Abstract
Hox transcript antisense intergenic RNA (HOTAIR) is a long non-coding RNA, its overexpression has been documented in various human solid tumor and it can be considered as a potential cancer biomarker. However, little is known about the role of HOTAIR in acute myeloid leukemia (AML). In this study, We evaluated HOTAIR expression in bone marrow of de novo AML patients, AML-CR patients and normal controls by real-time quantitative reverse transcription-PCR (qRT-PCR), then we inhibited hotair expression of two cell lines by siRNA and evaluated their proliferation by CCK, finally we analyzed its relationship with the clinicopathological parameters of AML. We found that HOTAIR is significantly upregulated in de novo AML patients compared with those of AML-CR patients and normal controls; the reduction of HOTAIR by small interfering RNA (siRNA) repressed the proliferation of HL-60 and K562; the higher expression level of HOTAIR in AML patients was significantly correlated with NCCN high risk group. In conclusion, our study indicated that hotair is highly expressed in AML patients, and hotair expression significantly correlates with clinicopathological prognostic stratification in AML.
Keywords: Acute myeloid leukemia, long non-coding RNA, HOTAIR, prognosis
Introduction
Acute myeloid leukemia (AML) is the most common acute leukemia in adults with an incidence of 3-4 per 100,000 persons per year. AML is a genetically heterogeneous disorder characterized by the somatically acquired genetic changes in hematopoietic progenitor cells altering normal mechanisms of self-renewal, proliferation, and differentiation. Greater insight into the genetic background of the disease fostered the extension of disease classification and pretreatment risk-categorization by gene mutations. In order to improve outcome in AML, multiple studies aimed at genetic characterization of AML have been performed in the hopes of furthering our understanding of AML pathogenesis and identifying new therapeutic approaches [1,2].
Long non-coding RNAs (lncRNAs) are a heterogeneous class of RNAs that are generally defined as non-protein-coding transcripts longer than 200 nucleotides. lncRNA which was considered as only transcriptional “noise” in the past decades can participate in various critical biological processes, such as chromatin remodeling, gene transcription, and protein transport and trafficking [3]. Recently, more and more studies have shown that lncRNAs are deregulated in a wide variety of cancers [4,5].
Hox transcript antisense intergenic RNA (HOTAIR) is a lncRNA with 2158 nucleotides in length which is expressed from HOXC locus on 12 chromosome [6]. It interacts with polycomb repressive complex 2 (PRC2) and causes targeting it to the HOXD locus, the transcriptional silencing of a40-kb region of the HOXD locus which encodes several transcription factors remodeled gene expression pattern of the cells [7,8]. HOTAIR can also interact with a second histone modification complex, the LSD1/CoREST/REST complex, which coordinates the targeting of PRC2 and LSD1 to chromatin for coupled histone H3K27 methylation and K4 demethylation [9].
Recently, HOTAIR has been determined to be a negative prognostic indicator in various solid-tumor patients [10-12]. Nevertheless, little is known about the impact of HOTAIR on AML. To understand the role of HOTAIR in AML, we investigated the expression level of HOTAIR in AML and analyzed its relationship to clinical pathological features.
Materials and methods
Patients and samples
A total of 34 AML cases were enrolled in our study including 21 de novo AML patients and 13 cases who had achieved complete remission (CR). 21 de novo AML patients were diagnosed in the Hematology Department of General Hospital Tianjin Medical University between February 2014 and November 2014 according to the “2008 WHO adult acute myeloid leukemia (non acute pro-myelocytic leukemia) diagnosis guidelines” and “2008 WHO adult acute pro-myelocytic leukemia diagnosis guidelines”. 13 cases achieved complement remission (CR) were enrolled as AML-CR group. 16 iron-deficiency anemia (IDA) patients diagnosed according to the international criteria of IDA in our department were enrolled as control group. We collected 2 ml EDTA anticoagulant fresh bone marrow samples from each patient and BM sample of de novo AML patients were collected before intervention. This study was approved by the Ethics Committee of Tianjin Medical University. Informed written consent was obtained from all patients in accordance with the Declaration of Helsinki.
Quantitative real-time PCR
Total RNA from human bone marrow was extracted using the TRIzol reagent (Invitrogen), and 1 µg of RNA was reverse-transcribed using the TIANScript RT Kit (TIANGEN, Beijing, China). The sequences of primers specific for HOTAIR (forward, 5’-GGTAGAAAAAGCAACCACGAAGC-3’; reverse, 5’-ACATAAACCTCTGTCTGTGAGTGCC-3’, 191-bpPCR product) were synthesized by GENEWIZ (Suzhou, China). Quantitative real-time PCR was performed using the BIO-RAD iQ5 Real-Time System (BIO-RAD, Hercules, CA, USA), and SYBR Green (TIANGEN) was used as a double strand DNA-specific dye. qRT-PCR cycling program: 95°C for 15 min, followed by 40 cycles at 95°C for 10 s and 62.4°C for 30 s. GAPDH was used as a housekeeping gene for standardizing the expression of targeted mRNA. After normalization of the data according to the expression of GAPDH mRNA, the levels of HOTAIR were calculated using the 2-ΔΔCt method ((Ct, hotair-Ct, GAPDH) sample-(Ct, hotair -Ct, GAPDH) control).
Cell lines and cell cultures
Human leukemia cell lines HL60 and K562 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium (Solarbio, Beijing, china) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (Solarbio, Beijing, China) in humidified air with 5% CO2 at 37°C.
SiRNA transfection
Three individual small interfering RNA (siRNAs) and scrambled negative control siRNA (si-NC) were purchased from GenePharma (GenePharma, Shanghai, China). The target sequences for HOTAIR siRNAs were as follows: (si-HOTAIR1, sense: 5’-GAACGGGAGUACAGAGAGAUU-3’ antisense: 3’-UUCUUGCCCUCAUGUCUCUCU-5’, si-HOTAIR2, sense: 5’-CCACAUGAACGCCCAGAGAUU-3’ antisense: 3’-UUGGUGUACUUGCGGGUCUCU-5’ andsi-HOTAIR3, sense: 5’-UAACAAGACCAGAGAGCUGUU-3’ antisense: 3’-UUAUUGUUCUGGUCUCUCGAC-5’ HL60 and K562 cells (2 × 105/well) were plated in 24-well plates overnight separately. Cells were then transfected with 20pM si-NC or 20pM siRNA against HOTAIR (siHOTAIR 1 siHOTAIR 2 and siHOTAIR 3) for 48 hours using Lipofectamine 2000 transfection reagent (invitrogen, CA, USA) according to the manufacturer’s protocol. Following the transfections, HL60 and K562 cells were then harvested for RT-PCR.
Cell proliferation assays
Cell proliferation was evaluated using the Transdetect Cell Counting Kit (Transgen, Beijing, China). Cells (2 × 103 cells/well) were seeded into 96-well flat-bottomed plates in 100 μl of complete medium. The cells were incubated overnight and were then transfected with siNC or siHOTAIR for 24, 48 and 72 h. CCK solution (10 μl) was added to each well, and the cells were incubated for an additional 2 h. Absorbance was measured at 450 nm using a microplate reader. Three independent experiments were performed in triplicate.
Statistical analysis
Mann-Whitney U test were conducted to analyze data using GraphPad Prism 5 Software. A P value of <0.05 was considered statistically significant.
Results
Clinical data of patients
There were 21 cases of de novo AML patients, male 11 cases, female 10 cases, mean age 50.61 ± 16.04 years old. Among them, 2 cases of acute myeloid leukemia differentiation type (M2), 7 cases of acute promyelocytic leukemia (M3), 7 cases of acute myelomonocytic leukemia (M4), 3 cases of acute monocytic leukemia (M5), and 2 cases of acute red leukemia (M6). According to the 2014 American National Comprehensive Cancer Network (NCCN) guidelines based on cytogenetic and molecular characteristics, all the patients were divided into low risk group, medial risk group and high risk group, including 15 cases of patients with low or medial risk group, 6 cases with high risk group. There were 13 cases of AML-CR patients, male 5 cases, female 8 cases, mean age 43.6 ± 16.5 years old. They included 3 cases of AML-M2, 8 cases of AML-M3 and 2 cases of AML-M4.
HOTAIR was upregu lated in de novo AML patients
The level of HOTAIR was detected in 21de novo AMLsamples, 13 AML-CR samples and 16 control samples by qRT-PCR, and normalized to GAPDH. HOTAIR expression was significantly up-regulated in de novo AML compared with AML-CR patients and controls. However, there were no significant differences between AML-CR patients and controls (Figure 1).
Figure 1.
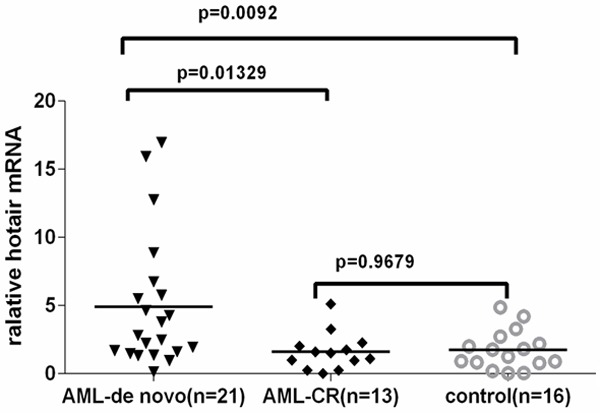
HOTAIR expression was significantly higher in AML-de novo patients (n=21) than in AML patients (n=13) and controls (n=16). Relative HOTAIR expression was determined using qRT-PCR with GAPDH as an internal control. Data are expressed as mean ± SD.
Correlations between elevated HOTAIR and clinicopathological characteristics
In order to examine the clinical importance of the HOTAIR overexpression, the correlation between clinicopathological parameters of de novo AML samples and level of HOTAIR expression was investigated. Analyses showed that the elevated HOTAIR level had significantly correlation with NCCN high risk group. However, there were not any relationships between HOTAIR and patients’ gender or leukemia subtype (Table 1).
Table 1.
Relationship between HOTAIR expression levels and clinicopathological parameters of de novo AMLsamples
| Clinicopathologic features | Number | Relative expression of HOTTIP (Mean ± SD) | P-value |
|---|---|---|---|
| Gender | 0.3091 | ||
| Male | 11 | 5.693 ± 5.005 | |
| Female | 10 | 4.064 ± 4.087 | |
| Subtype | 0.8149 | ||
| AML-M2 | 2 | 7.047 ± 8.082 | |
| AML-M3 | 7 | 5.020 ± 5.588 | |
| AML-M4 | 7 | 3.858 ± 3.023 | |
| AML-M5 | 3 | 7.176 ± 7.802 | |
| AML-M6 | 2 | 2.746 ± 1.482 | |
| NCCN groups | 0.0233* | ||
| Low and medium risk | 15 | 2.875 ± 1.966 | |
| High risk | 6 | 10.02 ± 6.235 |
P<0.05.
Manipulation of HOTAIR levels in leukemia cells
To investigate the functional effects of HOTAIR in leukemia cells, three individual HOTAIR siRNAs were transfected into HL-60 and K562 cells. qRT-PCR analysis of HOTAIR levels was performed 48 h post-transfection. When compared with control cells (si-NC), HOTAIR expression was knocked down by 77.84% in K562 and 71.8% in HL-60 by si-HOTAIR3 (Figure 2A). Thus, si-HOTAIR3, the most effective siRNA was used in subsequent experiments.
Figure 2.
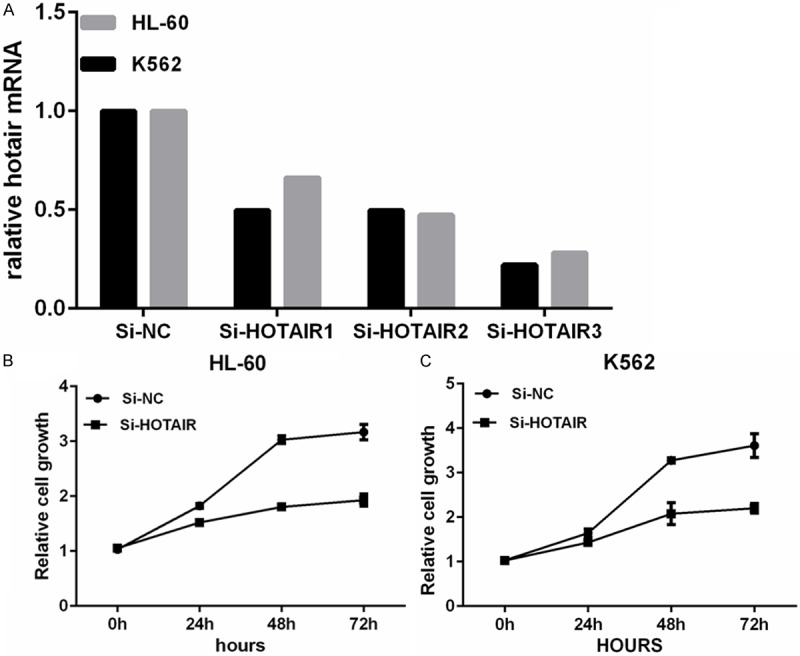
A. Knockdown of HOTAIR inhibits the cell proliferation of leukemia cells. CELLS were transfected with HOTAIR-specific siRNA and negative control siRNA (siNC), and knockdown efficiency was determined by qRT-PCR analysis. B. Knockdown of HOTAIR decreases cell proliferation in HL-60 cells. The proliferation of leukemia cells transfected with siHOTAIR and negative control siRNA (siNC) was determined using the CCK assay. Bars indicate mean ± SD of three independent experiments performed in triplicate. *P<0.05 vs. siNC. C. Knockdown of HOTAIR decreases cell proliferation in K562 cells. The proliferation of leukemia cells transfected with siHOTAIR and negative control siRNA (siNC) was determined using the CCK assay. Bars indicate mean ± SD of three independent experiments performed in triplicate. *P<0.05 vs. siNC.
HOTAIR knockdown decreases cell proliferation in leukemia cells
To determine the role of HOTAIR in leukemia cell growth, siHOTAIR-transfected cells were used in the CCK assay. siRNA-mediated knockdown of HOTAIR decreased cell proliferation by 39.07% and 39.0% at 72 h post-transfection in HL-60 and K562 cells respectively (Figure 2B, 2C). This finding indicates that HOTAIR is involved in the proliferation of leukemia cells.
Discussion
So far more than 10,000 lncRNAs have been identified in the human genome [3] and several lncRNAs have been associated with hematopoietic cancers [13]. HOTAIRM1 (HOX antisense intergenic RNA myeloid 1) plays a role in the myelopoiesis through modulation of gene expression in the HOXA cluster, Knockdown of HOTAIRM1 quantitatively blunted RA-induced expression of HOXA1 and HOXA4 during the myeloid differentiation of NB4 cells, and selectively attenuated induction of transcripts for the myeloid differentiation genes CD11b and CD18 [14]. In addition, the level of ANRIL was increased in many AML and ALL patients [15], and the abundance of MEG3 was decreased in myeloid leukemia [16] but BIC was increased in B cell lymphoma [17].
Until now, many studies have showed that increased expression of HOTAIR is associated with malignant progression and poor survival in various solid cancer types. Hence, HOTAIR may be considered as a potential target for diagnosis and treatment of various cancer types.
A study in non-small cell lung cancer (NSCLC) indicates that HOTAIR is significantly up-regulated in NSCLC tissues, and regulates NSCLC cell invasion and metastasis, partially via the down-regulation of HOXA5 [18]. Jung et al [19] indicated that the expression level of HOTAIR in cervical cancer tissues was higher than that in corresponding non-cancerous tissues. High HOTAIR expression correlated with lymph node metastasis, and reduced overall survival. A multivariate analysis showed that HOTAIR was a prognostic factor for predicting cervical cancer recurrence; knockdown of HOTAIR reduced cell proliferation, migration, and invasion in cervical cancer cell lines. Furthermore some studies indicated that epigenetic reprogramming as a result of changes in lncRNA abundance and PcG complex recruitment could be a general property of malignant cells [12,20]. Because PRC2 and the HOX cluster are essential factors during blood cell development [21,22], HOTAIR may also function in malignant hematopoiesis, although there are still no reports regarding the abundance of lncRNAs as a prognostic marker of leukemia. So we did preliminary research on the role of hot air in AML.
The results of our study indicated that the expression of HOTAIR was upregulated in de novo AML patients compared with normal controls and AML-CR patients. Moreover, HOTAIR expression was found to be significantly higher in NCCN high risk group. These findings indicate that HOTAIR may play a direct role in the modulation of AML progression and may be useful as a novel prognostic marker for AML.
To further assess the role of HOTAIR in AML, we investigated the effects of loss of function of HOTAIR on leukemia cell proliferation. We demonstrated that SiRNi-mediated suppression of HOTAIR in HL-60 and K562 cells led to a significant inhibition of cell proliferation, hence HOTAIR knockdown can inhibit the leukemia cells proliferation in vitro; thus, HOTAIR represents a new prognosis marker and a promising target for AML treatment. Our results are consistent with previous research about the effect of hot air on other cancers.
However, it should be noted that the sample size of this study is not very big, thus each leukemia subtype has few cases, so we cannot thoroughly compare whether there is any difference in hot air level between them. Moreover, although several lncRNAs have been identified the association with hematopoietic cancers, the mechanism of the effect of these lncRNA on hematopoietic malignances is not very clear. Our study also failed to answer this question and we will precede thorough research on it in the future.
In summary the expression of HOTAIR was upregulated in de novo AML patients: and highly expressed HOTAIR is associated with a poor clinicopathological prognostic stratification; HOTAIR knockdown can inhibit the leukemia cells proliferation in vitro but its exact function and mechanism remains to be further studied.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81170472, 81370607, 81400088 and 81400085), Tianjin Municipal Natural Science Foundation (12JCZDJC21500), Health Industry Research and Special Projects (201202017), Tianjin Science and Technology support key project plan (20140109), Health Industry Research and Special Projects (201202017), Tianjin Cancer Research of Major Projects (12ZCDZSY17900 and 12ZCDZSY18000) and Science Foundation of The Tianjin Education Commission (20140118).
Disclosure of conflict of interest
None.
References
- 1.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlenk RF. Post-remission therapy for acute myeloid leukemia. Haematologica. 2014;99:1663–1670. doi: 10.3324/haematol.2014.114611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao W, Chan JY, Wong TS. Long non-coding RNA deregulation in tongue squamous cell carcinoma. Biomed Res Int. 2014;2014:405860. doi: 10.1155/2014/405860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hämmerle M, Tornillo L, Heim MH, Diederichs S, Cillo C, Terracciano LM. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–23. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo CJ, Kingston RE. HOTAIR lifts noncoding RNAs to new levels. Cell. 2007;129:1257–9. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Wang Y, Yu J, Dong R, Qiu H. A high level of circulating HOTAIR is associated with progression and poor prognosis of cervical cancer. Tumour Biol. 2015;36:1661–5. doi: 10.1007/s13277-014-2765-4. [DOI] [PubMed] [Google Scholar]
- 11.Emadi-Andani E, Nikpour P, Emadi-Baygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135. doi: 10.4103/2277-9175.133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–25. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis. Sci Signal. 2013;6:re5. doi: 10.1126/scisignal.2004099. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad S U A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, Georgiou I, Bourantas KL. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34:148–53. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–30. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 21.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000–8. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 22.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]


