml) | Golgi region | Growth cones |
| Nontransfected | 249 ± 17 | 660 ± 24 |
| wt-LIMK1 | 1246 ± 68a | 1850 ± 48a |
| Δ-LIM | 312 ± 32 | 2412 ± 18a |
| Rhodamine-phalloidin (0.5 μg/ml) | Golgi region | Growth cones |
| Nontransfected | 383 ± 47 | 1256 ± 22 |
| LIMK1-kd | 52 ± 12a | 325 ± 18a |
| S3A-Cofilin | 65 ± 14a | 420 ± 36a |
Values represent the average fluorescent intensity expressed in pixels within a 5-μm area located in the central region of the Golgi apparatus or growth cones. 12-Bit images were used for this quantification. Scale, 0 (black) to 4095 (white). Each value is the mean ± SEM. At least 50 cells were measured for each experimental condition.
Values significantly different from those of nontransfected cells.
As a further evaluation of this idea, quantitative measurements were performed on several neuronal shape parameters. The results obtained confirmed our qualitative observations and clearly revealed that overexpression of wt-LIMK1, LIMK1-kd, or the Δ-LIM and Δ-PDZ mutants have significant and specific consequences on neurite length, branching, and growth cone morphology (Table 3).
Table 3.
Quantification of neuronal shape parameters in neurons overexpressing wt-LIMK1 and their mutants
| Groups | No. of primary neurites/cell | Total axonal length (μm) | No. of collateral branches (axon) | No. of growth cone like structures/axon | Growth cone area (μm2) |
|---|---|---|---|---|---|
| Nontransfected | 5.2 ± 0.4 | 295 ± 22 | 12 ± 2 | 1.2 ± 0.2 | 16 ± 4 |
| wt-LIMK1 | 5.6 ± 0.2 | 425 ± 65* | 26 ± 4* | 5.4 ± 0.6* | 46 ± 7* |
| LIMK1-kd | 5.2 ± 0.4 | 545 ± 44* | 45 ± 8* | 1.2 ± 0.2 | 9 ± 2* |
| Δ-LIM (wt) | 5.8 ± 0.4 | 220 ± 16* | 18 ± 6 | 4.8 ± 0.4* | 68 ± 8* |
| Δ-LIM (kd) | 5.2 ± 0.2 | 636 ± 32* | 56 ± 10* | 1.6 ± 0.4 | 8 ± 2 |
| Δ-PDZ (wt) | 4.8 ± 0.4 | 388 ± 14* | 22 ± 6* | 2.5 ± 0.5 | 22 ± 8 |
| Δ-PDZ (kd) | 5.1 ± 0.7 | 180 ± 26* | 8 ± 4 | 1.0 ± 0.3 | 10 ± 2 |
Each value represents the mean ± SEM. At least 100 cells were analyzed for each experimental condition. Two DIV hippocampal pyramidal cultures were transfected with each of the constructs, fixed 18 h later, and processed for immunofluorescence with a rabbit antibody against HA and a mAb against the class III β-tubulin isotype. Control nontransfected cells had similar neuronal shape parameters to those of cells transfected with GFP alone (6 μg DNA).
Values significantly different from those of nontransfected neurons.
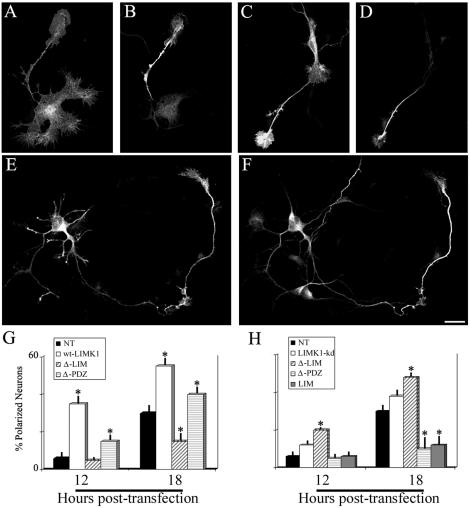
We then analyzed the involvement of LIMK1 in neurite initiation and axon formation. Cells were transfected 2 h after plating and examined 12 and 18 h later. Any neurite of at least 40-50 μm longer than the other minor neurites of the same cell and displaying a proximo-distal accumulation of Tau1 immunofluorescence (Shi et al., 2002) was considered to be an axon; the length and number of axons and minor processes also was quantitated. The results obtained showed that overexpression of LIMK1 accelerates the appearance of axon-like neurites (Figure 7, A-D); thus, 12 h after transfection we observed that many transfected neurons have already extended a Tau1 immunoreactive neurite, whereas the remaining processes still display large lamellipodia (Figure 7, A and B; Dehmelt et al., 2003); in other cases, the Tau1 immunoreactive neurite originates from the rim of a lamellipodial extension (Figure 7, C and D). In any case, by 20 h postplating, a considerable percentage (>50%) of neurons overexpressing wt-LIMK1 display an axon-like neurite of >70 μm with a distal accumulation of Tau1 immunofluorescence. Many of these axon-like neurites end in a prominent growth cone with a large lamellipodium. The effect of wt-LIMK1 was dependent on its targeting to the Golgi apparatus, because it was not observed in neurons expressing the Δ-LIM mutant and to a much lesser extent on its presence in growth cones (Figure 7, G and H). An increase in the percentage of neurons displaying axon-like neurite also was detected after transfection of LIMK1-kd; as in the case of older neurons, the stimulatory effect of LIMK1-kd on axon formation was not observed after deletion of the PDZ domain. As a matter of fact, expression of this LIMK1 mutant inhibits axon outgrowth; conversely, deletion of its LIM domain enhanced axon formation (Figure 7, G and H). Consistent with this, overexpression of the LIM domain alone also produced a similar inhibitory effect. One prediction of these observations (Birkenfeld et al., 2001) is that expression of the LIM domain might inhibit axon outgrowth by preventing the binding of endogenous LIMK1 to the Golgi apparatus. We test this possibility by examining the distribution of endogenous LIMK1 in neurons overexpressing the LIM domain. In these cells, endogenous LIMK1 and p-cofilin staining completely disappears from the Golgi region, but not from growth cones (our unpublished data). Finally, it is worth mentioning that none of the constructs tested affect either the time course of appearance, number, or length of minor processes.
Figure 7.
LIMK1 promotes axon formation. (A-F) For this experiment, the cells were transfected 2 h after plating with 6 μg of HA-tagged wt-LIMK1 and fixed 12 (A-D), or 18 (E-F) h later. The cells were stained with a rabbit polyclonal antibody against HA (A, C, and E) and the mAb Tau1 (B, D, and F). Note that 12 h after transfection the neurons have already extended a long neurite that stains intensely with the Tau1 mAb; note also the large size of the axonal growth cones of the transfected neurons. Bar, 10 μm. (G and H) Percentage of neurons displaying an axon-like neurite after transfection with wt-LIMK1 or its mutants. Cultures were transfected 2 h after plating and fixed 12 or 18 h later. Each value represents the mean ± SEM. At least 50 cells were measured for each transfection, as described in MATERIALS AND METHODS. Values significantly different from those of control nontransfected (NT) neurons are indicated by asterisk (*).
LIMK1 Promotes the Growth Cone Accumulation of Par3/Par6, Tyrosine Kinase Receptors, and Phospho-AKT during Axon Formation
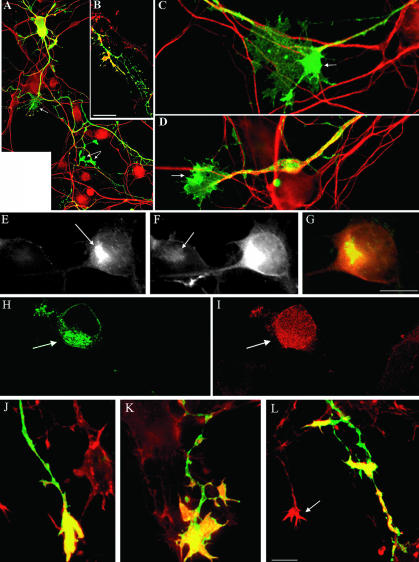
It has recently been shown that as cultured hippocampal neurons change from stage II to stage III by rapidly extending one of its neurites to form an axon, Par3/Par6 proteins and activated phosphatidylinositol 3-kinase (PI 3-kinase) accumulate at its tip (Shi et al., 2002). Because LIMK1 promotes axon outgrowth, we sought to determine whether it might regulate Par3/Par6 localization. As reported previously by Shi et al., (2002), we found that in control neurons Par3 and Par 6 antibodies (used at a dilution of 1/50) stain intensely the cell body and much faintly, but with high selectivity, the axonal growth cone of stage III hippocampal pyramidal neurons; when used at a dilution of 1/200 Par3/Par6 immunolabeling was basically found in the cell body. Neurons overexpressing wt-LIMK1 display a considerable and selective increase in Par3/Par 6 immunolabeling (antibody dilution 1/200) at the axonal growth cone (Figure 8, A-E). High-power views of the transfected neurons revealed that Par3/Par6 immunolabeling also was associated with LIMK1 in vesicle-like structures located along the main axonal shafts and in waves (Figure 8, F and G). Previous studies have shown that ectopic expression of Par3 leaves neurons with no axons specified (Shi et al., 2002). However, coexpression of wt-LIMK1 and Par3 results in the extension of a single, highly branched axon that contains large accumulations of Par3 at growth cones and branch points (Figure 8, H and K). The effects of wt-LIMK1 were abolished by deletion of the LIM domain or by coexpression of S3A cofilin (Table 4). In accordance with this, expression of LIMK1-kd or of the LIM domain alone considerable reduced Par3/Par6 accumulation at the axonal growth cone (Table 4).
Figure 8.
LIMK1 promotes the accumulation of Par3/Par6 at axonal growth cones and waves. (A and B) Confocal micrographs showing the distribution of HA-tagged LIMK1 (red) and Par3 (green) in cultured hippocampal pyramidal neurons. Cells were transfected with wt-LIMK1 2 h after plating, fixed 18 h later and stained with a mouse antibody against HA and a rabbit polyclonal antibody against Par3 (diluted 1/200). (C and D) Confocal micrographs showing the distribution of HA-tagged LIMK1 (green), F-actin (red), and Par6 (blue) in cultured hippocampal pyramidal neurons. Cells were transfected as described previously. Note that only the transfected neurons display intense Par3/Par6 immunofluorescence at the axonal growth cone (arrows). (F and G) Confocal micrographs showing colocalization and accumulation of HA-tagged LIMK1 (red) and Par3 (green) within an axonal shaft and a wave (arrows). (H and I) Confocal micrographs showing colocalization and accumulation of HA-tagged LIMK1 and transfected Par3 within the terminal axonal arbor of a cultured hippocampal pyramidal neuron. (J) Red-green overlay of the images shown previously. (K) Confocal image showing the axonal shaft of a neuron transfected with Par3 alone. For these experiments cultures were transfected 1 d after plating and fixed 18 h later; the Par3 antibody was used at a dilution of 1/500.
Table 4.
Quantification of Par3 immunofluorescence in neurons overexpressing LIMK1 or its mutants
| Groups | Antibody dilution | Cell body | Distal axon | Growth cone |
|---|---|---|---|---|
| NT | 1/200 | 1320 ± 46 | 125 ± 18a | 181 ± 23 |
| NT | 1/50 | 1854 ± 66 | 263 ± 52 | 560 ± 74 |
| WT | 1/200 | 1050 ± 28 | 340 ± 25a | 650 ± 64a |
| KD | 1/200 | 1710 ± 92 | 60 ± 32 | 110 ± 15a |
| KD | 1/50 | 2050 ± 124 | 140 ± 14a | 315 ± 17a |
| Δ-LIMb | 1/200 | 2023 ± 87 | 136 ± 22 | 176 ± 30 |
| WT + S3A | 1/200 | 1415 ± 39 | 158 ± 16a | 240 ± 34 |
| LIMc | 1/50 | 1950 ± 65 | 134 ± 16 | 205 ± 15a |
| Δ-PDZd | 1/200 | 1268 ± 18 | 186 ± 14 | 480 ± 26a |
Values represent the average fluorescent intensity expressed in pixels within a 5 μm area located in the cell body, distal axonal segment, and growth cones. 12-Bit images were used for this quantification. Scale, 0 (black) to 4095 (white). Each value is the mean ± S.E.M. Cultures were fixed and processed for immunofluorescence 18 h after transfection. At least 50 cells were analyzed for each experimental condition.
Values significantly different from those of the corresponding control groups.
This group corresponds to cells transfected with the Δ-LIM mutant of wt-LIMK1.
This group corresponds to cell transfected with the LIM domain alone.
This group corresponds cells transfected with the Δ-PDZ mutant of wt-LIMK1.
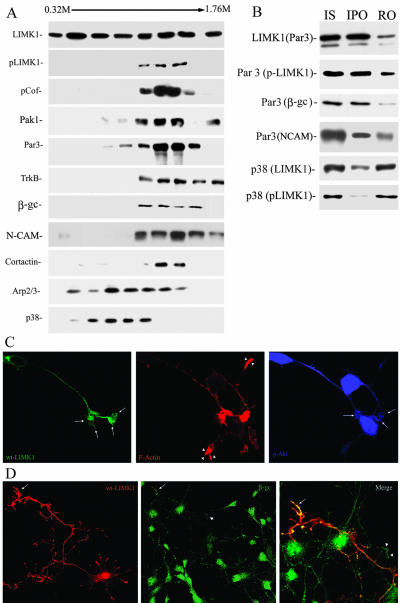
To further characterize the relationship between LIMK1-Par3/Par6, microsomal fractions from 1-d-old rat cerebral cortex were fractionated by ultracentrifugation across a continuous sucrose density gradient extending from 0.32 to 1.76 M sucrose (Peretti et al., 2000), and the distribution of LIMK1, p-LIMK1, cofilin, and p-cofilin, as well as of several membrane and actin-associated proteins analyzed by WB with specific antibodies. This analysis revealed that although LIMK1 has a widespread distribution throughout the gradient, its phosphorylated form is highly enriched in fractions extending from 1.12 to 1.52 M sucrose; interestingly, p-cofilin, PAK1, the actin regulatory protein cortactin, and Par3/Par6 displayed a striking codistribution with p-LIMK1 (Figure 9A). Besides, all of them codistributed with proteins directly implicated in axon extension, such as β-gc (Mascotti et al., 1997; Pfenninger et al., 2003), TrkB (Pfenninger et al., 2003), and the cell adhesion molecule L1 (Peretti et al., 2000). By contrast, p-LIMK1 only partially colocalizes with synaptophysin (Figure 9A), synapsin (our unpublished data), and the actin regulatory protein Arp 2 (Figure 9A). Together, these observations raise the possibility of activated LIMK1 interacting with a type of nonsynaptic vesicle containing Par3/Par6 as membrane-associated proteins and growth factor receptor tyrosine kinases, such as β-gc or TrkB, as integral membrane proteins. Therefore, to directly test this possibility, immunoisolation of organelles (Peretti et al., 2000) from the microsomal fractions was performed with the anti-Par3 antibody; the remaining organelles were recovered by pelleting from the supernatant fraction. Figure 9B shows that with this method, Par3-containing organelles were collected. More importantly, in this immunoisolated organelle fraction (Figure 9B) p-LIMK1, p-cofilin, and β-gc were quantitatively recovered. By contrast, synaptophysin and synapsin (our unpublished data) were not detectable in this fraction. They were quantitatively recovered in the remaining organelle fraction; very little if any p-LIMK1 staining was detected in this fraction. The p38 antibody also was used to immunoisolate fractions enriched in synaptophysin (0.4-0.92 M sucrose) and probed with antibodies against LIMK1 and p-LIMK1. LIMK1 was partially recovered in the p38-immunoprecipitated organelles; however, this fraction only contains a small amount of activated LIMK1 (Figure 9B).
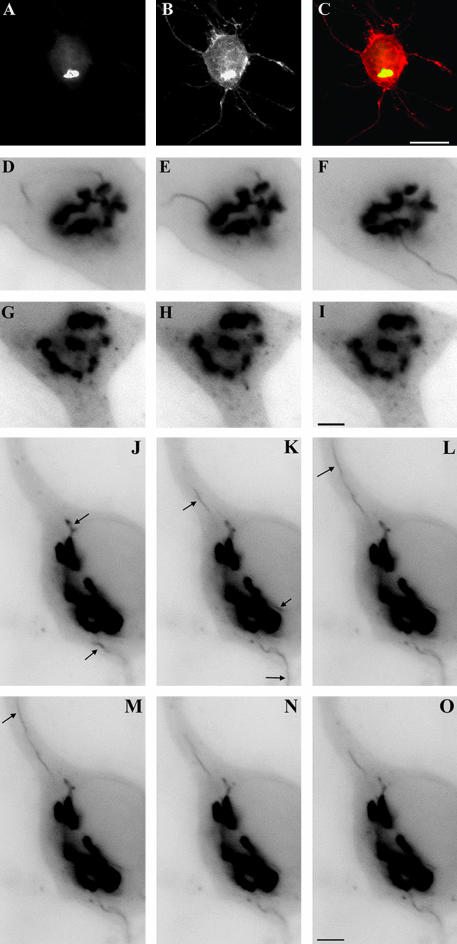
Figure 9.
Activated LIMK1 associates with Par3/Par6 containing vesicles. (A) Microsome fraction from developing rat cerebral cortex was fractionated by centrifugation across a continuous SDG and the same amount of protein (20 μg) from each fraction was applied to SDS-PAGE, stained with Coomassie Blue, or transferred to polyvinylidene difluoride membranes and analyzed by Western blot with antibodies against LIMK1, p-LIMK1, p-cofilin, Par3, TrkB, β-gc, NCAM, cortactin, Arp2, and synaptophysin (p38). Note the strong colocalization of p-LIMK1, p-cofilin, PAK1, Par3, TrkB, β-gc, and NCAM. (B) LIMK1 (Par3) Immunoisolation of Par3-containing organelles with anti-LIMK1 (dilution 1/50) from a microsomal fraction extending from 0.92 to 1.6 M sucrose; Par3 (LIMK1/p-LIMK1/β-gc/NCAM) Immunoisolation of p-LIMK1-, β-gc-, and NCAM-containing organelles with anti-Par3 (dilution 1/50). Note that all proteins are quantitatively recovered in the immunoprecipitated organelle fraction (IPO). Immunoisolation of LIMK1 and p-LIMK1-containing organelles with anti-p38 (dilution 1/50). Note that only a small amount of p-LIMK1 is associated with the immunoprecipitated organelle fraction. (C) Confocal images showing the distribution of HA-tagged wt-LIMK1 (green), F-actin (red), and p-Akt in a transfected hippocampal pyramidal neuron. Note the accumulation and intense labeling of p-Akt in the axonal growth cone (arrows) of the transfected neuron when compared with equivalent ones of nontransfected cells (arrowheads). (D) Confocal images showing the distribution of HA-tagged wt-LIMK1 (red) and β-gc (green) in a transfected hippocampal pyramidal neuron. Note the accumulation and intense labeling of β-gc in the axonal growth cone (arrows) of the transfected neuron when compared with nontransfected cells (arrowheads). Bar, 10 μm.
We next examined whether the accumulation of Par3/Par6 observed in neurons overexpressing wt-LIMK1 was paralleled by increased activity of PI 3-kinase. For such a purpose, we monitored the activation of endogenous Akt, a major downstream effector of PI 3-kinase, by using an antibody specific for activated Akt that is phosphorylated at Ser 476 (Shi et al., 2003). A considerable increase in activated Akt was detected within axonal growth cones of neurons over-expressing wt-LIMK1 (Figure 9C). Interestingly, an increase in β-gc immunostaining also was detected within axonal tips of neurons transfected with wt-LIMK1 (Figure 9D). As in the case of Par3/Par6, these effects were prevented by deletion of the LIM domain, or cotransfection with S3A cofilin (our unpublished data).
NCAM also colocalizes with p-LIMK1 across the microsomal sucrose gradient. Therefore, in an additional set of experiments we evaluated the effect of LIMK1 on NCAM trafficking. As reported previously (Silverman et al., 2001; Sampo et al., 2003) overexpression of NCAM-GFP results in the labeling of the Golgi complex, vesicular structures located in all processes, and growth cones (Figure 10, A and B). Coexpression of wt-LIMK1 did not alter this pattern but considerable increases NCAM-GFP fluorescent intensity within neurites and growth cones (Figure 10, C and D). High-resolution confocal microscopy confirmed these observations by revealing an increased number of NCAM-GFP-containing vesicles within neuritic processes (Figure 10E), and waves (Figure 10F) located along axonal shafts. Numerous NCAM-GFP containing vesicles also were detected in the neurites of cells coexpressing the Δ-PDZ mutant of LIMK1 (Figure 10, G and H). A completely different picture was detected in cells coexpressing LIMK1-kd (Figure 10, I-K). We also observed that coexpression of S3A-cofilin reduces the stimulatory effect of wt-LIMK1 on GFP-NCAM distribution (Figure 10, L and M); in these cells most of the labeling was found accumulated within the cell body and particularly the Golgi region (Table 5).
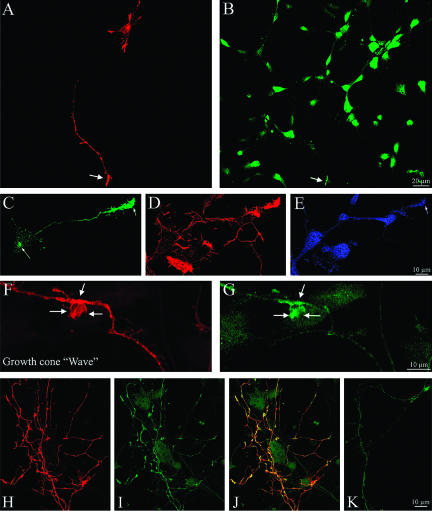
Figure 10.
LIMK1 regulation of the Golgi-derived export of NCAM-containing vesicles. (A and B) Double fluorescence micrographs showing the distribution of NCAM-GFP (A) and endogenous LIMK1 (B) in a cultured hippocampal pyramidal neuron. For this experiment, the cells were fixed 12 h after transfection and stained with the rabbit polyclonal antibody against LIMK1 used at a dilution of 1/1000. Note that the most intense NCAMGFP labeling is found within the cell body and at neuritic tips; light labeling is detected within minor neurites and along the axon (arrows) (C) Fluorescence micrograph showing the presence of NCAM-GFP along the axon and growth cones of a neuron cotransfected with HA-tagged wt-LIMK1. Note the intense NCAM-GFP labeling of the axon and growth cones. (D) Corresponding overlay image showing the distribution of NCAM-GFP (green) and HA-tagged wt-LIMK1 (red). The cells also were analyzed 12 h after transfection. (E) High-power confocal micrograph showing the presence of numerous NCAMGFP-containing vesicles within a neurite of a cell cotransfected with HA-tagged wt-LIMK1. (F) Abundant NCAM-GFP-containing vesicles (green) also are detected within growth cone-like structures (waves) located along the axonal shaft of a neuron cotransfected with HA-tagged wt-LIMK1 (red). (G and H) Series of confocal images showing the presence of numerous NCAM-GFP-containing vesicles in the axon of a neuron coexpressing HA-tagged Δ-PDZ LIMK1. (I and J) Immunofluorescence micrographs showing the distribution of NCAM-GFP in neurons cotransfected with HA-tagged LIMK1-kd. The cell shown in I was transfected with 2 μg/ml NCAM-GFP, whereas that shown in J with 4 μg of NCAM-GFP. Note that in both cases most of the labeling is localized to the cell body and absent from neurites and their tips. (K) Fluorescence micrograph showing the corresponding labeling of HA-tagged LIMK1-kd for the cell shown in J. Note that labeling is not only found in the cell body but also at neuritic tips. (L and N) Confocal images showing the distribution of NCAM-GFP (L) in a hippocampal neuron cotransfected with FLAG-tagged S3A cofilin (M) and HA-tagged wt-LIMK1 (N). Note that in this triple-transfected cell most of the NCAM-GFP labeling is found within the cell body. No labeling is detected in the middle or distal parts of neuritic processes, including growth cones. For all these experiments each LIMK1 cDNA was used at a concentration of 2 μg/ml. Bars, 10 μm (A-D, L and M) and 10 μm (E-K).
Table 5.
Quantification of synaptophysin-GFP and NCAM-GFP immunofluorescence in neurons overexpressing wt-LIMK1 and its mutants
| Group | Golgi region | Proximal neurites | Distal neurites | Growth cones |
|---|---|---|---|---|
| P38-GFP | ||||
| Control | 1450 ± 40 | 542 ± 38 | 640 ± 56 | 850 ± 65 |
| wt-LIMK1 | 1750 ± 20 | 160 ± 22a | 156 ± 24a | 132 ± 16a |
| LIMK1-kd | 1500 ± 35 | 656 ± 34 | 720 ± 62 | 1350 ± 60a |
| Δ-LIM | 1400 ± 30 | 528 ± 44 | 615 ± 27 | 920 ± 25 |
| NCAM-GFP | ||||
| Control | 1500 ± 65 | 364 ± 41 | 488 ± 46 | 1250 ± 80 |
| wt-LIMK1 | 1300 ± 45 | 725 ± 74a | 742 ± 38a | 1880 ± 56a |
| LIMK1-kd | 1780 ± 20 | 156 ± 32a | 131 ± 43a | 148 ± 28a |
| Δ-PDZ | 1430 ± 30 | 652 ± 64a | 728 ± 40a | 1640 ± 60a |
Control cells were transfected with 2 μg/ml of synaptophysin-GFP or NCAM-GFP cDNAs. Values represent the average fluorescent intensity expressed in pixels within a 5-μm area located in the central region of the Golgi apparatus or growth cones. 12-Bit images were used for this quantification. Scale, 0 (black) to 4095 (white). Values represent synaptophysin-GFP or NCAM-GFP fluorescence intensities within a 30 μm stretch located in the proximal or distal segments of individual neurites. Each value is the mean ± SEM. At least 50 cells were analyzed for each experimental condition.
Values significantly different from those of the corresponding control groups.
Finally, we also evaluated the effect of LIMK1 on synaptophysin trafficking. Previous studies have established that in cultured neurons synaptophysin-GFP localizes to vesicles or tubulo-vesicular structures inside neurites and that it becomes enriched at neuritic tips (Nakata et al., 1998; Kaether et al., 2000). In accordance with this, transfection of 2-3 DIV hippocampal pyramidal neurons with synaptophysin-GFP alone revealed the presence of synaptophysin-containing vesicles in the cell body within and around the Golgi area, as well as throughout neurites (our unpublished data). By contrast, when the neurons were additionally transfected with wt-LIMK1, synaptophysin-GFP-containing vesicles completely disappear from the neurites after 12-24 h; in these neurons, most of the synaptophysin-GFP labeling was found concentrated in the Golgi apparatus (Figure 11, A-C). On the other hand, cotransfection with LIMK1-kd (Figure 11, D-G) or S3A-cofilin (Figure 11, H-J) results in the appearance of numerous synaptophysin-GFP-containing vesicles within the cell body and along neurites; in most cases, these vesicles accumulate at neuritic tips. Quantification of synaptophysin-GFP fluorescence intensity confirmed these observations (Table 5), strongly suggesting a connection between changes in LIMK1 levels and/or activity and the Golgi-derived export of this type of synaptic membrane protein.
Figure 11.
LIMK1 regulation of the Golgi-derived export of synaptophysin-containing vesicles. (A and B) Double fluorescence micrographs showing the effect of the overexpression of wt-LIMK1 (A) on the distribution of synaptophysin-GFP fusion protein (B). The cells were fixed 12 h after transfection and stained with a rabbit polyclonal antibody against HA (dilution 1/1000). Note that most of the synaptophysin-GFP signal is localized to the region of the Golgi apparatus. (C) Red-green overlay of the images shown in A and B. (D and E) Double fluorescence micrographs showing the effect of the overexpression of HA-tagged LIMK1-kd (D) on the distribution of synaptophysin-GFP fusion protein (E). Note the presence of numerous synaptophysin-GFP vesicle-like structures along neurites (arrows). (F) Red-green overlay of the images shown in D and E. (G) A red-green overlay of another neuron cotransfected with LIMK1-kd (red) and synaptophysin-GFP (green). In this example, the HA-antibody was used at a dilution of 1/2500. Note the presence of intense synaptophysin-GFP labeling at neuritic tips (arrows). (H and I) Double fluorescence micrographs showing the effect of the overexpression of FLAG-tagged S3A-cofilin (H) on the distribution of synaptophysin-GFP (I). Note the presence of numerous synaptophysin-GFP vesicle-like structures along neurites. (J) Red-green overlay of the images shown in H and I. For these experiments each cDNAs was used at a concentration of 2 μg/ml. Bar, 7 μm.
DISCUSSION
LIMK1 Functioning in the Golgi Apparatus
The present results provide new evidence concerning the subcellular distribution and function of LIMK1 in developing neurons. Thus, one striking finding of this study was the discovery that LIMK1 is present in the Golgi apparatus. The LIM domain of LIMK1, recognized as a motif involved in protein-protein interactions (Dawid et al., 1998), seems to be responsible for this localization. On the other hand, the PDZ domain, which is found in various submembranous proteins and also functions as a protein binding domain (Ponting et al., 1997), is not required for the association of LIMK1 with the Golgi; however, a region containing this domain is essentially required for its localization to growth cones (this study) and for nuclear exclusion (Yang and Mizuno, 1999). All these observations are fully consistent with the idea that in LIM-containing proteins each domain functions as a module mediating a specific interaction.
The association of LIMK1 with Golgi membranes is in line with a series of recent experimental data showing that Rho family members are not only localized to vesicular compartments but also that play important roles in the trafficking of endocytic and exocytic vesicles (Ridley, 2001). This include the early demonstration that Cdc42 localizes primarily to the Golgi apparatus (Erikson et al., 1996; Michaelson et al., 2000) and that in epithelial cells, it is involved in the exit and sorting of proteins from the trans-Golgi network (Kroschewski et al., 1999; Cohen et al., 2001; Musch et al., 2001), as well as the more recent evidence showing that Citron-N, a Rho-binding protein, ROCK-II, and Profilin IIa localize to the Golgi apparatus (Camera et al., 2003). In addition, PAK4, a novel effector for Cdc42, not only localizes to the brefeldin A-sensitive compartment of the Golgi (Abo et al., 1998) but also interacts specifically with LIMK1 stimulating its ability to phosphorylate cofilin (Dan et al., 2001). Thus, various elements required for activation of LIMK1 and for the coupling of Rho-GTPase signaling to actin cystokeletal dynamics are present in the neuronal Golgi apparatus, raising the possibility of LIMK1 acting as a regulator of Golgi organization and/or trafficking.
In favor of this view, we have now presented evidence suggesting that a functional LIMK1 is present in the Golgi apparatus. Western blot analysis clearly revealed the presence of cofilin and phosphorylated-cofilin in Golgi membranes. Consistent with this, overexpression of wt-LIMK1 significantly increases the fluorescent signals generated by the phospho-cofilin antibody and rhodamine-phalloidin in the Golgi region. On the other hand, overexpression of LIMK1-kd, which is likely to act as a dominant-negative mutant for endogenous LIMK1 and LIMK2 (Sumi et al., 1999), has the opposite effect. Changes in the levels/activity of LIMK1 or cofilin also were paralleled by alterations in Golgi morphology. In this regard, the long-term decrease in cofilin phosphorylation, elicited by expressing LIMK1-kd or S3A cofilin for >20 h, result in Golgi fragmentation. Together, these observations show that regulation of actin turnover is critically involved in maintaining neuronal Golgi architecture. In agreement with this, overexpression of LIMK1-kd or S3A cofilin dramatically accelerates cytochalasin D-induced Golgi fragmentation, whereas that of wt-LIMK1 has the opposite effect.
Our results also suggest a more subtle involvement of LIMK1 and actin dynamics in the regulation of Golgi morphology and function. Thus, overexpression of wt-LIMK1 dramatically reduces the formation of GalT2-EYFP-containing tubules that emerge from the Golgi stacks, whereas that of LIMK1-kd or S3A-cofilin has a rather opposite effect. Several reasons suggest that the type of tubule that we have evaluated belongs to those emanating from the trans-Golgi network: 1) Gal-T2, the protein that was fused with EYFP, is a resident enzyme of the trans-Golgi network (Giraudo et al., 2001); 2) the Gal-T2 EYFP-containing tubules seem not to connect adjacent cisternae, and/or to label structures resembling the endoplasmic reticulum; and 3) the length, rate of elongation/retraction, and number of Gal-T2 EYFP-containing tubules emanating from the Golgi were very similar to those reported for nonneuronal cells transfected with the ts045 vesicular stomatitis virus G protein fused to GFP and visualized at the permissive temperature of 32°C (Hirschberg et al., 1998). Our observations showing that modifications of LIMK1 levels and/or activity alter trans-Golgi network-tubule formation are consistent with studies suggesting that actin-based cytoskeletal components could be involved in their formation. For example, Hirschberg et al. (1998) reported that Golgi-derived tubules formed in cytochalasin B-treated cells are significantly longer than in untreated ones; interestingly, expression of LIMK1-kd, which reduces cofilin phosphorylation and F-actin at the Golgi region, produces a similar effect. LIMK1 and cofilin also regulate the trafficking of Golgi-derived vesicles, a phenomenon that is likely to be important for explaining its participation in axon formation (see below).
LIMK1 Functioning during Axonal Development
The present observations confirm and extend previous studies showing the involvement of LIMK1 in neuronal development. For example, the alterations in growth cone morphology, cofilin phosphorylation, and actin organization here reported are fully consistent with those described in LIMK1 KO mice (Meng et al., 2002). Thus, the size of growth cones was greatly reduced in LIMK1-deficient neurons, as in the case of neurons expressing LIMK1-kd. By contrast, over-expression of wt-LIMK1 or its Δ-LIM mutant results in a complete different phenotype, with growth cones several times larger than control ones, and containing high levels of phosphorylated cofilin and F-actin. An inhibition of neurite elongation and growth cone motility has been observed in chick DRG cells overexpressing LIMK1 (Endo et al., 2003); in hippocampal pyramidal neurons, the long-term overexpression of wt-LIMK1 or its Δ-LIM mutant also results in growth cone collapse and neurite retraction (this study). The high levels of expression of exogenous LIMK1 detected in hippocampal pyramidal neurons after a 24 h posttransfection interval or the one reported for DRG neurons (>10-fold; Endo et al., 2003), and the dramatic increase in cofilin phosphorylation precludes a simple explanation for this phenomenon. However, one obvious possibility is that the large accumulation of F-actin present in the growth cones of transfected neurons affects process extension by preventing microtubule advance and decreasing the membrane vesicle content of neuritic tips. Alternatively, a decrease in actin turnover may allow myosin to generate sufficient contractile force to produce axon retraction (Gallo et al., 2002).
On the other hand, the axogenic effect of LIMK1-kd, ADF-cofilin, or S3A-cofilin in cortical (Meberg and Bamburg, 2000), DRG (Endo et al., 2003), or hippocampal pyramidal (this study) neurons fits well with current models suggesting that axon extension and the differentiation of a neuritic growth cone to become the axon growth cone requires an increase in actin turnover, with cofilin acting as a positive regulator and LIMK1 as a negative one (Bradke and Dotti, 1999; Kunda et al., 2001; Endo et al., 2003; Sarmiere and Bamburg, 2004).
However, it is likely that this view represents an oversimplification of LIMK1 participation in axon formation and that a full understanding of its functioning cannot rely exclusively on its presumptive inactivation at the growth cone. By contrast, our results suggest that active LIMK1 is required for axon formation; several aspects support this idea. First, a high fluorescent signal for activated LIMK1 was detected at the Golgi region, in axonal growth cones, and at the peripheral, actin-rich rim of the large growth cone of prospective axons. Second, an increase in the number of growth cone-like structures located along axons was observed in neurons overexpressing wt-LIMK1. This is an interesting observation, because previous studies have established that similar structures, nicknamed waves, exhibiting high lamellipodial activity and containing a marked concentration of F-actin, GAP-43, and ezrin (Ruthel and Banker, 1998), are directly related with bursts of axonal growth (Ruthel and Banker, 1999). LIMK1-induced waves also contain accumulations of molecules directly implicated in axon formation such as Par3/Par6 and NCAM (Lemmon et al., 1992; Doherty et al., 1995; Peretti et al., 2001; Shi et al., 2003). Third, and perhaps most importantly, overexpression of wt-LIMK1 accelerates axonal development, a phenomenon paralleled by an enhanced accumulation of Par3/Par6, β-gc, and p-Akt at the axonal growth cone. This function of LIMK1 involves cofilin phosphorylation because it is prevented by coexpression of S3A-cofilin and is not observed in neurons overexpressing LIMK1-kd.
In neurons, a signal transduction pathway linking growth factor tyrosine kinase receptors (e.g., IGF1 receptors), activation of PI 3-kinase, Par proteins, and atypical protein kinase C is essentially required for the establishment of neuronal polarity (Shi et al., 2003). In nonneuronal cells, Rho-GTPases and the cytoskeleton regulate the distribution of Par proteins, and positive feedback loops linking PI(3,4,5)P3 with Rho GTPases and/or polymerized actin have been shown to be important for neutrophil polarity during chemotaxis (Wang et al., 2002; Weiner et al., 2002). Thus, our results are consistent with activated LIMK1 being part of a positive feedback loop linking Rho GTPases and the regulation of actin dynamics with the asymmetric distribution of Par proteins in polarizing neurons. LIMK1-induced accumulation of p-Akt also may be important for regulating ADF-cofilin phosphocycling (Gungabissoon and Bamburg 2003); thus, it has been proposed that downstream of p-Akt is a specific phosphatase, possibly of the slingshot family, that can activate ADF-cofilin through dephosphorylation.
Inhibition of axon formation and reduced accumulation of Par3/Par6 at growth cones also were observed after expression of the Δ-LIM mutant of wt-LIMK1 or of the LIM domain alone; interestingly, inhibition of neurite extension by over-expression of LIM domains also has been detected in NGF-treated PC12 cells (Birkenfeld et al., 2001). Because the LIM domain is responsible for LIMK1 localization to the Golgi apparatus, these observations raise the interesting possibility of LIMK1 regulating the trafficking of Golgi-derived vesicles capable of interacting with Par3-Par6 proteins. In favor of this, analysis of microsomal fractions obtained by SDG centrifugation revealed a striking colocalization of p-LIMK1 with p-cofilin, Pak1, Par3/Par6, β-gc, and NCAM. Immunoisolation experiments allow us to isolate Par3-containing vesicles and directly demonstrate that they contain activated LIMK1, β-gc, NCAM, and phospho-cofilin.
The fact that other types of vesicles, like those containing synaptophysin, have low amounts of p-LIMK1 and p-cofilin, suggest that activation-deactivation of LIMK1 may differentially regulate the export and/or trafficking of Golgi-derived vesicles. In accordance with this, overexpression of LIMK1-wt dramatically reduces synaptophysin-GFP labeling from neuritic shafts, with most of the fluorescence signal found within the Golgi region; by contrast, it increases Par3 and GFP-NCAM labeling within neurites, growth cones, and waves. A complete different picture was observed after expression of LIMK1-kd. All these observations are in line with recent studies suggesting distinct roles for actin in the formation of different classes of Golgi-derived vesicles (Musch et al., 1997; Heimann et al., 1999; Fucini et al., 2000; Rozelle et al., 2000). The recent demonstration of β/γ-actin on vesicles budding from the Golgi by immunoelectron microscopy (Valderrama et al., 2000) and the existence of different actin-binding proteins, including multiple non-muscle myosins, on distinct populations of Golgi-derived vesicles (Heimann et al., 1999; this study) supports this proposal. Other proteins, such as dynamin also might provide a dynamic component to this actin-membrane matrix, thereby aiding in vesicle scission. Interestingly, a GTPase-deficient dynamin2-GFP prevents vesiculation and induces the formation of long membrane tubules terminated with a prominent bulb (Orth and McNiven, 2003) that resemble those observed after expression of LIMK1-kd or S3A cofilin. It will be now important to precisely define the role of LIMK1 and cofilin in membrane protrusion, tubulation, and scission.
Supplementary Material
Acknowledgments
We are greatly indebted to Carlos Dotti (European Molecular Biology Laboratory), Claudio Giraudo and Hugo Maccioni (CIQUIBIC-Consejo Nacional de Investigaciones CientíficasyTécnicas), Enrique Rodriguez-Boulan (Cornell Medical College), Gery Kreitzer (Cornell Medical College), and Tony Pawson (Mt. Sinai Hospital, Toronto, ON Canada) for providing some of the constructs used in this study. We specially thank Dr. James Bamburg for the critical reading of the article, helpful discussions, and for providing a phospho-cofilin rabbit polyclonal antibody. This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (PICT-PIP4906), Fondo Nacional Ciencia y Tecnia (PICT 05-01697), Beca Ministerio Salud, and a Fogarty International Research Collaborative Award grant (to A.F. and A.C.). It also was supported by a Howard Hughes Medical Institute grant to A.C. awarded under the International Research Scholars Program.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-05-0328. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0328.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Abo, A., Qu, J., Cammarano, M., Dan, C., Fritsch, A., Baud, A., Belisle, B., and Minden, A. (1998). PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 22, 6527-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa, H., et al. (2001). Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat. Neurosci. 4, 367-373. [DOI] [PubMed] [Google Scholar]
- Arber, S., Barbayannis, F., Hanser, H., Schneider, C., Stanyon, C., Bernard, O., and Caroni, P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805-812. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185-230. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R., and Bray, D. (1987). Distribution and cellular localization of actin-depolymerizing factor. J. Cell Biol. 105, 2817-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluji, U., Lichtenberger, L., Galaburda, A., and Korenberg, J.R. (1999). Bridging cognition, the brain and molecular genetics: evidence from Williams syndrome. Trends Neurosci. 22, 187-207. [DOI] [PubMed] [Google Scholar]
- Birkenfeld, J., Betz, H., and Roth, D. (2001). Inhibition of neurite extension by overexpression of individual domains of LIMK1. J. Neurochem. 78, 924-927. [DOI] [PubMed] [Google Scholar]
- Bishop, A., and Hall, A. (2000). Rho GTPases and their effector proteins. Biochem. J. 248, 241-255. [PMC free article] [PubMed] [Google Scholar]
- Bradke, F., and Dotti, C.G. (1999). The role of local actin instability in axon formation. Science 283, 1931-1934. [DOI] [PubMed] [Google Scholar]
- Camera, P., Santos Da Silva, J., Griffiths, G., Giuffrida, M., Ferrara, L., Schubert, V., Imarisio, S., Silengo, L., Dotti, C., and Di Cunto, F. (2003). Citron-N is a neuronal Rho-associated protein involved in Golgi organization through actin cytoskeletal organization. Nat. Cell Biol. 5, 1071-1078. [DOI] [PubMed] [Google Scholar]
- Cohen, D., Musch, A., and Rodriguez-Boulan, E. (2001). Selective control of basolateral membrane protein polarity by Cdc42. Traffic 2, 556-564. [DOI] [PubMed] [Google Scholar]
- Dan, C., Kelly, A., Bernard, O., and Minden, A.,. (2001). Cytoskeletal changes regulated by the PAK4 serine-threonine kinase are mediated by LIMK1 and cofilin. J. Biol. Chem. 276, 32115-32121. [DOI] [PubMed] [Google Scholar]
- Daniels, R. H., Hall, P., and Bokoch, G. (1998). Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBOJ. 17, 754-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid, I., Bren, J., and Toyama, R. (1998). LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14, 156-162. [DOI] [PubMed] [Google Scholar]
- Dehmelt, L., Smart, F., Ozer, R., and Halpain, S. (2003). The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J. Neurosci. 23, 9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, P., Williams, E., Walsh, F. (1995). A soluble chimeric form of the L1 glycoprotein stimulates neurite outgrowth. Neuron. 14, 57-66. [DOI] [PubMed] [Google Scholar]
- Endo, M., Ohashi, K., Sasaki, Y., Goshima, Y., Niwa, R., Uemura, T., and Mizuno, K. (2003). Control of growth cone motility and morphology by LIM kinase and slingshot via phosphorylation and dephosphorylation of cofilin. J. Neurosci. 23, 2527-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, J., Zhang, C., Kahn, R., Evans, T., and Cerione, P. (1996). Mammalian Cdc42 is a brefeldin A-sensitive component of the Golgi apparatus. J. Biol. Chem. 271, 26850-26854. [DOI] [PubMed] [Google Scholar]
- Fucini, R., Navarrete, A., Vadakkan, C., Lacomis, L., Erdjument-Bromage, H., Tempts, P., and M. Stamnes, M. (2000) Activated ADP-ribosylation factor assembles distinct pools of actin in Golgi membranes. J. Biol. Chem. 275, 18824-18829. [DOI] [PubMed] [Google Scholar]
- Gallo, G., Yee, H., and Letourneau, P.C. (2002). Actin turnover is required to prevent retraction driven by endogenous actomyosin contractility. J. Cell Biol. 158, 1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo, C., Daniotti, J., and Maccioni, H. (2001). Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 98, 1625-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungabissoon, R., and Bamburg, J. (2003). Regulation of growth cone actin dynamics by ADF/Cofilin. J. Histochem. Cytochem. 51, 411-420. [DOI] [PubMed] [Google Scholar]
- Heimann, K., Percival, J., Weiberger, R., Gunning, P., and Stow. J. (1999) Specific isoforms of actin-binding protein on distinct populations of Golgi-derived vesicles. J. Biol. Chem. 274, 10743-10750. [DOI] [PubMed] [Google Scholar]
- Hirschberg, K., Miller, C., Ellenberg, J., Presley, J., Siggia, E., Phair, R., and Lippincott-Schwartz, J. (1998). Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J. Cell Biol. 143, 1485-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaether, C., Skebel, P., and Dotti, C.G. (2000). Axonal membrane proteins are transported in distinct carriers: a two-color video microscopy study in cultured hippocampal neurons. Mol. Biol. Cell 11, 1213-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khun, T., Meberg, P., Brown, M., Bernstein, B., Minamide, L., Jensen, J., Okada, K., Soda, E., and Bamburg, J.R. (2000). Regulating actin dynamics in neuronal growth cones by ADF/cofilin and Rho family GTPases. J. Neurobiol. 44, 126-144. [PubMed] [Google Scholar]
- Kroschewski, R., Hall, A., and Mellman, I. (1999). Cdc42 controls secretion and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1, 8-13. [DOI] [PubMed] [Google Scholar]
- Kunda, P., Paglini, P., Kosik, K., Quiroga, S., and Cáceres, A. (2001). Evidence for the involvement of Tiam1 in axon formation. J. Neurosci. 21, 2361-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, V., Burden, S., Payne, H., Elslie, G., and Havin, M. (1992). Neurite growth on different substrates: permissive versus instructive influences and the role of adhesive strength. J. Neurosci. 12, 818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascotti, F., Cáceres, A., Pfenninger, K., and Quiroga, S. (1997). Expression and distribution of IGF-1 receptors containing a b-subunit variant (b-gc) in developing neurons. J. Neurosci. 15, 1447-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg, P., and Bamburg, J.R. (2000). Increase in neurite outgrowth mediated by overexpression of actin depolymerizing factor. J. Neurosci. 20, 2459-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg, P., Ono, S., Minamide, L., Takahashi, M., and Bamburg, J.R. (1998). Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell. Motil. Cytoskeleton 39, 172-190. [DOI] [PubMed] [Google Scholar]
- Meng, Y., Zhang, Y., Tregoubov, V., Janus, C., Cruz, L., MacDonald, J., Wang, J., Falls, D., and Jia, Z. (2002). Abnormal spine morphology and enhanced LTP in LIMK1 knockout mice. Neuron 35, 121-133. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., Silletti, J., Murphy G., D'Eustachio, P., Rush, M., and M. Phillips, M. (2000) Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musch, A., Cohen, D., Kreitzer, G., and Rodriguez-Boulan, E. (2001). Cdc42 regulates the exit of apical and basolateral proteins from the trans-Golgi network. EMBO J. 20, 2171-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musch, A., Cohen, D., and Rodriguez-Boulan, E. (1997). Myosin II is involved in the production of constitutive transport vesicles from the trans-Golgi network. J. Cell Biol. 138, 291-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata, T., and N. Hirokawa, N. (1987). Cytoskeletal reorganization of human platelets after stimulation revealed by the quick-freeze deep-etch technique. J. Cell Biol. 106, 1771-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata, T., Terada, S., and Hirokawa, N. (1998). Visualization of the dynamics of synaptic vesicle and plasma membrane proteins in living axons. J. Cell Biol. 140, 659-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, J., and McNiven, M. (2003). Dynamin at the actin-membrane interface. Curr. Op. Cell Biol. 15, 31-39. [DOI] [PubMed] [Google Scholar]
- Paglini, G., Kunda, P., Quiroga, S., Kosik, K., and Cáceres, A. (1998). Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J. Cell Biol. 143, 443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglini, G., Peris, L., Quiroga, S., and Cáceres, A. (2001). The Cdk5/p35 kinase associates with the Golgi apparatus and regulates membrane traffic. EMBO Rep. 2, 1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti, D., Peris, L., Rosso, S., Quiroga, S., and Caceres, A. (2001). Evidence for the involvement of KIF4 in the anterogrado transport of L1-containing vesicles. J. Cell Biol. 149, 141-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger, K.H., Ellis, L., Johnson, M., Friedman, L., and Somlo, S. (1983). Nerve growth cones isolated from fetal rat brain: subcellular fractionation and characterization. Cell 35, 573-584. [DOI] [PubMed] [Google Scholar]
- Pfenninger, K.H., Laurino, L., Peretti, D., Wang, X., Rosso, S., Morfini, G., Caceres, A., and Quiroga, S. (2003). Regulation of membrane expansion at the nerve growth cone. J. Cell Sci. 16, 1209-1217. [DOI] [PubMed] [Google Scholar]
- Ponting, C., Phillips, C., Davies, K., and Blake, D. (1997). PDZ domains: targeting signaling molecules to sub-membranous sites. Bioessays 19, 469-479. [DOI] [PubMed] [Google Scholar]
- Proschel, C., Blouin, M-I. Gutowski, N., Ludwig, R., and Noble, M. (1995). LIMK1 is predominantly expressed in neural tissue and phosphorylates serine threonine and tyrosine residues in vitro. Oncogene 11, 1271-1281. [PubMed] [Google Scholar]
- Ridley, A. (2001). Rho proteins: linking signaling with membrane trafficking. Traffic 2, 303-310. [DOI] [PubMed] [Google Scholar]
- Rozelle, A.L., Machesky, L., Yamamoto, Y., Driessens, M., Insall, R., Roth, M., Luby-Phelps, K., Marriot, G., Hall, A., and Yin, E. (2000). Phosphatidylinositol 4, 5 bisphosphate induces actin-based movements of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311-320. [DOI] [PubMed] [Google Scholar]
- Ruthel, G., and Banker, G. (1998). Actin-dependent anterograde movements of growth-cone like structures along growing hippocampal axons: a novel form of axonal transport? Cell Motil. Cytoskeleton 40, 160-173. [DOI] [PubMed] [Google Scholar]
- Ruthel, G., and Banker, G. (1999). Role of moving growth cone-like waves structures in the outgrowth of cultured hippocampal axons and dendrites. J. Neurobiol. 39, 97-106. [DOI] [PubMed] [Google Scholar]
- Sampo, B., Kaech, S., Kunz, S., and Banker, G. (2003). Two distinct mechanisms target membrane proteins to the axonal surface. Neuron 37, 611-624. [DOI] [PubMed] [Google Scholar]
- Sarmiere, P., and Bamburg, J. (2004). regulation of the neuronal actin cytoskeleton by ADF/Cofilin. J. Neurobiol 58, 103-117. [DOI] [PubMed] [Google Scholar]
- Shi, S.H., Jan, L.Y., and Jan, Y.N. (2003). Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and Pi3-kinase activity. Cell 112, 63-75. [DOI] [PubMed] [Google Scholar]
- Silverman, M.A., Kaech, S., Jareb, M., Burack, M., Vogt, L., Sonderegger, P., and Banker, G. (2001). Sorting and directed transport of membrane proteins during development of hippocampal neurons in culture. Proc. Natl. Acad. Sci. USA. 98, 7051-7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi, T., Matsumoto, K., Takai, Y., and Nakamura, T. (1999). Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147, 1519-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama, F., Luna, A., Babia, T., Martinez-Menarquez, J., Ballesta, J., Barth, H., Chaponnier, C., Renau-Piqueras, J., and Egea, G. (2000). The Golgi-associated COP1-coated buds and vesicles contain β/γ-actin. Proc. Natl. Acad. Sci. USA 97, 1560-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Herzmark, P., Weiner, O.D., Srinivasan, S., Servant, G., and Bourne, H.R. (2002). Lipids products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 4, 509-512. [DOI] [PubMed] [Google Scholar]
- Weiner, O.D., Neilsen, P., Prestwich, G., Kirschner, M., Cantley, L., and Bourne, H.R. (2002). A PtdInsP3- and Rho GTPase-mediated positive feedback loop regulates neutophil plarity. Nat. Cell Biol. 4, 509-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E., and Mizuno, K. (1998). Cofilin phosphorylation by LIMK1 and its role in Rac-mediated actin reorganization. Nature 393, 809-812. [DOI] [PubMed] [Google Scholar]
- Yang, N., and K. Mizuno, K. (1999). Nuclear export of LIM-kinase 1 mediated by two leucine-rich nuclear-export signals within the PDZ domain. Biochem. J. 338, 793-796. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.