Abstract
Recent studies indicated the predominance of Toxoplasma gondii haplogroup 12 in wildlife in USA. But still little is known of the genetic diversity of this parasite circulating in wildlife. In the present study, we tested coyotes (Canis latrans), red foxes (Vulpes vulpes), white-tailed deer (Odocoileus virginianus), and geese (Branta canadensis) from the state of Pennsylvania for T. gondii infection. Antibodies to T. gondii were found in 160 of 367 animals, including 92 (34.5%) of 266 coyotes, 49 (62.0%) of 79 white-tailed deer, 17 (85.0%) of 20 red fox, and two of two Canada geese tested by the modified agglutination test (cut off titer 1:25). Tissues from 105 seropositive animals were bioassayed in mice, and viable T. gondii was isolated from 29 animals, including 10 of 53 coyotes, 11 of 16 foxes, 7 of 49 deer, and one of one goose. DNA isolated from culture-derived tachyzoites of these isolates was characterized initially using multilocus PCR-RFLP markers. Nine genotypes were revealed, including ToxoDB PCR-RFLP #1 (4 isolates), #2 (2 isolates), #3 (4 isolates), #4 (6 isolates), #5 (4 isolates), #54 (1 isolate), #141 (1 isolate), #143 (1 isolate), and #216 (6 isolates), indicating high genetic diversity of T. gondii in wildlife in Pennsylvania. Pathogenicity of six T. gondii isolates (5 of #216 and #141) was determined in outbred Swiss Webster mice. Three of #216 and the #141 isolates were acute virulent to mice, and the other 2 #216 isolates were intermediate virulent. To determine the extent of genetic variation of these as well as a few recently reported virulent isolates from wildlife in North America, intron sequences were generated. Analysis of intron sequences and PCR-RFLP genotyping results indicated that the #216 isolates are likely derived from recombination of the clonal type I and III lineages. To determine if T. gondii virulence can be predicted by typing, we genotyped a collection of strains using PCR-RFLP markers for polymorphic genes ROP5, ROP16, ROP18 and GRA15, which are known to interact with host immune response. The results showed that there is an association of genotypes of ROP5 and ROP18 with mouse-virulence, however, additional gene(s) may also contribute to virulence in distinct T. gondii genotypes.
Keywords: Toxoplasma gondii, Wildlife, Seroprevalence, Isolation, Genotyping, Pennsylvania, USA
1. Introduction
The protozoan Toxoplasma gondii infects virtually all warm-blooded animals, including birds, humans, livestock, and marine mammals (Dubey, 2010). In the USA, various surveys have found that 10–50% of the adult human population has antibodies to T. gondii (reviewed in Dubey and Jones, 2008). Humans become infected postnatally by ingesting tissue cysts from undercooked meat, or by consuming food or drink contaminated with oocysts. However, only a small percentage of seropositive adult humans or other animals develop clinical signs of disease. It is unknown whether the severity of toxoplasmosis in immunocompetent hosts is due to the parasite strain, host variability, or to other factors. Recently, attention has been focused on the genetic variability among T. gondii isolates from apparently healthy and sick hosts (Grigg and Sundar, 2009). Severe cases of toxoplasmosis have been reported in immunocompetent patients in association with atypical T. gondii genotypes (Ajzenberg et al., 2004; Demar et al., 2007; Elbez-Rubinstein et al., 2009; Grigg and Sundar, 2009; Vaudaux et al., 2010; Wendte et al., 2011; Pomares et al., 2011; Sobanski et al., 2013). However, little is known of the association of genotype and clinical disease in animals and humans in the USA (Dubey, 2010). An atypical T. gondii genotype was isolated from a lamb aborted from a chronically infected sheep from Texas (Edwards and Dubey, 2013). A variant of Type II (NE-II) was recently found associated with prematurity and severe disease at birth in congenitally infected children in the USA (McLeod et al., 2012). Type II strains are the most prevalent in Europe, and are also abundant in North America, and cause congenital toxoplasmosis in children (Howe and Sibley, 1995).
Historically, T. gondii was considered to be clonal with low genetic diversity and grouped into 3 types, namely I, II, III (Howe and Sibley, 1995; Sibley and Ajioka, 2008). However, recent studies have revealed a greater genetic diversity of T. gondii, particularly isolates from Brazil (Khan et al., 2011; Su et al., 2012; Dubey et al., 2012, 2013a). A fourth clonal lineage (Type 12) was recently described, predominantly from wildlife (Dubey et al., 2011; Khan et al., 2011). This genotype includes the Type X and Type A T. gondii strains reported in sea otters from California and Washington State (Miller et al., 2004; Sundar et al., 2008). Although Type 12 has been identified from pigs and sheep in the USA, the frequency is low, and the dominant genotype in these domestic animals is the Type II (Dubey et al., 2008a; Velmurugan et al., 2008). It is not clear why there is difference in genotype distribution among wildlife versus domestic animals, unless there is exclusive sylvatic cycling. Also, this trend was not seen in isolates from wild animals in France (Richomme et al., 2009; Aubert et al., 2010), and Norway (Prestrud et al., 2008). It may be due to sampling variation or adaptation of biological traits in different genotypes. Isolation of T. gondii from wildlife is time consuming, expensive, and difficult. Most of the data are derived from opportunistic sampling that may have limited availability due to the species habitat use, range, or season the species is available due to harvest season regulations. Although investigating wildlife it is not clear how a genotype will become established in a particular host.
In the present study we had an opportunity to survey different wildlife species in Pennsylvania. The species we examined are found statewide, across habitats, and have different exposure potentials due to their behavior, and may be important in the spread of T. gondii in both wildlife and human populations.
2. Materials and methods
2.1. Naturally infected animals
Wildlife samples were obtained through various methods with the majority of animals collected through hunter harvest or those taken to resolve wildlife damage issues conducted by USDA-APHIS-Wildlife Services. Heart, tongue, or brain (goose only) samples as well as blood samples were collected from each animal. Samples were collected within 48 h of mortality and refrigerated until they could be submitted for testing. Hunter harvested samples were taken using legal means and during approved seasons. All sampling or control activities were conducted under permits assigned to USDA-APHIS-Wildlife Services (USFWS permit MB 068253-0, Pennsylvania Special Use Permits 141-2010, 131-2011, 153-2012). All wildlife species taken to resolve wildlife damage conflicts were euthanized using approved AVMA methods (https://www.avma.org/KB/Policies/Documents/euthanasia.pdf). During 2007 and 2008 samples of blood, heart, and tongue were collected from hunted coyotes by one us (M. Weaver) as part of her Master’s degree thesis; these samples were treated as the other samples described above. All samples were submitted to the Animal Parasitic Diseases Laboratory (APDL), United States Department of Agriculture, Beltsville, Maryland for T. gondii examination. Samples were derived from most counties in Pennsylvania.
2.2. Serology
Sera from animals were tested for antibodies to T. gondii by the modified agglutination test (MAT) as described by Dubey and Desmonts (1987). Sera were screened at 1:25, 1:50, 1:100, and 1:200 dilutions. Selected samples were titrated further.
2.3. Bioassay in mice
Tissues were homogenized in saline, digested in acidic pepsin, centrifuged, and aliquots of homogenates were inoculated subcutaneously into two to five outbred SW mice and/or one or two KO mice (Dubey, 2010). Tissue imprints of lungs and brains of inoculated mice that died were examined for T. gondii tachyzoites or tissue cysts. Survivors were bled on day 45 days p.i. and a 1:25 dilution of serum was tested for T. gondii antibodies by MAT. Mice were killed 46 days p.i. and brains of all mice were examined for tissue cysts as described (Dubey, 2010). The inoculated mice were considered infected with T. gondii when tachyzoites or tissue cysts were found in tissues.
2.4. Pathogenicity of oocysts of T. gondii strains in mice
Pathogenicity of oocysts of the T. gondii isolates from seven isolates was done in SW mice. For this, T. gondii-free cats (Dubey, 1995) were fed tissues of infected mice and oocysts collected from the feces of cats (Dubey, 2010). Oocysts were sporulated in 2% sulfuric acid for a week on a shaker at room temperature, washed, counted, and diluted 10-fold from 10−1 to 10−7 to reach an end point of ≅1 oocyst. Aliquots from each dilution of oocysts were fed to five SW mice and the recipient mice examined for T. gondii infection. Mortality was recorded, and after two months mice were tested for T. gondii infection as described above.
To determine pathogenicity of tachyzoites from six isolates, SW mice fed oocysts were killed five to seven days p.i.; their mesenteric lymph nodes were homogenized in saline, filtered through 5-µm membrane filter to remove host cells, tachyzoites were counted, and diluted 10-fold to reach end point with <1 tachyzoite. Aliquots from each dilution were inoculated subcutaneously into five SW mice for each dilution.
2.5. In vitro cultivation
Infected mouse tissues were seeded on to CV1 cell culture flasks and tachyzoites were harvested from the medium as described (Dubey et al., 2013a).
2.6. Genetic characterization
T. gondii DNA was extracted from cell-cultured tachyzoites and strain typing was performed using the genetic markers SAG1, 5′- and 3′-SAG2, alt.SAG2, SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1, and Apico as described previously (Su et al., 2010). To further reveal the extent of genetic diversity of T. gondii isolates, four introns from three unlinked genes UPRT, EF1 and HP2 were sequenced as reported previously (Khan et al., 2007). Sequencing of introns was conducted by Genewiz (Genewiz Inc, NJ, USA). Sequences were aligned using ClustalX (Higgins et al., 1996) and nexus file was imported into Molecular Evolutionary Genetic Analysis (MEGA) Version 4.1 to identify all variable sites (Kumar et al., 2001). All variable sites from intron sequences and RFLP markers were concatenated and incorporated into SplitsTree v4.4 to generate unrooted neighbor-net analysis using 1000 bootstrap replicates (Huson and Bryant, 2006).
To determine if T. gondii virulence can be predicted by genotyping polymorphic genes ROP5, ROP16, ROP18 and GRA15, which are known to interact with the host immune response (Melo et al., 2011), we conducted PCR-RFLP typing of the above loci for T. gondii strains with known virulent phenotypes. The typing was performed following previously reported method (Su et al., 2010). The primer sequences and condition for restriction digestion of PCR products for this assay are summarized in Table 1. Network analysis of these typing data was performed using SplitsTree4.4 as above.
Table 1.
PCR primes for genotyping ROP5, ROP16, ROP18 and GRAG15.
| Markers | External primers for multiplex PCR |
Internal primers for nested PCR | NEB Restriction enzymes, buffers, incubation temperature and time, and gel electropheresisa |
|---|---|---|---|
| ROP5 | ROP5-Fext GGACAGACGCAGGCTTTTAC ROP5-Rext TCAAACGTCCTGACACTTCG |
ROP5-Fint: TGTGGCAGTTCAGTCTCAGC ROP5-Rint: TCGAAGTTGAGGAACCGTCT |
FspBI digestion: 37 °C, 1 h, 2.5% agarose gel |
| ROP18 | ROP18-DelFext: CTCGTCGACCACACAGCTAA ROP18-UPSFext: TTTTATCGACATCCCGCTTC ROP18-UPSRext: GAGTGCTTTCTGTCGCTCCT |
Rop18-UPSFint: CACAGCATGAGCTTAAGAGTTG ROP18-UPSRint: CACCGCAAGACAGGCTGTCTTC |
No enzyme treatment. Type III has positive PCR, Type I and II are negative. 1.5% agarose gel. |
| ROP18-DelFint: AGTTCCCTTCCCTGGTGTCT ROP18-DelRint: ACAAACTGGACTGGGGTGAG |
Type III has no PCR products, others have PCR products for RFLP analysis. ScrFI + MfeI, NEB4, BSA, 37 °C, 1 h, 2.5% gel |
||
| ROP16 | ROP16-Fext ATCTGCTTATCCGGCGACTA ROP16-Rext TCCGTTGGCATTTATCATCA |
ROP16-Fint: TACCAAACCCAGCTTTCACC ROP16-Rint: TCGTCAACAGCTGACTCCAC |
NlaIII + TaqαI, NEB4, BSA, 37 °C 30 min, 65 °C 30 min, 2.5% agarose gel |
| GRA15 | GRA15-Fext CACGTACACAACCCATCTCG GRA15-Rext2 CCTTTGAACGGGTAATGGAA |
GRA15-Fint: GGACCACCCAGAACAGAAAA GRA15-Rint2: CCCTTATCGGTTTTTGGTCA |
No restriction enzyme needed. 1.5% agarose gel |
Restriction enzyme FspBI was purchased from Fisher Scientific, Pittsbergh, PA, all other enzymes were purchased from the New England BioLab.
2.7. Ethical considerations
All experiments were performed according to approved protocols by all institutions involved.
3. Results
Between 2010 and 2012, tissues and serum from 265 animals, including 164 coyotes, 20 foxes, 79 deer, and two geese were collected by one of us (Van Why). The rest 102 samples were collected from coyotes in 2007. Antibodies to T. gondii were found in 160 of 367 animals with highest prevalence in red fox and geese (Table 2).
Table 2.
Prevalence of T. gondii in wildlife in Pennsylvania.
| Host | No. tested | No. seropositive (%) | No. of animals with MAT titers of: | No. bioassayed | No. T. gondii isolated | |||
|---|---|---|---|---|---|---|---|---|
| 25 | 50 | 100 | ≥200 | |||||
| Coyote | 266 | 92 (34.5) | 25 | 33 | 18 | 16 | 53a | 10b |
| Fox | 20 | 17 (85.0) | 1 | 2 | 2 | 12 | 16 | 11 |
| Deer | 79 | 49 (62.0) | 4 | 11 | 8 | 26 | 35 | 7 |
| Goose | 2 | 2 (100.0) | 0 | 1 | 0 | 1 | 1 | 1 |
| Total | 367 | 160 (43.6%) | 30 | 47 | 28 | 55 | 105 | 29 |
22 coyotes from 2007 survey.
2 from 2007 survey.
Viable T. gondii was isolated from 10 coyotes, 11 foxes, seven deer, and one goose (Tables 2 and 3). Of these, some or all SW mice inoculated with one isolate from coyote, four isolates from foxes, and two isolates from deer died of acute toxoplasmosis. Oocysts were obtained from seven mouse virulent isolates either by feeding tissues of mice that died after inoculation with the host tissues or by feeding mice sub-inoculated with the isolated strains (Table 3).
Table 3.
Isolates of viable T. gondii from wildlife in Pennsylvania.
| Host | County | Date | MAT | Bioassaya | Isolate ID | Cat oocysts | ToxoDB-PCR-RFLP genotype # |
|
|---|---|---|---|---|---|---|---|---|
| SWb | KOc | |||||||
| Coyote | ||||||||
| 23 | Susquehanna | 2/8/2012 | >200 | 1/5 | 0/0 | TgCoPa1 | 1 | |
| 42 | Sullivan | 2/8/2012 | ≥200 | 5/5 | 0/0 | TgCoPa2 | 5 | |
| 53 | Wyoming | 2/10/2011 | ≥200 | 2/4 (29) | 1/1 | TgCoPa3 | Cat #50 | 5 |
| 76 | Clearfield | 2/16/2011 | 50 | 2/2 | 1/1 | TgCoPa4 | 5 | |
| 137 | Clearfield | 3/3/2011 | 50 | 4/4 | 1/1 | TgCoPa5 | 3 | |
| 164 | Mercer | 3/3/2011 | 50 | 4/4 | 1/1 | TgCoPa6 | 4 | |
| 198 | Erie | 3/3/2011 | 50 | 1/4 | 1/1 | TgCoPa7 | 4 | |
| 201 | Lycoming | 3/3/2011 | 100 | 4/4 | 0/1 | TgCoPa8 | 1 | |
| 45 | Potter | 2/23/2008 | 100 | 2/2 | Not done | TgCoPa9 | 5 | |
| 78 | Bradford | 2/23/2008 | 100 | 1/2 | Not done | TgCoPa10 | 1 | |
| Fox | ||||||||
| 13 | Adams | 2/3/2011 | ≥200 | 1/1 | 1/1 | TgFoxPa1 | 3 | |
| 89 | Adams | 2/16/2011 | ≥200 | 3/3 | 1/1 | TgFoxPa2 | 54 | |
| 90 | Adams | 2/16/2011 | ≥200 | 0/2 | 1/1 | TgFoxPa3 | 2 | |
| 91 | Bradford | 2/16/2011 | 50 | 2/2 | 1/1 | TgFoxPa4 | 216 | |
| 95 | Adams | 2/16/2011 | 50 | 3/3 | 1/1 | TgFoxPa5 | 1 | |
| 96 | Adams | 2/16/2011 | 100 | 3/3 (16, 16, 20) | 1/1 | TgFoxPa6 | Cat #53 | 216 |
| 238 | Philadelphia | 4/22/2011 | 200 | 4/4 (14, 14, 13, 9) | 1/1 | TgFoxPa7 | Cat #70 | 216 |
| 239 | Philadelphia | 4/22/2011 | 100 | 1/4 (13) | 1/1 | TgFoxPa8 | Cat #74 | 216 |
| 240 | Philadelphia | 4/22/2011 | 400 | 2/4 (19, 20) | 1/1 | TgFoxPa9 | Cat #42 | 141 |
| 241 | Philadelphia | 4/22/2011 | 800 | 2/4 | 1/1 | TgFoxPa10 | 4 | |
| 242 | Philadelphia | 4/22/2011 | 400 | 4/4 | 1/1 | TgFoxPa11 | 2 | |
| Goose 9 | Philadelphia | 7/6/2011 | 50 | 1/3 | 0/2 | TgGoosePa1 | 143 | |
| White-tailed deer | ||||||||
| 28 | St. Clair | 2/9/2011 | ≥200 | 3/3 | 2/2 | TgWTDPa1 | 3 | |
| 111 | Allegheny | 2/24/2011 | ≥200 | 3/3 | 0/0 | TgWTDPa2 | 3 | |
| 114 | Allegheny | 2/24/2011 | ≥200 | 1/3 | 0/0 | TgWTDPa3 | 4 | |
| 118 | Philadelphia | 2/24/2011 | ≥200 | 3/3 (18, 18, 19) | 0/0 | TgWTDPa4 | Cat #64 | 216 |
| 119 | Philadelphia | 2/24/2011 | ≥200 | 1/3 (41) | 0/0 | TgWTDPa5 | Cat #58 | 216 |
| 122 | Philadelphia | 2/24/2011 | ≥200 | 3/3 | 0/0 | TgWTDPa6 | 4 | |
| 129 | Philadelphia | 2/24/2011 | ≥200 | 3/3 | 0/0 | TgWTDPa7 | 4 |
No of mice infected/No. of mice inoculated. SW = Swiss Webster, KO = knockout. MAT = modified agglutination test.
Day of death is parenthesis.
All infected KO mice died of toxoplasmosis.
Oocysts of four T. gondii isolates from foxes were most virulent for SW mice (Table 4). All mice fed oocysts died of toxoplasmosis and the mortality was dose dependent. Most mice died of enteritis and pneumonia within 21 days p.i. Tachyzoites of three of these four isolates were also lethal for all mice; tachyzoites of the fourth isolate were not titrated when it was discovered that all isolates were the same genotype. Oocysts and tachyzoites of two isolates from deer were also virulent for SW mice (Table 5). Oocysts from the coyote isolate were only mildly pathogenic; lethal dose was more than 1000 oocysts (data not shown). This strain belongs to PCR-RFLP genotype #5.
Table 4.
Pathogenicity of oocysts of T. gondii isolates from wildlife in Pennsylvania to Swiss Webster micea.
| Dosec | TgFoxPa9 (Cat #42, Fox 240) | TgFoxPa7 (Cat #70, Fox 238) | TgFoxPa8 (Cat #74, Fox 239) | TgFoxPa6 (Cat #53, Fox 96)f |
|||
|---|---|---|---|---|---|---|---|
| Oocysts | Tachyzoites | Oocysts | Tachyzoites | Oocysts | Tachyzoites | Oocysts | |
| 100,000 | 5 (5, 5, 5, 5, 5)b | Not done | Not done | Not done | Not done | Not done | 5 (6, 6, 6, 9, 10) |
| 10,000 | 5 (7, 7, 7, 7, 9) | Not done | Not done | Not done | Not done | Not done | 5 (6, 6, 8, 8, 9) |
| 1000 | 5 (8, 8, 8, 8, 8) | Not done | Not done | Not done | 5 (8, 8, 8, 8, 10) | 5 (17, 17, 17, 17, 17) | 5 (8, 8, 9, 9, 9) |
| 100 | 5 (9, 9, 10, 10, 10) | 5 (15, 20, 20, 20, 21) | 5 (8, 10, 10, 10, 12) | 5 (18, 19, 19, 20, 20) | 5 (10, 10, 10, 10, 10) | 5 (17, 18, 18, 20, 26) | 5 (9, 9, 10, 10, 10) |
| 10 | 4 (11, 11, 14, 17) | 3 (24, 24, 28) | 5 (10, 11, 11, 11, 11) | 5 (21, 21, 25, 26, 39d) | 2 (11, 12) | 5 (20, 20, 21, 25, 25) | 3 (12, 12, 12) |
| 1 | 2 (11, 11) | 1 (28) | 4 (13, 16, 16, 21) | 1 (25) | 1 (11) | 1 (25) | 1 (122e) |
| <1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Five mice per group. Oocysts were inoculated orally, tachyzoites were inoculated subcutaneously.
No. of mice infected with T. gondii of five inoculated (day of death of each mouse is in parenthesis). S = survived, infected-killed day 39.
Based on estimation that the last infective dilution has 1 infective organism.
Killed day 39. Many tissue cysts.
Killed day 122. Many tissue cysts.
Tachyzoites not titrated
Table 5.
Pathogenicity of oocysts of T. gondii isolates from 2 WTD in Pennsylvania to Swiss Webster micea.
| Doseb | TgWTDPa5 (Cat #58, WTD #119) | TgWTDPa4 (Cat #64, WTD #118) | ||
|---|---|---|---|---|
| Oocysts | Tachyzoites | Oocysts | Tachyzoites | |
| 100,000 | Not done | Not done | Not done | Not done |
| 10,000 | 5 (6, 7, 7, 7, 7)c | Not done | 5 (7, 7, 7, 7, 7) | Not done |
| 1000 | 5 (11, 11, 11, 11, 11) | 5 (18, 18, 18, 19, 19) | 5 (8, 8, 8, 8, 9) | 5 (15, 18, 20, 20, 20) |
| 100 | 5 (11, 11, 11, 11, 11) | 5 (18, 19, 21, 21, 21) | 5 (9, 9, 9, 10, 10) | 5 (19, 20, 21, 21, 25) |
| 10 | 5 (11, 11, 11, 11, 12) | 5 (18, 19, 19, 19, 20) | 4 (10, 12, 12, 17) | 5 (19, 19, 22, 54) |
| 1 | 4 (11, 17, 21, 32) | 2 (27, 27) | 3 (10, 12, 12) | 3 (19, 25, 32) |
| <1 | 0 | 0 | 0 | Not done |
Five mice per group. Oocysts were inoculated orally and tachyzoites were inoculated subcutaneously.
Based on estimation that the last infective dilution has 1 infective organism.
No. of mice dead/infected with T. gondii of five inoculated (day of death of each mouse is in parenthesis).
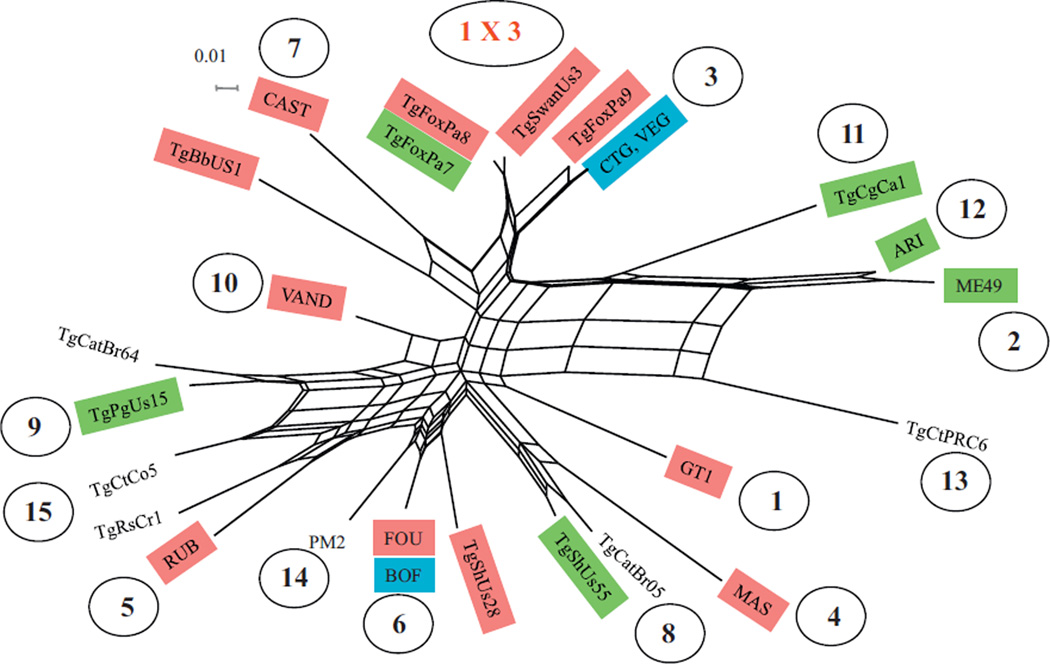
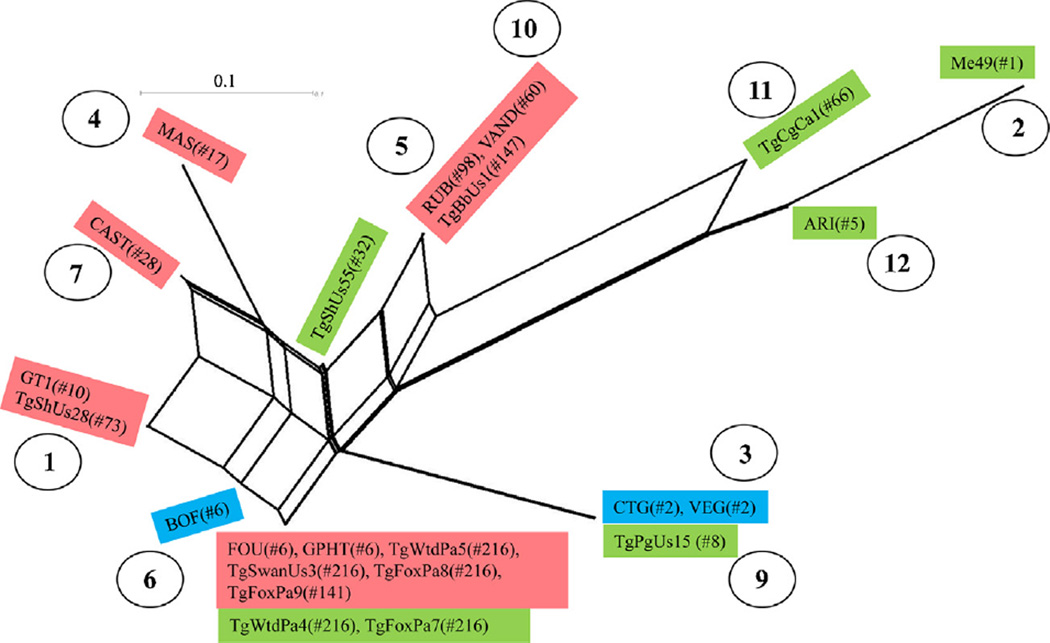
PCR-RFLP analysis of 29 T. gondii isolates from wildlife in Pennsylvania by the genetic markers SAG1, 5′- and 3′-SAG2, alt.SAG2, SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1, and Apico revealed nine genotypes (Table 6). Network analysis of concatenated intron sequences for three Pennsylvania isolates belonging to the genotype #216 and other 23 reference stains revealed that the genotype #216 is closely related to haplogroup 3 strains (Fig. 1). For 23 T. gondii strains with known virulence phenotypes (including 5 Pennsylvania isolates in this study), PCR-RFLP analysis of loci ROP5, ROP16, ROP18 and GRA15 are summarized in Table 7 and Fig. 2. The markers ROP5, ROP16, ROP18 and GRA15 revealed 5, 4, 2 and 2 alleles, respectively (Table 7). Eleven genotypes were revealed from the 23 T. gondii strains (Fig. 2). Due to low allele diversity for markers ROP16 and GRA15, they contribute little to the genotyping of these T. gondii strains, whereas the combinations of ROP5 and ROP18 alleles identified 10 of the 11 genotypes. In addition, markers ROP16 and GRA15 alleles were not associated with virulence phenotypes. There are clusters of haplogroups: 2, 11 and 12; 3 and 9; 5 and 10; and 1, 4 and 7 that associated well with virulence phenotypes. The cluster related to haplogroup 6 has a mixed collection of virulent, intermediate virulent and non-virulent T. gondii strains.
Table 6.
Genotyping of T. gondii isolates from Wildlife in Pennsylvania.
| Strain ID (Number of isolates) | Genotypes (ToxoDB PCR-RFLP genotypes) |
Genetic markers (Su et al., 2010) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAG1 | (5′+3′) SAG2 | alt. SAG2 | SAG3 | BTUB | GRA6 | c22-8 | c29-2 | L358 | PK1 | Apico | ||
| GT1 | Type I (#10) | I | I | I | I | I | I | I | I | I | I | I |
| PTG | Type II (#1) | II/III | II | II | II | II | II | II | II | II | II | II |
| CTG | Type III (#2) | II/III | III | III | III | III | III | III | III | III | III | III |
| MAS | Atypical (#17) | u-1 | I | II | III | III | III | u-1 | I | I | III | I |
| TgCgCa1 | Atypical (#66) | I | II | II | III | II | II | II | u-1 | I | u-2 | I |
| TgCtBr5 | Atypical (#19) | I | III | III | III | III | III | I | I | I | u-1 | I |
| TgCtBr64 | Atypical (#111) | I | I | u-1 | III | III | III | u-1 | I | III | III | I |
| TgRsCr1 | Atypical (#52) | u-1 | I | II | III | I | III | u-2 | I | I | III | I |
| Present study | ||||||||||||
| TgCoPa1, 8, TgFoxPa5, TgCoPa10 (N = 4) | Type II (#1) | II/III | II | II | II | II | II | II | II | II | II | II |
| TgFoxPa3, 11 (N = 2) | Type III (#2) | II/III | III | III | III | III | III | III | III | III | III | III |
| TgCoPa5, TgFoxPa1 TgWTDPa1, 2 (N = 4) | Type II variant (#3) | II/III | II | II | II | II | II | II | II | II | II | I |
| TgCoPa6, 7, TgFoxPa10, TgWTDPa3, 6, 7 (N = 6) | Type 12 (#4) | II/III | II | II | II | II | II | II | II | I | II | I |
| TgCoPa2, 3, 4, 9 (N = 4) | Type 12 (#5) | u-1 | II | II | II | II | II | II | II | I | II | I |
| TgFoxPa2 (N = 1) | Atypical (#54) | II/III | II | II | III | III | III | III | III | III | III | II |
| TgFoxPa9 (N = 1) | Atypical (#141) | II/III | III | III | III | III | III | III | III | III | I | III |
| TgGoosePa1 (N = 1) | Atypical (#143) | I | I | I | III | I | III | III | I | III | I | III |
| TgFoxPa4, 6, 7, 8, TgWTDPa,4, 5 (N = 6) | Atypical (#216) | I | I | I | III | III | I | III | III | III | I | III |
Fig. 1.
Neighbor-net analysis using concatenated 4 intron sequence markers comprising 1775 bp and 10 RFLP markers. The, wildlife isolates (TgFoxPa7, 8, 9) of T. gondii from Pennsylvania are closely related to haplogroup 3 strains. T. gondii haplogroups are shown in the circle numbers as designated previously (Su et al., 2012). Mouse-virulent strains are in red boxes, intermediate virulent in, green, and non-virulent strains in blue, the unknowns are in black. Samples TgFoxPa7, TgFoxPa8 and TgFoxPa9 are from this study. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Table 7.
Genotyping of T. gondii isolates with polymorphic loci of ROP5, ROP16, ROP18 and GRAG15.
| Strain ID | ToxoDB# | ROP5 | ROP18a | GRA15 | ROP16 | Virulenceb | Accumulative mortality% | References for virulence determination in mice |
|---|---|---|---|---|---|---|---|---|
| FOU | 6 | III | I (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| GPHT | 6 | III | I | I/III | I/III | Vir | 100 | Khan et al. (2007) |
| GT1 | 10 | I | I (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| TgShUs28 | 73 | I | I | I/III | I/III | Vir | 100 | Dubey et al. (2008a) |
| MAS | 17 | u-1 | u1 (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| CAST | 28 | I | u1 (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| VAND | 60 | u-2 | u1 (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| RUB | 98 | u-2 | u1 (I*) | I/III | I/III | Vir | 100 | Khan et al. (2007) and Khan et al. (2009) |
| TgBbUs1 | 147 | u-2 | u1 | I/III | I/III | Vir | 100 | Dubey et al. (2010) |
| TgFoxPa9 | 141 | III | I | I/III | I/III | Vir | 100 | This study |
| TgFoxPa8 | 216 | III | I | I/III | I/III | Vir | 100 | This study |
| TgSwanUs3 | 216 | III | I | I/III | I/III | Vir | 100 | Dubey et al. (2013a) |
| TgWtdPa5 | 216 | III | I | I/III | I/III | Vir | 100 | This study |
| TgWtdPa4 | 216 | III | I | I/III | I/III | Int | 83 | This study |
| TgFoxPa7 | 216 | III | I | I/III | I/III | Int | 91 | This study |
| P89 (TgPgUs15) | 8 | III | III (III*) | I/III | I/III | Int | 76 | Khan et al. (2007) and Khan et al. (2009) |
| TgShUs55 | 32 | III | u1 | I/III | I/III | Int | 80 | Edwards and Dubey (2013) |
| ARI | 5 | II | II | I/III | II | Int | 60 | Khan et al. (2011) |
| Me49 | 1 | II | II (II*) | II | II | Int | 40 | Khan et al. (2007) and Khan et al. (2009) |
| TgCgCa1 | 66 | u-2 | II (II*) | I/III | II | Int | 90 | Khan et al. (2007) and Khan et al. (2009) |
| BOF | 6 | nd | I (I*) | I/III | I/III | Non | 8 | Khan et al. (2007) and Khan et al. (2009) |
| CTG | 2 | III | III (III*) | I/III | I/III | Non | 0 | Khan et al. (2007) and Khan et al. (2009) |
| VEG | 2 | III | III (III*) | I/III | I/III | Non | 13 | Khan et al. (2007) and Khan et al. (2009) |
ROP18 alleles in the parenthesis are based on intron sequence data. The three distinct allele groups were designated as I*, II* and III*, respectively (Khan et al., 2009).
Virulence of T. gondii is determined based on accumulative mortality of infected outbred mice. Mice were infected with a series of low does (10, 100 and 1000 tachyzoites) by intraperitoneal injection. Mortality of mice was determined at day 30 post infection. T. gondii strains cause 100% mortality in mice are considered acute virulent (Vir), 99–30% are intermediately virulent (Int), and <30% are non-virulent (Non) (Su et al., 2002).
Fig. 2.
Genetic relationship of 23 T. gondii strains based on ROP5, ROP16, ROP18 and GRA15 PCR-RFLP polymorphisms. Ten, genotypes were identified among these strains. Mouse-virulent strains are in red boxes, intermediate virulent in green, and non-virulent, strains in blue. The number in the parenthesis is ToxoDB PCR-RFLP genotypes. T. gondii haplogroups are shown in the circle numbers, as designated previously (Su et al., 2012). Isolates TgWtdPa4, TgWtdPa5, TgFoxPa7, TgFoxPa8 and TgFoxPa9 are from current study. The result shows potential association of genotypes with virulence phenotype. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
4.1. Coyotes
Coyotes are considered resistant species to clinical toxoplasmosis, and we are not aware of any report of a clinical case of toxoplasmosis in this animal. Laboratory-reared coyotes fed T. gondii oocysts or tissue cysts became infected but remained asymptomatic (Dubey, 1982). In the present study, T. gondii antibodies were found in 34.5% of 266 coyotes, which is lower than 51.4% of 35 coyotes from Wisconsin (Dubey et al., 2007), 59% of 222 coyotes collected in Kentucky, Ohio, and Indiana (Dubey et al., 1999), and 62% of 52 coyotes from Texas (Lindsay et al., 1996). In the present study viable T. gondii was isolated from 10 (32.3%) of 53 seropositive coyotes. The majority of isolates that have been genotyped (18 in total) in the present or previous studies (Dubey et al., 2004a, 2007, 2011) comprise genotypes #1 and #3 are also known as the Type II, genotypes #4 and #5 are known as Type 12, both are dominant in wildlife in North America (Dubey et al., 2011).
Coyotes may be ideal as sentinels of T. gondii due to their habitat use and behavior (Aguirre, 2009). Coyotes are generalists that are able to exploit a variety of habitats and resources. Their populations have increased in many areas over the past 20 years, and they have colonized much of the eastern United States, using both rural and urban habitats (Anonymous, 1997; Bekoff and Gese, 2003; Gehrt and Riley, 2010; Mastro et al., 2012). Due to their diversity of diet and habitat use, the potential for coyotes to contract toxoplasmosis is high (Dubey et al., 1999). Because coyotes prey on or scavenge many species that may be harboring T. gondii, they could be the indicator of possible presence and exposure levels in an area.
4.2. Foxes
Very little information is available concerning T. gondii infection in red foxes in the USA (Dubey et al., 1999; Dubey, 2010). In the present study 85% of 20 foxes were seropositive, similar to results from other states (Dubey et al., 1999), and viable T. gondii was isolated from 11 (68.7%) of 16 seropositive foxes. T. gondii was previously isolated from a red fox from Georgia (Dubey et al., 2004a) and one fox from Kansas (Smith and Frenkel, 1995); the isolate from Georgia was Type II based on SAG2 and the isolates from Kansas were not genotyped. Two viable T. gondii from two red foxes from Alaska were genotype #3 (Type II with type I allele at the Apico locus) (Dubey et al., 2011). In the present study seven genotypes were identified from the 11 fox isolates, including genotype #1 of one isolate, #2 of two, #3 of one, #4 of one, #54 of one, #141 of one and #216 of four. It is interesting to see the relatively high frequency of #216 from foxes. Genotype #216 has been limited in North America, with one isolate from Grenada. Based on genotyping of ROP5, 16, 18 and GRA5, it is close to haplogroup 6 (Fig. 2). However, based on the 11 PCR-RFLP markers and intron sequences, it is closely related to haplogroup 3 (Fig. 1). It is likely a recombinant of the Type I and III strains.
Although it is difficult to determine the role that red foxes play in the spread of T. gondii, this species is more closely associated with human environments and resources than coyotes are (Soulsbury et al., 2010). Similar to coyotes, this species consumes prey and carrion that has been identified as carriers of T. gondii as well as preying on feral cats (Korschgen, 1959; Crossett and Elliott, 1991; Golightly et al., 1994) and possibly also exhibit coprophagy. Red fox may have more exposure potential to T. gondii due to a closer association with domestic/feral cat populations in rural, suburban, and urban environments (Soulsbury et al., 2010), providing a source for monitoring toxoplasmosis in the environment in areas where coyotes may not be available to sample.
4.3. Deer
In the present study, 62% of 79 deer were seropositive, indicating high seroprevalence. Humphreys et al. (1995) reported 60% of 593 deer from Pennsylvania sampled in 1991 had T. gondii antibodies. Samples collected in this study were from highly urbanized locations where deer densities are considered higher than normal and encounters with humans and domestic animals could be considered high. The areas where Humphreys et al. (1995) collected samples are from counties with much lower human density and with deer being collected from hunter harvest, it is likely that most deer were not found within a close proximity to a human occupied area. This observation indicates that deer densities or habitat may not be a limiting factor in exposure of deer to T. gondii.
In the present study, T. gondii was isolated from seven (20%) of 35 seropositive deer. T. gondii has been isolated previously from WTD in the USA (Lindsay et al., 1991, 1997; Dubey et al., 2004b, 2008b, 2013b; Yu et al., 2013), and some of these isolates have been genotyped using the 11 markers used in the present study (Dubey et al., 2011; Yu et al., 2013). Of the 44 T. gondii isolates (6 from Iowa, 9 from Minnesota, 19 from Mississippi, 9 from New Jersey, 1 Alabama), four were ToxoDB PCR-RFLP genotype #1 (Type II clonal), five were #2 (Type III), one was #3 (Type II variant), nine were #4 (Type 12), 18 were #5 (Type 12), one was #54, and one was #74, one was #216, one was #220, and three were #221. It is intriguing that 18 of 19 isolates from WTD from Mississippi State were genotype #5 (Type 12) although these deer were from six sites over a broad geographic area (Dubey et al., 2004a). Thus, there was a greater genetic variability than previously realized, although type 2 and 12 still predominate as seen in other wildlife species in North America.
4.4. Pathogenicity of natural isolates
Historically, T. gondii strains were grouped as virulent or avirulent, based on mortality in outbred mice. Pathogenicity is dependent on the stage of the parasite, dose, and the route of inoculation. Oocyst induced infections are more pathogenic, irrespective of the dose (Dubey, 2010). Therefore, we determined pathogenicity of the strains isolated here using both tachyzoites and oocysts, and inocula were titrated to reach an end point. In the present study, two of the seven T. gondii isolate from WTD were mild to high virulent for mice, irrespective of the dose or the stage inoculated (Tables 4 and 5). We further examined their genetic relationship with a collection of well characterized strains, using a set of intron markers that together with RFLP genotypes, has been used to cluster related strains into haplotypes (Khan et al., 2007; Su et al., 2012). Three representatives (TgFoxPa7, TgFoxPa8 – genotype #216; and TgFoxPa9 – genotype #141), together with four virulent strains reported recently, were sequenced for four introns at three genetic loci as described (Khan et al., 2007). The four recently reported virulent strains are TgSwanUs3 (genotype #216) (Dubey et al., 2013a), TgBbUs1 (genotype #147) (Dubey et al., 2010), TgShUs28 (genotype #73) (Dubey et al., 2008a), and TgShUs55 (genotype #32) (Edwards and Dubey, 2013). The analysis of composite data of intron sequences and PCR-RFLP showed that, the Pennsylvania isolates TgFoxPa7, TgFoxPa8, TgFoxPa9 and the isolate from mute swan TgSwanUs3 have the combination of type I and III alleles at different PCR-RFLP loci and the type III sequences for the four introns. All these isolates are more closely related to haplopgroup 3 (also called Type III) than haplogroup 1 (also called Type I, ToxoDB PCR-RFLP genotype #2) (Fig. 1).
To determine if T. gondii virulence can be predicted by typing, we genotyped a number of T. gondii strains using markers for polymorphic genes ROP5, ROP16, ROP18 and GRA15, which are known to interact with host immune response (Melo et al., 2011; Hunter and Sibley, 2012). The serine/threonine kinase, ROP18, has been identified as a major determinant of virulence (Saeij et al., 2007; Taylor et al., 2006). ROP18 phosphorylates and inactivates a family of host derived immunity-related p47 GTPases (IRGs), thereby protecting the parasite from clearance by innate immune effectors (Fentress et al., 2010; Fentress and Sibley, 2011). The ROP18 alleles in Type I and II strains are important for virulence, whereas the Type III allele is not expressed in Type III strains, therefore making these strains non-virulent. ROP5 locus encodes a pseudokinase, it consists of a family of 6–10 tandem repeats. The alleles in Type I and III strains are important for virulence, whereas Type II allele is non-virulent (Behnke et al., 2011; Reese et al., 2011). ROP5 controls virulence in T. gondii by facilitating ROP18 to phosphorylate IRG protein (Behnke et al., 2012; Fleckenstein et al., 2012). Therefore, the interaction of different alleles of ROP5 and ROP18 may have differential consequence of virulence. The results of genotyping T. gondii strains with different virulent phenotypes by markers ROP5, ROP18, ROP16 and GRA15 indicate that allele types of ROP16 and GRA15 have little contribution to virulence, whereas there is association of allele types of ROP5 and ROP18 with virulence (Table 7 and Fig. 2). However, additional gene(s) may also contribute to virulence in distinct T. gondii genotypes. Future study with a large sample size is needed to verify this finding.
4.5. Significance for human toxoplasmosis
Results of the present study, and other recent studies, indicate that a variety of T. gondii genotypes circulate in the food animal chain in the USA. Some of these genotypes may have potential to be highly virulent. T. gondii infection in wildlife is important because people can become infected directly by eating undercooked game meat, occasionally with serious consequences (Dubey, 2010). Both deer and geese are important game species, so animals harvested through recreational activities in addition to animals obtained through depredation activities and road kill collection provides considerable human exposure opportunities. Study of T. gondii infection rate in hunters or people consuming games may provide useful information to evaluate such risk.
Acknowledgement
In part supported by NIH AI059176, AI036629 grants awarded to David Sibley.
Footnotes
Conflict of interest
None.
References
- Anonymous. Coyote (Canis latrans) food habits in three urban habitat types of western Washington. Northwest Sci. 1997;71:1–5. [Google Scholar]
- Aguirre AA. Wild canids as sentinels of ecological health: a conservation medicine perspective. Parasites Vectors. 2009;2(Suppl 1):S7. doi: 10.1186/1756-3305-2-S1-S7. http://dx.doi.org/10.1186/1756-3305-2-SI-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajzenberg D, Bañnuls AL, Su C, Dumètre A, Demar M, Carme B, Dardé ML. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int. J. Parasitol. 2004;34:1185–1196. doi: 10.1016/j.ijpara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Aubert D, Ajzenberg D, Richomme C, Gilot-Fromont E, Terrier ME, de Gevigney C, Game Y, Maillard D, Gibert P, Dardé ML, Villena I. Molecular and biological characteristics of Toxoplasma gondii isolates from wildlife in France. Vet. Parasitol. 2010;171:346–349. doi: 10.1016/j.vetpar.2010.03.033. [DOI] [PubMed] [Google Scholar]
- Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Fentress SJ, Mashayekhi M, Li LX, Taylor GA, Sibley LD. The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog. 2012;8:e1002992. doi: 10.1371/journal.ppat.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekoff M, Gese EM. Coyote (Canis latrans) In: Feldhamer GA, Thompson BC, Chapman AJ, editors. Wild Mammals of North America: Biology, Management and Conservation. 2nd ed. Baltimore, Maryland: Johns Hopkins University Press; 2003. pp. 467–481. [Google Scholar]
- Crossett RL, Elliott CL. Winter food habits of red foxes and coyotes in Central Kentucky. Proceedings of the Annual Conference, South-eastern Association of Fish and Wildlife Agencies. 1991;45:97–103. [Google Scholar]
- Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, Punwasi W, Valery N, Peneau C, Daigre JL, Aznar C, Cottrelle B, Terzan L, Dardé ML, Carme B. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin. Infect. Dis. 2007;45:e88–e95. doi: 10.1086/521246. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Induced Toxoplasma gondii, Toxocara canis, and Isospora canis infection in coyotes. J. Am. Vet. Med. Assoc. 1982;181:1268–1269. [PubMed] [Google Scholar]
- Dubey JP, Desmonts G. Serological responses of equids fed Toxoplasma gondii oocysts. Equine Vet. J. 1987;19:337–339. doi: 10.1111/j.2042-3306.1987.tb01426.x. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Duration of immunity to shedding of Toxoplasma gondii oocysts by cats. J. Parasitol. 1995;81:410–415. [PubMed] [Google Scholar]
- Dubey JP, Storandt ST, Kwok OCH, Thulliez P, Kazacos KR. Toxoplasma gondii antibodies in naturally exposed wild coyotes, red foxes, and gray foxes and serologic diagnosis of toxoplasmosis in red foxes fed T. gondii oocysts and tissue cysts. J. Parasitol. 1999;85:240–243. [PubMed] [Google Scholar]
- Dubey JP, Graham DH, de Young RW, Dahl E, Eberhard ML, Nace EK, Won K, Bishop H, Punkosdy G, Sreekumar C, Vianna MCB, Shen SK, Kwok OCH, Sumners JA, Demarais S, Humphreys JG, Lehmann T. Molecular and biologic characteristics of Toxoplasma gondii isolates from wildlife in the United States. J. Parasitol. 2004a;90:67–71. doi: 10.1645/GE-110R. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Parnell PG, Sreekumar C, Vianna MCB, de Young RW, Dahl E, Lehmann T. Biologic and molecular characteristics of Toxoplasma gondii isolates from striped skunk (Mephitis mephitis), Canada goose (Branta canadensis), black-winged lory (Eos cyanogenia), and cats (Felis catus) J. Parasitol. 2004b;90:1171–1174. doi: 10.1645/GE-340R. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Nolden CA, Samuel MD, Velmurugan GV, Bandini LA, Kwok OCH, Bodenstein B, Su C. Characterization of Toxoplasma gondii from raccoons (Procyon lotor), coyotes (Canis latrans), and striped skunks (Mephitis mephitis) in Wisconsin identified several atypical genotypes. J. Parasitol. 2007;93:1524–1527. doi: 10.1645/GE-1245.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Jones JL. Toxoplasma gondii infection in humans and animals in the United States. Int. J. Parasitol. 2008;38:1257–1278. doi: 10.1016/j.ijpara.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Hill D, Velmurugan GV, Bandini LA, Kwok OCH, Majumdar D, Su C. High prevalence and abundant atypical genotypes of Toxoplasma gondii isolated from lambs destined for human consumption in the USA. Int. J. Parasitol. 2008a;38:999–1006. doi: 10.1016/j.ijpara.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Velmurugan GV, Ulrich V, Gill J, Carstensen M, Sundar N, Kwok OCH, Thulliez P, Majumdar D, Su C. Transplacental toxoplasmosis in naturally-infected white-tailed deer: isolation and genetic characterisation of Toxoplasma gondii from foetuses of different gestational ages. Int. J. Parasitol. 2008b;38:1057–1063. doi: 10.1016/j.ijpara.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Rajendran C, Ferreira LR, Kwok OCH, Sinnett D, Majumdar D, Su C. A new atypical highly mouse virulent Toxoplasma gondii genotype isolated from a wild black bear in Alaska. J. Parasitol. 2010;96:713–716. doi: 10.1645/GE-2429.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Toxoplasmosis of Animals and Humans. 2nd ed. Boca Raton, Florida: CRC Press; 2010. pp. 1–313. [Google Scholar]
- Dubey JP, Velmurugan GV, Rajendran C, Yabsley M, Thomas NJ, Beckman KB, Sinnett D, Ruid D, Paul W, Hart J, Fair PA, McFee WE, Shearn-Bochsler V, Kwok OCH, Ferreira L, Choudhary S, Faria EB, Zhou H, Felix TA, Su C. Genetic characterization of Toxoplasma gondii in wildlife from North America revealed widespread and high prevalence of the fourth clonal type. Int. J. Parasitol. 2011;41:1139–1147. doi: 10.1016/j.ijpara.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lago EG, Gennari SM, Su C, Jones JL. Toxoplasmosis in humans and animals in Brazil: high prevalence, high burden of disease, and epidemiology. Parasitology. 2012;139:1375–1424. doi: 10.1017/S0031182012000765. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Choudhary S, Kwok OCH, Ferreira LR, Oliveira S, Verma SK, Marks DR, Pedersen K, Mickley RM, Randall AR, Arsnoe D, Su C. Isolation and genetic characterization of Toxoplasma gondii from mute swan (Cyngus olor) from the USA. Vet. Parasitol. 2013a;195:42–46. doi: 10.1016/j.vetpar.2012.12.051. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Randall AR, Choudhary S, Ferreira LR, Oliveira S, Verma SK, Kwok OCH, Su C. Serologic prevalence, isolation and genetic characterization of Toxoplasma gondii from white tailed deer (Odocoileus virginianus) in New Jersey. J. Parasitol. 2013b doi: 10.1645/13-209.1. (in press). [DOI] [PubMed] [Google Scholar]
- Edwards JF, Dubey JP. Toxoplasma gondii abortion storm in sheep on a Texas farm and isolation of mouse virulent atypical genotype T. gondii from an aborted lamb from a chronically infected ewe. Vet. Parasitol. 2013;192:129–136. doi: 10.1016/j.vetpar.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Elbez-Rubinstein A, Ajzenberg D, Dardé ML, Cohen R, Dumètre A, Yera H, Gondon E, Janaud JC, Thulliez P. Congenital toxoplasmosis and reinfection during pregnancy: case report, strain characterization, experimental model of reinfection, and review. J. Infect. Dis. 2009;199:280–285. doi: 10.1086/595793. [DOI] [PubMed] [Google Scholar]
- Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, Bzik DJ, Taylor GA, Turk BE, Lichti CF, Townsend RR, Qiu W, Hui R, Beatty WL, Sibley LD. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host. Microbe. 2010;8:484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentress SJ, Sibley LD. The secreted kinase ROP18 defends Toxoplasma’s border. Bioessays. 2011;33:693–700. doi: 10.1002/bies.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, Steinfeldt T. A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLOS Biol. 2012;10:e1001358. doi: 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrt SD, Riley PD. Coyotes (Canis latrans) In: Gehrt SD, Riley PD, Cypher BL, editors. Urban Carnivores; Ecology, Conflict, and Conservation. Baltimore, Maryland: The Johns Hopkins University Press; 2010. pp. 78–95. [Google Scholar]
- Golightly RT, Faulhaber MR, Sallee KL, Lewis JC. Food habits and management of introduced red fox in southern California. Proceedings of the Sixteenth Vertebrate Pest Conference, Paper 21. 1994;21 http://digitalcommons.unl.edu/vpc16/21. [Google Scholar]
- Grigg ME, Sundar N. Sexual recombination punctuated by outbreaks and clonal expansions predicts Toxoplasma gondii population genetics. Int. J. Parasitol. 2009;39:925–933. doi: 10.1016/j.ijpara.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J. Infect. Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- Humphreys JG, Stewart RL, Dubey JP. Prevalence of Toxoplasma gondii antibodies in sera of hunter-killed white-tailed deer in Pennsylvania. Am. J. Vet. Res. 1995;56:172–173. [PubMed] [Google Scholar]
- Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 2012;10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM, Sibley LD. Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14872–14877. doi: 10.1073/pnas.0702356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Taylor S, Ajioka JW, Rosenthal BM, Sibley LD. Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are virulence in mice. PLoS Genet. 2009;5:e1000404. doi: 10.1371/journal.pgen.1000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. Genetic analyses of atypical Toxoplasma gondii strains reveals a fourth clonal lineage in North America. Int. J. Parasitol. 2011;41:645–655. doi: 10.1016/j.ijpara.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korschgen LJ. Food habits of the red fox in Missouri. J. Wildlife Manage. 1959;23:168–176. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics Appl. Note. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lindsay DS, Blagburn BL, Dubey JP, Mason WH. Prevalence and isolation of Toxoplasma gondii from white-tailed deer in Alabama. J. Parasitol. 1991;77:62–64. [PubMed] [Google Scholar]
- Lindsay DS, Kelly EJ, McKown RD, Stein FJ, Plozer J, Herman J, Blagburn BL, Dubey JP. Prevalence of Neospora caninum and Toxoplasma gondii antibodies in coyotes (Canis latrans) and experimental infections of coyotes with Neospora caninum. J. Parasitol. 1996;82:657–659. [PubMed] [Google Scholar]
- Lindsay DS, Sundermann CA, Dubey JP, Blagburn BL. Update on Toxoplasma gondii infections in wildlife and exotic animals from Alabama. J. Alabama Acad. Sci. 1997;68:246–254. [Google Scholar]
- Mastro LL, Gese EM, Young JK, Shivik JA. Coyote (Canis latrans), 100+ years in the east: a literature review. Addendum to the Proceedings of the 14th Wildlife Damage Management Conference (2012) 2012 www.aphis.usda.gov/wildlife_damage/nwrc/publications/
- Melo MB, Jensen KDC, Saeij JPJ. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol. 2011;27:487–495. doi: 10.1016/j.pt.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Members of the panel on euthanasia. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition. American Veterinary Medical Association. 2013 https://www.avma.org/KB/Policies/Documents/euthanasia.pdf. [Google Scholar]
- McLeod R, Boyer KM, Lee D, Mui E, Wroblewski K, Karrison T, Noble AG, Withers S, Swisher CN, Heydemann PT, Sautter M, Babiarz J, Rabiah P, Meier P, Grigg ME the Toxoplasmosis Study Group. Prematurity and severity are associated with Toxoplasma gondii alleles (NCCCTS, 1981–2009) Clin. Infect. Dis. 2012;54:1595–1605. doi: 10.1093/cid/cis258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Grigg ME, Kreuder C, James ER, Melli AC, Crosbie PR, Jessup DA, Boothroyd JC, Brownstein D, Conrad PA. An unusual genotype of Toxoplasma gondii is common in California sea otters (Enhydra lutris nereis) and is a cause of mortality. Int. J. Parasitol. 2004;34:275–284. doi: 10.1016/j.ijpara.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Pomares C, Ajzenberg D, Bornard L, Bernardin G, Hasseine L, Dardé ML, Marty P. Toxoplasmosis and horse meat, France. Emerg. Infect. Dis. 2011;17:1327–1328. doi: 10.3201/eid1707.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestrud KW, Åsbakk K, Mørk T, Fuglei E, Tryland M, Su C. Direct high-resolution genotyping of Toxoplasma gondii in arctic foxes (Vulpes lagopus) in the remote arctic Svalbard archipelago reveals widespread clonal Type II lineage. Vet. Parasitol. 2008;158:121–128. doi: 10.1016/j.vetpar.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Reese ML, Zeiner GM, Saeij JPJ, Boothroyd JC, Boyle JP. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9625–9630. doi: 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richomme C, Aubert D, Gilot-Fromont E, Ajzenberg D, Mercier A, Ducrot C, Ferté H, Delorme D, Villena I. Genetic characterization of Toxoplasma gondii from wild boar (Sus scrofa) in France. Vet. Parasitol. 2009;164:296–300. doi: 10.1016/j.vetpar.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Saeij JPJ, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu. Rev. Microbiol. 2008;62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- Smith DD, Frenkel JK. Prevalence of antibodies to Toxoplasma gondii in wild mammals of Missouri and east central Kansas: biologic and ecologic considerations of transmission. J. Wildl. Dis. 1995;31:15–21. doi: 10.7589/0090-3558-31.1.15. [DOI] [PubMed] [Google Scholar]
- Sobanski V, Ajzenberg D, Delhaes L, Bautin N, Just N. Severe toxoplasmosis in immunocompetent hosts: be aware of atypical strains. Am. J. Respir. Crit. Care Med. 2013;187:1143–1145. doi: 10.1164/rccm.201209-1635LE. [DOI] [PubMed] [Google Scholar]
- Soulsbury CD, Baker PJ, Iossa G, Harris S. Red foxes (Vulpes vulpes) In: Gehrt SD, Riley PD, Cypher BL, editors. Urban Carnivores; Ecology, Conflict, and Conservation. Baltimore, Maryland: The Johns Hopkins University Press; 2010. pp. 62–77. [Google Scholar]
- Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137:1–11. doi: 10.1017/S0031182009991065. [DOI] [PubMed] [Google Scholar]
- Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, Dardé ML, Zhu XQ, Ajioka JW, Rosenthal BM, Dubey JP, Sibley LD. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5844–5849. doi: 10.1073/pnas.1203190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar N, Cole RA, Thomas NJ, Majumdar D, Dubey JP, Su C. Genetic diversity among sea otter isolates of Toxoplasma gondii. Vet. Parasitol. 2008;151:125–132. doi: 10.1016/j.vetpar.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, El Hajj H, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- Vaudaux JD, Muccioli C, James ER, Silveira C, Magargal SL, Jung C, Dubey JP, Jones JL, Doymaz MZ, Bruckner DA, Belfort R, Jr, Holland GN, Grigg ME. Identification of an atypical strain of Toxoplasma gondii as the cause of a waterborne outbreak of toxoplasmosis in Santa Isabel do Ivaí. Braz. J. Infect. Dis. 2010;202:1226–1233. doi: 10.1086/656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan GV, Dubey JP, Su C. Genotyping studies of Toxoplasma gondii isolates from Africa revealed that the archetypal clonal lineages predominate as in North America and Europe. Vet. Parasitol. 2008;155:314–318. doi: 10.1016/j.vetpar.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Wendte JM, Gibson AK, Grigg ME. Population genetics of Toxoplasma gondii: New perspectives from parasite genotypes in wildlife. Vet. Parasitol. 2011;182:96–111. doi: 10.1016/j.vetpar.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shen J, Su C, Sundermann CA. Genetic characterization of Toxoplasma gondii in wildlife from Alabama, USA. Parasitol. Res. 2013;112:1333–1336. doi: 10.1007/s00436-012-3187-0. [DOI] [PubMed] [Google Scholar]