Abstract
In the event of a nuclear accident or radiological terrorist attack, there will be a pressing need for biodosimetry to triage a large, potentially exposed population and to assign individuals to appropriate treatment. Exposures from fallout are likely, resulting in protracted dose delivery that would, in turn, impact the extent of injury. Biodosimetry approaches that can distinguish such low-dose-rate (LDR) exposures from acute exposures have not yet been developed. In this study, we used the C57BL/6 mouse model in an initial investigation of the impact of low-dose-rate delivery on the transcriptomic response in blood. While a large number of the same genes responded to LDR and acute radiation exposures, for many genes the magnitude of response was lower after LDR exposures. Some genes, however, were differentially expressed (P < 0.001, false discovery rate < 5%) in mice exposed to LDR compared with mice exposed to acute radiation. We identified a set of 164 genes that correctly classified 97% of the samples in this experiment as exposed to acute or LDR radiation using a support vector machine algorithm. Gene expression is a promising approach to radiation biodosimetry, enhanced greatly by this first demonstration of its potential for distinguishing between acute and LDR exposures. Further development of this aspect of radiation biodosimetry, either as part of a complete gene expression biodosimetry test or as an adjunct to other methods, could provide vital triage information in a mass radiological casualty event.
INTRODUCTION
Ongoing concerns about the potential for terrorist attacks using either radiological “dirty bombs” or improvised nuclear devices, as well as accidents similar to that of the Fukushima Daiichi nuclear power plant, have made the improvement of radiation biodosimetry a top national priority for radiological preparedness (1). After such a large-scale radiological event, radiation dose estimates would be needed for medical triage to assign individuals to appropriate treatment. This would also help in the timely reassurance of many individuals not requiring treatment (the “worried well”), who might otherwise overwhelm health resources. Several promising approaches to radiation biodosimetry have been pursued in the past few years, including automation or streamlining of classical cytogenetic assays (2, 3) and more recently, the development of proteomic (4, 5), transcriptomic (6–9) or metabolomic signatures (10, 11). The majority of such studies have focused on single acute exposures, but depending on the situation, protracted low-dose-rate (LDR) exposures (e.g., fallout or ground shine) may be expected to constitute much of the total dose. As protraction of an exposure over time can significantly decrease the extent of injury compared with an acute dose (12), understanding the rate of exposure could be very important for triage and selection of the most appropriate treatment. For instance, protraction of dose rate has been shown to increase the LD50 for both acute and late radiation syndromes in several mouse strains (13–15). Current biodosimetry approaches, however, have not been developed to distinguish between acute and LDR exposures for assessment of potential radiological injury. While cytogenetic approaches have demonstrated an ability to effectively reconstruct an individual’s total dose from chronic occupational or environmental exposures (16, 17), cytogenetic methods cannot distinguish these from acute radiation exposures.
Several groups are developing gene expression signatures as an approach for emergency biodosimetry (7–9), while others are developing devices for implementing low-cost, rapid measurements in the field (18–21). Because of the diversity of gene expression responses to stresses such as ionizing radiation, it may be possible to develop such signatures to discriminate between acute and LDR exposures, either as part of a complete gene expression-based solution, or as an adjunct to a separate dose-determining approach such as high-throughput cytogenetics (22) or electron paramagnetic resonance measurements (23). In support of this idea, studies of LDR-irradiated mice with various radioisotopes administered as internal emitters have shown dramatically different gene expression response patterns from those typically seen in acute external beam studies (24–27), although the relative contributions of dose, dose-rate and radiation quality are not yet fully understood. An earlier study in a human myeloid leukemia cell line identified genes that responded to dose independent of dose rate, and other genes that showed strong dose-rate dependence (28), further suggesting that gene expression may be able to distinguish a LDR component of exposure to inform triage.
Given the potential importance that the rate of radiation exposure has to radiological triage, we wanted to determine if gene expression measurements could provide dose rate information to supplement basic biodosimetry. As an initial study in this area, we irradiated male C57BL/6 mice with a range of X-ray doses delivered as either acute (1.03 Gy/min) or LDR (3.09 mGy/min) exposures, and compared global gene expression after 24 h in exposed or sham-exposed control mice. Many genes were found to be regulated by both acute and LDR exposures when compared with controls, with the majority of these showing a larger magnitude of response after acute exposures. Some genes did respond similarly regardless of the rate of exposure, however, and some genes even showed a greater response after protracted exposures. Our results suggest that gene expression signatures could be developed to provide rate-of-exposure information to supplement estimates of total absorbed dose.
METHODS
Mouse Housing and Irradiation
All animal husbandry and related experimental procedures were conducted in accordance with applicable federal and state guidelines and approved by the Animal Care and Use Committee of Columbia University Medical Center. Male C57BL/6 mice (7 weeks of age) were purchased from Charles River Laboratories (Frederick, MD) and acclimated under standard vivarium conditions with free access to water and standard rodent chow for one week before irradiation. The mice were exposed to either acute or low-dose-rate (LDR) radiation using an X-Rad 320 X-ray machine (Precision X-Ray Inc., Branford, CT) with a custom Thoraeus filter (1.25 mm Sn, 0.25 mm Cu, 1.5 mm Al). The filter facilitated the use of the same quality of X rays in both the acute (1.03 Gy/min at 320 keV, 12.5 mA) and LDR (3.09 mGy/min at 320 keV/0.1 mA X) exposures. For LDR irradiation, mice were housed in a specially designed plastic cage with individual compartments, each 6 cm (w), 12.5 cm (l) and 12 cm (h). Each compartment was supplied with bedding, water and food. Mice were acclimated to this housing with one training session of 24 h prior to the start of irradiations. After this acclimation session, the mice no longer engaged in active exploration of the LDR housing environment when they were returned to it, but settled into sleep immediately after being placed in the irradiator. The mice were exposed at a dose rate of 3.09 mGy/min for 6 h (1.1 Gy), 12 h (2.2 Gy) or 24 h (4.4 Gy). During irradiation, standard mouse housing conditions were maintained inside the X-ray machine with a 12 h light/dark cycle, optimum temperature (22°C ± 0.5°C) and humidity (30–40%) maintained by a portable custom air conditioning system, and in excess of 10 air exchanges per hour achieved by mounting a NuLine tube axial fan (cat. no. 04424883; MSC Industrial Supply Co., Melville, NY) over one of the X-ray machine access ports. Mice were monitored during irradiation and they followed usual patterns of sleeping, feeding and activity. For acute irradiation, mice were placed in a custom-made plastic box (6 cm × 12.5 cm × 8 cm) inside the X-ray machine and exposed to 1.1, 2.2 or 4.4 Gy at a dose rate of 1.03 Gy/min. Sham-irradiated controls, for both acute and LDR exposures, were treated exactly the same as the corresponding exposed animals but without radiation exposure. Two separate experiments were performed, each using 3 mice per point and the data were pooled for analysis.
Blood Collection and RNA Preparation
Blood was collected from the mice 24 h after the onset of irradiation. Mice were rapidly euthanized by CO2 asphyxia and blood (approximately 0.5 ml) was collected by syringe (BD Vacutainer; Becton, Dickinson, Franklin Lakes, NJ) via cardiac puncture, mixed in PAXgene (Becton, Dickinson) blood RNA solution (1:4 ratio) and stored overnight at 4°C before further processing. The whole blood RNA was extracted using the PAXgene blood RNA kit (QIAGEN®, Valencia, CA) according to manufacturer’s instruction, followed by on-column DNase I treatment, and removal of globin transcripts using the GLOBINclear™ kit (Ambion®, Austin, TX) to remove both α- and β-globin transcripts. The RNA was quantified with a NanoDrop-1000 spectrophotometer, and quality was determined with the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). All RNA samples had RNA integrity numbers >9.0.
Quantitative Real-Time PCR
For each sample, 500 ng total RNA was reverse transcribed with the High-Capacity cDNA Archive Kit (Applied Biosystems®, Foster City, CA) according to the manufacturer’s instructions. Primer Express® software (Applied Biosystems) and the online TaqMan Primer Design software (GenScript Corp., Piscataway Township, NJ) were used to design gene-specific primers and probes (Table 1), and probes with 6-carboxyfluorescein (FAM) at the 5′ end and BHQ1 quencher at the 3′ end were synthesized by Operon Biotech, Inc. (Huntsville, AL). Alternately, predesigned primer-probe sets were purchased from Life Technologies (Grand Island, NY) (Table 1). Standard curves were generated to optimize the amount of input cDNA for each gene (5 or 10 ng). Real-time PCR reactions were performed with Universal PCR Master Mix (Applied Biosystems®, Carlsbad, CA), according to the manufacturer’s recommendations and were run on the Applied Biosystems 7900 Real-Time PCR System. All samples were performed in duplicate and repeated a second time on a different day for each gene. Relative fold inductions were calculated using the ΔΔ CT method with averaged relative levels of Actb used for normalization.
TABLE 1.
Primers and Probes Used for qRT-PCR
| Gene name | Primer/probe/part number | Sequence or source |
|---|---|---|
| Ccr6 | Forward primer | CTGTGTGGCAGTGTGGTTCA |
| Reverse primer | CGTGATGGGCTCTGAGACAG | |
| Probe | TGGCTCACAGACATCACGATCCTGC | |
| Faim3 | Forward primer | CTGGGCTTTCTCTTGCTGGT |
| Reverse primer | CTCGCCTCCTCATCGCTAGT | |
| Probe | CTCCTCCAGACGTGCGGGCC | |
| Ccng1 | Forward primer | TGGGAAGTCAGGGAAGATGG |
| Reverse primer | TGCGAGCTGCTAAAGGTGAA | |
| Probe | CCCGCAGGAAATACATGATCATGCG | |
| Phlda3 | Forward primer | GGGCCTGGTCAAGTTCAAGA |
| Reverse primer | CACATGTAGCCAGGTCCCAA | |
| Probe | ATCCAGACTGTGCGGGCCCG | |
| Rab17 | Forward primer | GGGCTGCCTCTTTGTCCAT |
| Reverse primer | CCTCCAGGGCTCAAGCATA | |
| Probe | TCATGTTCTGTGTCGGAGATGGC | |
| Rad51l1 | Forward primer | GCCACGAAAGACCATAGGGA |
| Reverse primer | TGTTGGCTTCCGTTTCCTTT | |
| Probe | TCCAAGCAGCGCAGTCCTGTTTCA | |
| Actb | Mm00607939_s1 | Life Technologies |
| Cd300lf | Mm00467508_m1 | Life Technologies |
| Ddx25 | Mm00600256_m1 | Life Technologies |
Microarrays and Data Analysis
The One-Color Quick Amp labeling kit (Agilent Technologies, Santa Clara, CA) was used to prepare Cyanine-3 (Cy3)-labeled cRNA from 100 ng input RNA according to the manufacturer’s instructions, followed by purification of the labeled cRNA on an RNAeasy column (QIAGEN). The specific activity of all cRNA samples ranged from 16.7 to 25.3 (mean 22.1), determined using the NanoDrop ND-1000 Spectrophotometer. For each sample, 1.65 μg labeled cRNA was fragmented and hybridized to mouse GE 4×44k v2 microarrays (part no. G4846A; Agilent Technologies) at 65°C for 17 h with rotation, followed by washing as recommended by the supplier. Microarray slides were immediately scanned (scanner no. G2404B; Agilent Technologies) using default settings. The resulting images were analyzed with Feature Extraction software, v10.5 (Agilent Technologies) using default parameters for background correction and flagging of nonuniform features.
Background corrected hybridization intensities were imported into BRB-ArrayTools, v. 4.3.2 (NCI, Biometric Research Branch, Bethesda, MD) (29) log2-transformed and median normalized. Nonuniform outliers or features not significantly above background intensity in 50% or more of the samples, as well as features not changing at least 1.5-fold in 20% or more of the samples were filtered out, giving 26,715 features that were used in subsequent analyses. The microarray data is available through the NCBI Gene Expression Omnibus (series no. GSE62623; http://1.usa.gov/1HkAtJR).
BRB-ArrayTools was used to identify genes that were differentially expressed under different exposure conditions. Genes with P < 0.001 were considered significantly differentially expressed, and the false discovery rate was also estimated for each gene using the method of Benjamini and Hochberg (30) to help control false positive results. The same software was also used to create heatmaps of clustered genes, and to build and test algorithms for classifying samples by radiation dose level using leave-one-out cross validation.
Gene Ontology Enrichment Analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) web-based tools were used to analyze gene ontology term enrichment of differentially expressed genes (31, 32). Bonferroni corrected P values < 0.05 were considered significant.
RESULTS
Microarray Experiments
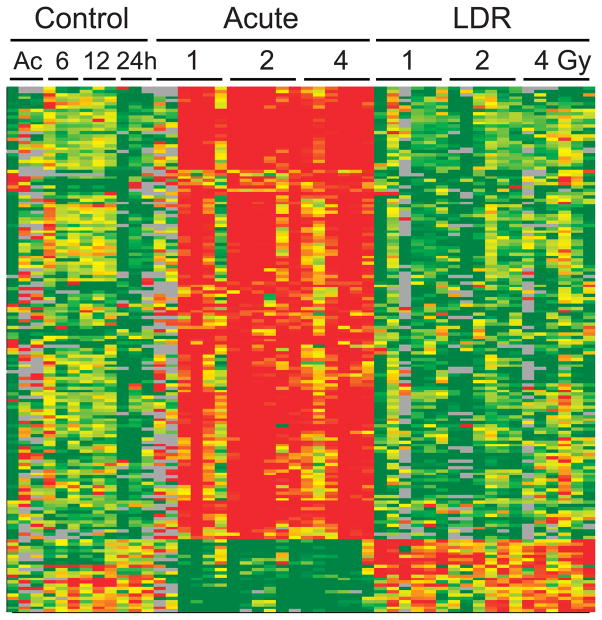
Total RNA was purified from the blood of mice sacrificed 24 h after the beginning of exposure to X rays delivered at either acute (1.03 Gy/min) or low dose rate (LDR; 3.09 mGy/min). Six mice were used for each dose and exposure rate, including sham-exposed controls (3 for each exposure time), and global gene expression was measured for each mouse using whole genome microarrays (Agilent Technologies). Genes with significantly different expression (P < 0.001, false discovery rate < 5%) across all doses including controls were identified using BRB-ArrayTools (29). Mice exposed to acute X rays showed 958 differentially expressed microarray features (Supplementary Table S1; http://dx.doi.org/10.1667/RR14044.1.S1), corresponding to 869 genes. Mice exposed to LDR X rays showed 975 differentially expressed microarray features (Supplementary Table S1), corresponding to 922 genes. Of these differentially expressed genes, 205 (22%) were common to both the acute and LDR data sets (Fig. 1). When mice exposed to any dose of LDR X rays were compared with acutely exposed mice, 1,021 features (Supplementary Table S1) representing 1,001 genes were found to be significantly differentially expressed. To test for effects of potential stress that might have been induced by the different mouse housing necessary during the LDR irradiations, we also looked for differentially expressed genes among control mice as a function of time in the LDR housing compared to the acute irradiation controls, which saw only standard housing. Three genes (Copa, Ccdc130 and Ldb1) were differentially expressed, suggesting that the difference in LDR housing during the exposure does not represent a significant source of stress or potential confounding of the radiation analysis.
FIG. 1.
Heatmap illustrating expression of the genes that responded to both acute and LDR irradiation. Microarray expression levels of the 205 genes, differentially expressed after both acute and LDR irradiation, were clustered to visualize their expression patterns after the different exposures. Red indicates high gene expression and green indicates low expression. Each column represents an individual mouse and each row an individual gene. Unexposed controls are indicated as either mock acute exposed (Ac) or by the number of hours of mock exposure in the LDR apparatus. Annotation of the genes in clustered order is provided in Supplementary Table S2 (http://dx.doi.org/10.1667/RR14044.1.S2).
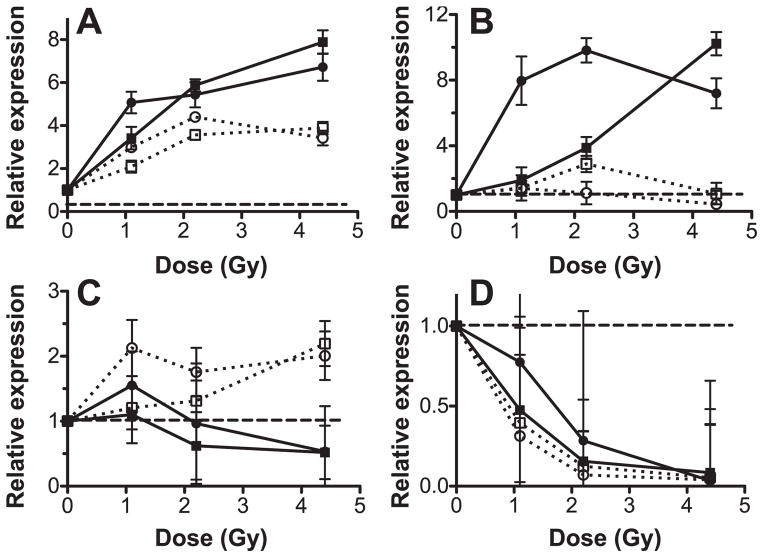
The patterns of expression found in the microarray data were confirmed for a selection of differentially expressed genes using quantitative real-time RT-PCR (qRT-PCR; Fig. 2). The Tp53 regulated genes Phlda3 and Ccng1 were overexpressed after both acute and LDR exposures (Fig. 2A), but with a lower magnitude of response to the protracted exposure. Rad51l1 and Rab17 were overexpressed after acute exposure, but did not respond to the LDR exposure (Fig. 2B). Ddx25 and Cd3001f were overexpressed after LDR but not acute exposures (Fig. 2C). Faim3 and Cdr6 were both strongly underexpressed after exposure to either dose rate, with no significant difference in the response to acute versus LDR exposure (P >0.05; Fig. 2D).
FIG. 2.
Gene expression measured by qRT-PCR. Measurements were made on RNA extracted from blood 24 h after exposure of mice to X rays at acute (closed symbols) or low-dose rate (open symbols) relative to levels in unexposed controls (dashed lines). Points represent the mean of measurements from six animals, error bars are SEM. Panel A: Tp53 regulated genes, Phlda3 (squares) and Ccng1 (circles). Panel B: Genes induced by acute exposure, Rab17 (circles) and Rad51l1 (squares). Panel C: Genes induced by LDR exposure, Cd300lf (circles) and Ddx25 (squares). Panel D: Genes with reduced expression after irradiation, Ccr6 (circles) and Faim3 (squares).
Sample Classification by Gene Expression
BRB-ArrayTools was used to build algorithms for assigning samples to a defined class and leave-one-out cross validation. Following an approach similar to that reported earlier (8), we used the genes that were differentially expressed in response to both the acute and LDR exposures to classify all samples by dose without regard for the rate of exposure. The 3-nearest neighbors (3NN) algorithm performed the best of the algorithms tested, classifying 85% of the samples as belonging to the correct dose group (Table 2). As differences in the rate of exposure can modify the extent of radiological injury expected, we also attempted to classify samples simultaneously by both dose and dose rate. In this case the correct classification rate of the 3NN algorithm was only 55% (Table 3). However, classification of all exposed samples as either acute or LDR exposures was 97% correct using a support vector machine algorithm operating on the full data set (Table 4). This classifier used 164 genes (Fig. 3), all of which were included in the set of genes identified as differentially expressed by rate of exposure.
TABLE 2.
Performance of the 3NN Classifier on Dose
| Dose (Gy) | Sensitivity | Specificity |
|---|---|---|
| 0 | 1 | 1 |
| 1.1 | 0.833 | 0.972 |
| 2.2 | 0.833 | 0.861 |
| 4.4 | 0.75 | 0.972 |
TABLE 3.
Performance of the 3NN Classifier on Dose and Dose Rate
| Class | Sensitivity | Specificity |
|---|---|---|
| 1.1 Gy Acute | 0.333 | 0.929 |
| 1.1 Gy LDR | 0.667 | 0.905 |
| 2.2 Gy Acute | 0.667 | 0.857 |
| 2.2 Gy LDR | 0.833 | 0.786 |
| 4.4 Gy Acute | 0.667 | 0.881 |
| 4.4 Gy LDR | 0.333 | 0.714 |
| 0 Gy Control | 0.25 | 0.861 |
TABLE 4.
Performance of the SVM Classifier on Irradiated Samples
| Class | Sensitivity | Specificity |
|---|---|---|
| Acute | 0.944 | 1 |
| LDR | 1 | 0.944 |
FIG. 3.
Heatmap of genes used in the LDR vs. acute exposure classifier. Microarray measurements of the 164 genes in the LDR vs. acute classifier were clustered to visualize their expression patterns after the different exposures. Red indicates high gene expression and green indicates low expression. Each column represents an individual mouse and each row an individual gene. Unexposed controls are indicated as either mock acute-exposed (Ac) or by the number of hours of mock exposure in the LDR apparatus. Annotation of the genes in clustered order is provided in Supplementary Table S2 (http://dx.doi.org/10.1667/RR14044.1.S2).
Gene Ontology Analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (31) was used to look for enrichment of KEGG pathways or gene ontology terms for biological processes that were over-represented among the genes responding to acute or LDR radiation exposure. Among upregulated genes, only two KEGG pathways were significantly enriched after both LDR and acute exposure: regulation of actin cytoskeleton (mmu04810, Benjamin-corrected P = 0.001 acute and 0.05 LDR) and p53 signaling pathway (mmu04115, P = 0.03 acute and 0.04 LDR). Additional biological processes and KEGG pathways were significantly enriched among genes overexpressed only after acute exposures. Among downregulated genes, most of the significantly over-represented biological processes and pathways were involved with hematopoietic cell functions or immune response. This was the case for all the processes significantly enriched for genes responding to both LDR and acute exposures (Table 5). Regulation of cell cycle was significantly enriched among downregulated genes responding to acute but not low-dose-rate exposure, while RNA processing functions were significantly over-represented among genes responding to LDR but not acute exposure (Table 5).
TABLE 5.
DAVID Gene Ontology Analysis of Downregulated Genes
| Term | Acute | LDR | TBIa |
|---|---|---|---|
| GO:0001775, cell activation | 6.74 × 10−06 | 4.72 × 10−04 | 1.72 × 10−05 |
| GO:0006955, immune response | 7.36 × 10−06 | 0.0054 | 0.0064 |
| GO:0045321, leukocyte activation | 9.10 × 10−05 | 6.26 × 10−04 | 9.91 × 10−07 |
| GO:0046649, lymphocyte activation | 1.19 × 10−04 | 9.56 × 10−04 | 2.53 × 10−07 |
| GO:0050863, regulation of T cell activation | 2.30 × 10−04 | 0.028 | 0.0053 |
| GO:0051249, regulation of lymphocyte activation | 0.002 | 0.039 | 0.0063 |
| GO:0002520, immune system development | 6.77 × 10−06 | 0.0018 | |
| GO:0048534, hemopoietic or lymphoid organ development | 1.10 × 10−05 | 0.005 | |
| GO:0030097, hemopoiesis | 8.34 × 10−05 | 0.0053 | |
| mmu05340, primary immunodeficiency | 1.54 × 10−04 | 5.19 × 10−04 | |
| GO:0002684, positive regulation of immune system process | 2.32 × 10−04 | 0.0057 | |
| GO:0050870, positive regulation of T cell activation | 5.96 × 10−04 | 0.027 | |
| GO:0042113, B cell activation | 0.0087 | 3.10 × 10−04 | |
| GO:0030098, lymphocyte differentiation | 0.0023 | 0.002 | |
| GO:0002694, regulation of leukocyte activation | 0.0029 | 0.006 | |
| GO:0050865, regulation of cell activation | 0.0029 | 0.00686 | |
| GO:0002521, leukocyte differentiation | 0.0073 | 0.00746 | |
| GO:0030217, T cell differentiation | 0.008 | 0.0063 | |
| GO:0042110, T cell activation | 0.0109 | 2.14 × 10−05 | |
| GO:0051251, positive regulation of lymphocyte activation | 0.0041 | ||
| GO:0002696, positive regulation of leukocyte activation | 0.0053 | ||
| GO:0050867, positive regulation of cell activation | 0.0057 | ||
| GO:0002377, immunoglobulin production | 0.012 | ||
| GO:0002440, production of molecular mediator of immune response | 0.015 | ||
| GO:0051726, regulation of cell cycle | 0.019 | ||
| GO:0042035, regulation of cytokine biosynthetic process | 0.022 | ||
| GO:0042108, positive regulation of cytokine biosynthetic process | 0.038 | ||
| GO:0010564, regulation of cell cycle process | 0.039 | ||
| GO:0002768, immune response–regulating cell surface receptor signaling pathway | 0.042 | ||
| mmu04640, hematopoietic cell lineage | 5.02 × 10−05 | ||
| GO:0016071, mRNA metabolic process | 0.026 | ||
| GO:0006396, RNA processing | 0.026 | ||
| mmu04672, intestinal immune network for IgA production | 0.028 | ||
| GO:0006397, mRNA processing | 0.042 | ||
| GO:0044265, cellular macromolecule catabolic process | 0.047 | ||
| hsa04660, T-cell receptor signaling pathway | 1.27 × 10−05 | ||
| GO:0022403, cell cycle phase | 5.73 × 10−04 | ||
| GO:0006968, cellular defense response | 0.0012 | ||
| GO:0000279, M phase | 0.0037 | ||
| GO:0000278, mitotic cell cycle | 0.0056 | ||
| GO:0022402, cell cycle process | 0.0061 | ||
| GO:0000280, nuclear division | 0.0067 | ||
| GO:0007067, mitosis | 0.0067 | ||
| GO:0000087, M phase of mitotic cell cycle | 0.0067 | ||
| GO:0048285, organelle fission | 0.0077 | ||
| GO:0002684, positive regulation of immune system process | 0.011 | ||
| hsa04650, natural killer cell mediated cytotoxicity | 0.011 | ||
| GO:0007059, chromosome segregation | 0.021 |
Total-body irradiation data from Paul et al. (51).
DISCUSSION
In this study we profiled the gene expression response in mice exposed to acute and LDR radiation, and tested the potential of the differentially expressed genes to classify blood samples by dose and dose rate of exposure. We also evaluated the biological processes implicated by gene ontology analysis using DAVID. Although several groups are working towards the development of gene expression as an approach for radiation biodosimetry (6–9, 33, 34), such studies, as well as the development of other radiation biodosimetry methods, have focused on the classification of acute exposures. Among studies using C57BL/6 mice for acute biodosimetry development, Filiano et al. (35) reported that only two genes, Cdkn1a and Ccng1, were needed to discriminate between unexposed mice and mice exposed to doses of 1 Gy and above. We have found that while the response of Cdkn1a was similar after acute and LDR exposure, Ccng1 showed a muted response to LDR (Fig. 2A). In more recent work, Lucas et al. (34) reported a 15-gene signature that could predict γ-ray doses up to 10.5 Gy in C57BL/6 mice. Although some of the genes in their signature were not significantly responsive in our study, perhaps reflecting their use of higher doses or platform-specific effects, we did find a strong acute induction of Ifit3, which was completely ablated by LDR exposure. Conversely, expression of Cnn3 and Tnfrsf13b was significantly enhanced by LDR exposure. Taken together these comparisons emphasize the potential for dose rate to affect the result of a biodosimetric test, and the need to develop radiation biodosimetry signatures specific to the type of exposure scenario anticipated as a means of more accurately capturing this information.
The potential usefulness of gene expression for bio-dosimetry in a situation also involving LDR exposure, such as from fallout, has not previously been investigated directly. Although some LDR gene profiling studies have been reported, these have been aimed towards the effects of occupational or environmental exposures, and have used mice exposed to much lower dose rates and/or total doses not of concern for radiological triage (36–39). Studies of humans occupationally exposed over many years to low total doses (40, 41) have also been published.
In comparing acute and LDR exposures in the current study, we found several general patterns of gene expression response. Many genes showed a muted response to LDR exposure, as shown in Fig. 2A, and other genes responded only to acute (Fig. 2B) or to LDR (Fig. 2C) exposures. The genes with a greater response to acute exposures echo the protective effect of exposure protraction reported for end points from cell mutation (42) to short- and long-term survival of mice (13–15). We have also investigated physiological responses in peripheral blood cells in mice exposed to the same doses and dose rates that were used here for gene expression studies (43). In that work, we found that acute exposures were more effective than LDR exposures in reducing T-cell numbers in peripheral blood, and in the induction of unrepaired double-strand breaks as monitored by γ-H2AX levels.
Other genes observed responded similarly regardless of the rate of exposure (Fig. 2D), although the dynamic nature of gene expression processes suggests that there will be a lower limit on the dose-rate range where this applies. We have also recently reported a similar dose-rate-independent induction of micronuclei in the peripheral blood of mice exposed to the same doses and dose rates as in this study (43). Similar findings of different sets of genes that are either dose-rate dependent or dose-rate independent have been reported in human and mouse cell lines (28, 44).
These broad patterns suggest that it might be possible to define gene expression signatures capable of classifying blood samples by both the total dose and the rate of exposure. Although we found that a classifier attempting to perform both operations simultaneously did not perform well, samples could be accurately classified as exposed to acute or LDR radiation (97% correct), or classified by dose (85% correct) without regard to dose rate. More detailed studies will be needed to determine the range of doses and dose rates where this approach can be applied, and if it will be possible to distinguish between different LDR ranges.
The performance of dose and dose-rate determination using gene expression is complicated by natural variation between individuals (45) and variations in individual responses to radiation exposure (46). While this may be a larger issue in an outbred human population with many lifestyle differences, natural variation also exists within a controlled inbred mouse population (e.g., Fig. 1). Slight variations in animal handling and behavioral differences, epigenetic or stochastic differences in gene expression and response, as well as variations in the measurement platform and sample processing may all contribute to such observed variation. Additional testing, including potential confounding factors, will be needed to more fully assess the robustness of any biodosimetry approach.
Gene ontology analysis of differentially expressed genes was used to compare the biological pathways potentially affected by acute and LDR radiation exposures. Using DAVID (31), we found that the KEGG p53 signaling pathway (mmu04115) was the top enriched pathway among genes upregulated by LDR exposures. Not surprisingly, this pathway was also significantly enriched among genes upregulated in response to acute exposures. Tp53-regulated genes are among the top responders reported in both acute and LDR studies (28, 44, 47, 48), although there have been suggestions that its transcriptional activity may be differentially regulated in a dose-rate-dependent manner (28, 49,50).
A greater number of biological functions were found to be significantly enriched among the genes that were expressed below control levels after LDR exposure. Among these functions, all that were also enriched among downregulated genes after acute exposure were related to activation of T cells, B cells or other aspects of immune response (Table 5). This would suggest a general suppression of the specialized functions of blood cells in which gene expression was measured, possibly contributing to impaired immune response after exposure to either acute or LDR radiation. Additional functions related to immune activation, cytokine production, and cell cycle regulation were also significantly enriched among downregulated genes after acute, but not LDR, exposures.
In a previous study in which a human myeloid leukemia cell line was exposed to a range of dose rates, we reported that genes involved with apoptosis responded to radiation in a dose-rate-dependent manner, while cell cycle regulatory genes showed dose-rate independence in their response (28). In contrast, in the current study, apoptosis genes were not significantly enriched after exposure to either dose rate, and we found enrichment of cell cycle genes only after the acute exposure, suggesting dose-rate dependence in the human cell line. While it is possible that this represents a fundamentally different response between mouse cells and cells of human origin, it may well be that the earlier finding resulted from the use of rapidly dividing transformed cells, where cell cycle regulatory genes may be more sensitive than in the relatively quiescent cells normally circulating in the blood.
To broadly compare our mouse results with those from a human exposure, we performed the same gene ontology analysis on the set of genes significantly downregulated in patients 24 h after the start of total-body irradiation (TBI) (51). The patients were exposed to three 1.25 Gy fractions at a dose rate of 0.1 Gy/min, a rate in between the acute (1.03 Gy/min) and LDR (3.09 mGy) exposures delivered to the mice in this study. All the significantly enriched processes that were common to TBI, acute and LDR exposures were related to immune response and lymphocyte activation, with additional immune response processes common to human TBI and mouse acute, but not LDR, exposures (Table 5). No other processes were shared between TBI and LDR exposures, although additional immune response and cell cycle processes were found to be enriched only in the TBI data set. The human TBI response was thus more similar to the response to acute exposure in the mice, consistent with the TBI dose rate being nearer the acute dose rate than the protracted dose rate. From this analysis, the biological processes significantly affected by radiation in humans appear to be generally well conserved in mice.
In conclusion, a large number of genes are differentially expressed in the blood of mice exposed to either acute or low-dose-rate radiation. The biological functions most impacted by these genes are consistent with activation of Tp53 and a general suppression of immune response functions, and are conserved between mouse and human responses. Different exposure rates result in both common and distinct patterns of response. These different gene expression patterns have the potential to provide both dosimetric information independent of the rate of exposure, and a novel means of distinguishing between acute and low-dose-rate exposures. The latter could be extremely important for the assessment of injury potential in a mass radiation casualty situation, and could be used in conjunction with dosimetry methods that cannot distinguish exposure rate, such as cytogenetics or electron paramagnetic resonance. Thus, studies using a wider range of dose rates and assay times should be undertaken to further develop this approach.
Supplementary Material
Differentially expressed genes. This Excel file contains information on the genes found to be differentially expressed across dose after acute (tab “Acute”) or LDR exposure (tab “LDR”). It also lists the genes differentially expressed between acute- and LDR-exposed mice (tab “LDR v Acute”).
Genes in clustered order. This Excel file contains the gene annotations in clustered order corresponding to the heat maps shown in Fig. 1 (tab “Fig 1 Intersect cluster”) and Fig. 3 (tab “Fig 3 LDR vs Acute Prediction”).
Acknowledgments
The authors would like to thank Mr. Gary Johnson and his team for fabrication of the mouse housing and custom X-ray filter and Dr. Congju Chen for assistance with animal handling and sample collection. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant no. U19 AI067773.
References
- 1.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat Res. 2005;163:115–23. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 2.Garty G, Chen Y, Turner HC, Zhang J, Lyulko OV, Bertucci A, et al. The rabit: A rapid automated biodosimetry tool for radiological triage. II. Technological developments. Int J Radiat Biol. 2011;87:776–90. doi: 10.3109/09553002.2011.573612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flegal FN, Devantier Y, Mcnamee JP, Wilkins RC. Quickscan dicentric chromosome analysis for radiation biodosimetry. Health Phys. 2010;98:276–81. doi: 10.1097/HP.0b013e3181aba9c7. [DOI] [PubMed] [Google Scholar]
- 4.Ivey RG, Subramanian O, Lorentzen TD, Paulovich AG. Antibody-based screen for ionizing radiation-dependent changes in the mammalian proteome for use in biodosimetry. Radiat Res. 2009;171:549–61. doi: 10.1667/RR1638.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma M, Moulder JE. The urine proteome as a radiation biodosimeter. Adv Exp Med Biol. 2013;990:87–100. doi: 10.1007/978-94-007-5896-4_5. [DOI] [PubMed] [Google Scholar]
- 6.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–6. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabacik S, Mackay A, Tamber N, Manning G, Finnon P, Paillier F, et al. Gene expression following ionising radiation: Identification of biomarkers for dose estimation and prediction of individual response. Int J Radiat Biol. 2011;87:115–29. doi: 10.3109/09553002.2010.519424. [DOI] [PubMed] [Google Scholar]
- 10.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Brenner DJ, Brown TR. Identification of urinary biomarkers from x-irradiated mice using NMR spectroscopy. Radiat Res. 2011;175:622–30. doi: 10.1667/RR2388.1. [DOI] [PubMed] [Google Scholar]
- 12.Hall EJ, Giaccia A. Radiobiology for the radiologist. 7. Philadelphia: Lippincott Williams & Wilkins; 2012. [Google Scholar]
- 13.Travis EL, Peters LJ, McNeill J, Thames HDJ, Karolis C. Effect of dose-rate on total body irradiation: Lethality and pathologic findings. Radiother Oncol. 1985;4:341–51. doi: 10.1016/s0167-8140(85)80122-5. [DOI] [PubMed] [Google Scholar]
- 14.Van Os R, Thames HD, Konings AW, Down JD. Radiation dose-fractionation and dose-rate relationships for long-term repopulating hemopoietic stem cells in a murine bone marrow transplant model. Radiat Res. 1993;136:118–25. [PubMed] [Google Scholar]
- 15.Safwat A, Nielsen OS, El-Badawy S, Overgaard J. Effect of radiation dose rate and cyclophosphamide on pulmonary toxicity after total body irradiation in a mouse model. Int J Radiat Oncol Biol Phys. 1996;34:85–91. doi: 10.1016/0360-3016(95)02078-0. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh WA, Lucas JN, Hwang JJ, Chan CC, Chang WP. Biodosimetry using chromosomal translocations measured by FISH in a population chronically exposed to low dose-rate 60Co gamma-irradiation. Int J Radiat Biol. 2001;77:797–804. doi: 10.1080/09553000110053873. [DOI] [PubMed] [Google Scholar]
- 17.Beinke C, Meineke V. High potential for methodical improvements of FISH-based translocation analysis for retrospective radiation biodosimetry. Health Phys. 2012;103:127–32. doi: 10.1097/HP.0b013e31824645fb. [DOI] [PubMed] [Google Scholar]
- 18.Huang TC, Paul S, Gong P, Levicky R, Kymissis J, Amundson SA, et al. Gene expression analysis with an integrated CMOS microarray by time-resolved fluorescence detection. Biosens Bioelectron. 2011;26:2660–5. doi: 10.1016/j.bios.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badie C, Kabacik S, Balagurunathan Y, Bernard N, Brengues M, Faggioni G, et al. Laboratory intercomparison of gene expression assays. Radiat Res. 2013;180:138–48. doi: 10.1667/RR3236.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brengues M, Smith S, Estes M, Zenhausern F. Integrated microfluidic system for gene expression based biodosimetry. In: Christensen DM, Sugarman SL, Fm O, editors. The medical basis for radiation accident preparedness V: Advances in diagnosis and management of ionizing radiation injuries and illnesses. Oak Ridge: Oak Ridge Associated Universities; 2013. pp. 363–6. [Google Scholar]
- 21.Kim D, Marchetti F, Chen Z, Zaric S, Wilson RJ, Hall DA, et al. Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Sci Rep. 2013;3:2234. doi: 10.1038/srep02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garty G, Karam A, Brenner DJ. Infrastructure to support ultra high throughput biodosimetry screening after a radiological event. Int J Radiat Biol. 2011;87:754–65. doi: 10.3109/09553002.2011.583317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams BB, Flood AB, Salikhov I, Kobayashi K, Dong R, Rychert K, et al. In vivo EPR tooth dosimetry for triage after a radiation event involving large populations. Radiat Environ Biophys. 2014;53:335–46. doi: 10.1007/s00411-014-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokolov MV, Smirnova NA, Camerini-Otero RD, Neumann RD, Panyutin IG. Microarray analysis of differentially expressed genes after exposure of normal human fibroblasts to ionizing radiation from an external source and from DNA-incorporated iodine-125 radionuclide. Gene. 2006;382:47–56. doi: 10.1016/j.gene.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Rudqvist N, Parris TZ, Schuler E, Helou K, Forssell-Aronsson E. Transcriptional response of BALB/c mouse thyroids following in vivo astatine-211 exposure reveals distinct gene expression profiles. EJNMMI Res. 2012;2:32. doi: 10.1186/2191-219X-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuler E, Rudqvist N, Parris TZ, Langen B, Spetz J, Helou K, et al. Time- and dose rate-related effects of internal (177)Lu exposure on gene expression in mouse kidney tissue. Nucl Med Biol. 2014;41:825–32. doi: 10.1016/j.nucmedbio.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, et al. Gene expression response of mice after a single dose of 137Cs as an internal emitter. Radiat Res. 2014;182:380–9. doi: 10.1667/RR13466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, et al. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1:445–52. [PubMed] [Google Scholar]
- 29.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Inform. 2007;2:11–7. [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blakely WF, Miller AC, Luo L, Lukas J, Hornby ZD, Hamel CJ, et al. Nucleic acid molecular biomarkers for diagnostic biodosimetry applications: use of the fluorogenic 5′-nuclease polymerase chain reaction assay. Mil Med. 2002;167:16–9. [PubMed] [Google Scholar]
- 34.Lucas J, Dressman HK, Suchindran S, Nakamura M, Chao NJ, Himburg H, et al. A translatable predictor of human radiation exposure. PLoS One. 2014;9:e107897. doi: 10.1371/journal.pone.0107897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filiano AN, Fathallah-Shaykh HM, Fiveash J, Gage J, Cantor A, Kharbanda S, et al. Gene expression analysis in radiotherapy patients and C57BL/6 mice as a measure of exposure to ionizing radiation. Radiat Res. 2011;176:49–61. doi: 10.1667/RR2419.1. [DOI] [PubMed] [Google Scholar]
- 36.Taki K, Wang B, Nakajima T, Wu J, Ono T, Uehara Y, et al. Microarray analysis of differentially expressed genes in the kidneys and testes of mice after long-term irradiation with low-dose-rate gamma-rays. J Radiat Res (Tokyo) 2009;50:241–52. doi: 10.1269/jrr.09011. [DOI] [PubMed] [Google Scholar]
- 37.Gridley DS, Rizvi A, Luo-Owen X, Makinde AY, Pecaut MJ. Low dose, low dose rate photon radiation modifies leukocyte distribution and gene expression in CD4(+) T cells. J Radiat Res (Tokyo) 2009;50:139–50. doi: 10.1269/jrr.08095. [DOI] [PubMed] [Google Scholar]
- 38.Gridley DS, Pecaut MJ, Rizvi A, Coutrakon GB, Luo-Owen X, Makinde AY, et al. Low-dose, low-dose-rate proton radiation modulates CD4(+) T cell gene expression. Int J Radiat Biol. 2009;85:250–61. doi: 10.1080/09553000902748609. [DOI] [PubMed] [Google Scholar]
- 39.Uehara Y, Ito Y, Taki K, Nenoi M, Ichinohe K, Nakamura S, et al. Gene expression profiles in mouse liver after long-term low-dose-rate irradiation with gamma rays. Radiat Res. 2010;174:611–7. doi: 10.1667/RR2195.1. [DOI] [PubMed] [Google Scholar]
- 40.Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Ghilardi-Netto T, Donadi EA, et al. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J Radiat Res (Tokyo) 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- 41.Abend M, Azizova T, Muller K, Dorr H, Senf S, Kreppel H, et al. Independent validation of candidate genes identified after a whole genome screening on Mayak workers exposed to prolonged occupational radiation. Radiat Res. 2014;182:299–309. doi: 10.1667/RR13645.1. [DOI] [PubMed] [Google Scholar]
- 42.Amundson SA, Chen DJ. Inverse dose-rate effect for mutation induction by gamma-rays in human lymphoblasts. Int J Radiat Biol. 1996;69:555–63. doi: 10.1080/095530096145562. [DOI] [PubMed] [Google Scholar]
- 43.Turner HC, Shuryak I, Taveras M, Bertucci A, Perrier JR, Chen C, et al. Effect of dose rate on residual gamma-H2AX levels and frequency of micronuclei in X-irradiated mouse lymphocytes. Radiat Res. 2015;183:315–24. doi: 10.1667/RR13860.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugihara T, Murano H, Tanaka K, Oghiso Y. Inverse dose-rate-effects on the expressions of extra-cellular matrix-related genes in low-dose-rate gamma-ray irradiated murine cells. J Radiat Res (Tokyo) 2008;49:231–40. doi: 10.1269/jrr.07074. [DOI] [PubMed] [Google Scholar]
- 45.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 46.Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75:885–90. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burns TF, El-Deiry WS. Microarray analysis of p53 target gene expression patterns in the spleen and thymus in response to ionizing radiation. Cancer Biol Ther. 2003;2:431–43. doi: 10.4161/cbt.2.4.478. [DOI] [PubMed] [Google Scholar]
- 48.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the national cancer institute anticancer drug screen. Cancer Res. 2008;68:415–24. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 49.Sugihara T, Magae J, Wadhwa R, Kaul SC, Kawakami Y, Matsumoto T, et al. Dose and dose-rate effects of low-dose ionizing radiation on activation of Trp53 in immortalized murine cells. Radiat Res. 2004;162:296–307. doi: 10.1667/rr3223. [DOI] [PubMed] [Google Scholar]
- 50.Sugihara T, Murano H, Nakamura M, Ichinohe K, Tanaka K. P53-mediated gene activation in mice at high doses of chronic low-dose-rate gamma radiation. Radiat Res. 2011;175:328–35. doi: 10.1667/RR2446.1. [DOI] [PubMed] [Google Scholar]
- 51.Paul S, Barker CA, Turner HC, Mclane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–65. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes. This Excel file contains information on the genes found to be differentially expressed across dose after acute (tab “Acute”) or LDR exposure (tab “LDR”). It also lists the genes differentially expressed between acute- and LDR-exposed mice (tab “LDR v Acute”).
Genes in clustered order. This Excel file contains the gene annotations in clustered order corresponding to the heat maps shown in Fig. 1 (tab “Fig 1 Intersect cluster”) and Fig. 3 (tab “Fig 3 LDR vs Acute Prediction”).