Summary
The Cricket paralysis virus (CrPV) uses an internal ribosomal entry site (IRES) to hijack the ribosome. In a remarkable RNA-based mechanism involving neither initiation factor nor initiator tRNA, the CrPV IRES jump starts translation in the elongation phase from the ribosomal A-site. Here we present cryo-EM maps of 80S•CrPV-STOP•eRF1•eRF3•GMPPNP and 80S•CrPV-STOP•eRF1 complexes revealing a previously unseen binding state of the IRES and directly rationalizing that an eEF2-dependent translocation of the IRES is required to allow the first A-site occupation. During this unusual translocation event the IRES undergoes a pronounced conformational change to a more stretched conformation. At the same time our structural analysis provides information about the binding modes of eRF1•eRF3•GMPPNP and eRF1 in a minimal system. It shows that neither eRF3 nor ABCE1 are required for the active conformation of eRF1 at the intersection between eukaryotic termination and recycling.
Introduction
During protein synthesis the information encoded in mRNA is translated into a polypeptide chain by the ribosome. The translation process is subdivided into four phases: initiation, elongation, termination and recycling. During initiation functionally competent ribosomes are assembled on the messenger RNA (mRNA) with initiator transfer RNA (tRNA Met-tRNAMeti) positioned in the ribosomal P-site and base-paired with the AUG codon of the mRNA. Canonical translation initiation in eukaryotes requires at least 12 initiation factors and a cap structure at the 5′ end of the mRNA (Aitken and Lorsch, 2012; Hinnebusch and Lorsch, 2012; Jackson et al., 2010). However, alternative pathways of internal initiation exist that are cap and end independent and require a reduced set of initiation factors (Jackson et al., 2010). Internal initiation is driven by structured RNA elements present in the 5′-untranslated region (UTR) of the mRNAs, which are known as internal ribosome entry sites (IRESs). Internal initiation via IRES elements is used by many viruses.
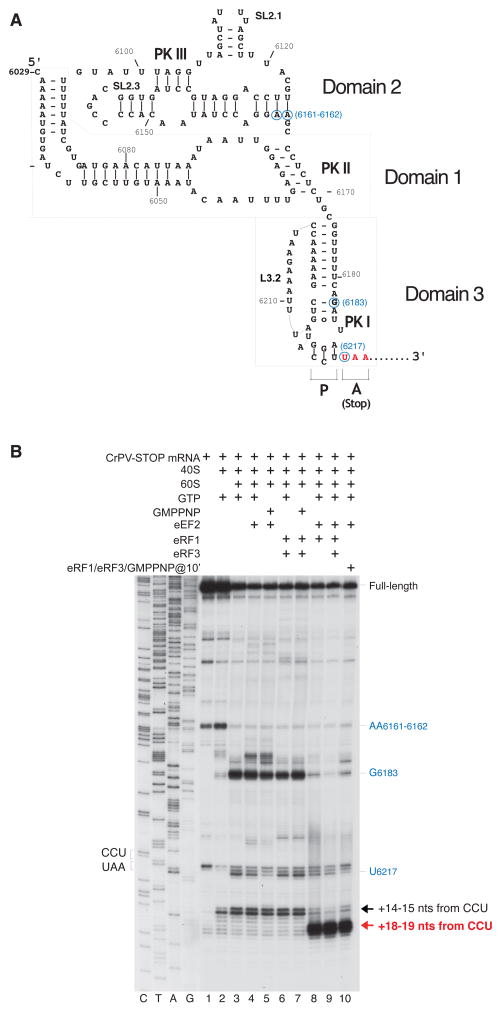
IRESs can be classified into four major types depending on their secondary structure, factor requirements and initiation site (Jackson et al., 2010). A particularly simple mechanism of translation initiation is used by type IV IRESs present in the intergenic region (IGR) of the genome of dicistroviruses such as Cricket paralysis virus (CrPV) (Wilson et al., 2000a). The IGR IRESs assemble functionally active 80S ribosomes without any initiation factor, initiator tRNA and AUG start codon, but jumpstart translation directly in the elongation phase from the A site (Pestova and Hellen, 2003; Sasaki and Nakashima, 2000; Wilson et al., 2000b). All IGR IRESs characterized so far share a highly conserved secondary structure comprising three domains, each characterized by a pseudoknot element (PK I to PK III) (Fig. 1A; Kanamori and Nakashima, 2001; Pfingsten et al., 2007). The first sense codon present at the 3′ edge of the PK I structure is alanine-encoding GCU.
Figure 1. eEF2-dependent association of eRF1 and eRF1/eRF3 with 80S ribosomal complexes assembled on CrPV-STOP mRNA.
(A) Secondary structure of the CrPV IRES with the first Ala codon mutated to an UAA stop codon (in red). Blue circles indicate the positions of toe-prints (panel B) caused by the contacts of the IRES with the 40S and 60S ribosomal subunits. (B) Toe-printing analysis of binding of eRF1 and eRF3 to 80S ribosomal complexes assembled on CrPV-STOP mRNA, depending on the presence of eEF2. Toe-prints corresponding to the pre-translocated ribosomal complexes (+14–15 nts from the CCU codon) are indicated by a black arrow. Toe-prints corresponding to the eRF1- or eRF1/eRF3-associated post-translocated ribosomal complexes (+18–19 nts from the CCU codon) are indicated by a red arrow. The +4 nt toe-print shift in post-translocated complexes includes the +2nt shift due to the presence of eRF1 (Alkalaeva et al., 2006). Additional toe-prints caused by the contacts of the IRES with the 40S subunit (at AA6161–6162) and with the 60S subunit (U6217 and G6183) are consistent with previous reports (Wilson et al., 2000b; Hellen and Pestova, 2003) and are shown in blue.
To fulfill their functional tasks, members of the IGR IRES family adopt a complex tertiary fold to facilitate specific interactions with the 40S subunit and the 80S ribosome in the intersubunit space (Schüler et al., 2006; Spahn et al., 2004). Domains 1 and 2 of the IGR IRES - containing PK II and PK III, respectively – tightly bind the 40S subunit and fold independently of domain 3 and can be therefore combined into a ribosome-binding domain (Costantino and Kieft, 2005; Jan and Sarnow, 2002; Nishiyama et al., 2003). The CrPV IRES structure has been derived independently by X-ray crystallography (Pfingsten et al., 2006) and by cryo-EM based de novo RNA modeling (Schüler et al., 2006). Crucial for the recruitment of the 40S subunit are the two RNA stem loops SL2.1 and SL2.3 of domain 2 of the IGR IRES (Jan and Sarnow, 2002) interacting with ribosomal proteins eS25 (rpS25 ; for a new nomenclature of ribosomal protein names see (Ban et al., 2014)) and uS7 (rpS5), respectively, at the head of the 40S subunit (Landry et al., 2009; Muhs et al., 2011; Schüler et al., 2006). Domain 3 containing PK I in turn is responsible for placing the start of the coding sequence into the ribosomal decoding center. A part of PK I mimics a tRNA anticodon stem loop (ASL) undergoing codon-anticodon interactions with a mRNA triplet (Costantino et al., 2008).
In the current model of IGR IRES mediated translation (for review see Thompson, 2012), the first Ala-tRNA is brought to the ribosome as a ternary complex with elongation factor 1A (eEF1A) and GTP once the binary 80S•IRES complex has been assembled from a 40S•IRES complex and a 60S subunit. Subsequently, the tRNA is translocated from the A-site into the P-site by elongation factor 2 (eEF2). However, while initial toe-print analysis (Jan et al., 2003; Pestova and Hellen, 2003; Wilson et al., 2000b) suggested that PK I is positioned in the ribosomal P site and the first GCU codon in the ribosomal A site, cryo-EM reconstructions of binary 80S•CrPV IRES complexes depicted the apical part of PK I in the A-site overlapping with the ASL of an A-site tRNA and forming A-site specific interactions with helices h18 and h34 of 18S rRNA (Schüler et al., 2006). Moreover, very recent cryo-EM reconstructions revealed that the apical part of PK I mimics tRNA/mRNA interaction in the decoding center of the A site (Fernández et al., 2014; Koh et al., 2014).
Thus, it follows that the PK I has to be moved out of the A-site, before the first Ala-tRNA containing ternary complex can decode the GCU codon (Schüler et al., 2006; Fernández et al., 2014; Koh et al., 2014). Indeed, it has been shown that stable binding of the first tRNA to 80S•CrPV IRES complexes requires not only eEF1A but also eEF2 (Yamamoto et al., 2007; Fernández et al., 2014). During this combined event, toe-printing indicates a movement of the 80S ribosome relative to the CrPV IRES RNA by 6 nucleotides (Jan et al., 2003) consistent with a movement of PK I (domain 3) of the IRES from the A-site to the E-site via the P-site. The need for an initial eEF2-dependent translocation of the CrPV IRES preceding the first tRNA binding has been recently reemphasized by the finding that the binary 80S•CrPV IRES complex co-exists in ribosome states with classical and rotated subunit rearrangements and therefore functionally mimics an elongation pre-translocational (PRE) state (Fernández et al., 2014; Koh et al., 2014). However, structural information about a potential post-translocational (POST) state of the CrPV IRES remains elusive.
Here, we have used a minimal system to study binding of release factor eRF1 to the ribosome using a stop codon-containing CrPV IRES (CrPV-STOP) mRNA. In agreement with a previous study (Jan et al., 2003) we see eEF2-dependent binding of eRF1 and also of eRF1•eRF3•GMPPNP. Interestingly, our visualization of these complexes using cryo-EM shows the IRES in a previously unseen, POST state and directly rationalizes the need for an unusual eEF2-dependent translocation of the CrPV IRES as a prerequisite for binding of A-site ligands during internal initiation. At the same time our structural analysis provides information about the binding modes of eRF1•eRF3•GMPPNP and eRF1 and suggests how a conformational change of eRF1 links the eukaryotic termination and recycling phases (Pisarev et al., 2010).
RESULTS
The binding of eRF1 to 80S•CrPV-STOP is dependent on eEF2-action
Direct structural investigations of CrPV IRES translocation is difficult, because binary 80S•CrPV IRES complexes are present predominantly in the PRE state and eEF2 dependent translocation may be followed by a spontaneous back-translocation unless the achieved POST state is stabilized by an A-site ligand. In the presence of the canonical aminoacyl-tRNA ligand however, not one but two eEF2 dependent translocation events can occur moving the aminoacyl-tRNA further from the A-site to the P-site. Interestingly, a modified CrPV IRES with the first alanine codon mutated to a stop codon (CrPV-STOP) allows the binding of eukaryotic release factor 1 (eRF1). In this case a change of the toeprint by +4 nucleotides was observed, but the binding of eRF1 was nevertheless dependent on eEF2 (Jan et al., 2003). We reasoned that in this system eRF1 contacts the stop codon in the ribosomal A-site of a translocated CrPV-STOP and, in contrast to a tRNA molecule, cannot be translocated to the P-site of the ribosome. Thus, eRF1 as an A-site ligand can be used to disentangle a possible first IRES translocation step from a second one.
To test this hypothesis, we assembled 80S•CrPV-STOP•eRF1 and 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complexes in vitro and analyzed the complexes by means of toe-printing and cryo-EM experiments. In order to obtain the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex, ribosomal subunits were first incubated with CrPV-STOP mRNA and eEF2 in the presence of GTP followed by addition of preformed eRF1•eRF3•GMPPNP complex. For the 80S•CrPV-STOP•eRF1 complex, ribosomal subunits were incubated with eEF2, eRF1 and GTP, simultaneously. Complex formation was monitored by toe-printing in situ (Fig. 1B). Toe-printing is a method to determine the position of e.g. the ribsoome on mRNA and involves primer extension by reverse transcriptase. The reverse transcriptase is blocked by the bound ribosome resulting in a cDNA, whose specific length indicates the 3′-border the ribosome-bound mRNA sequence. Incubation of 80S•CrPV-STOP complexes with eRF1 or with eRF1•eRF3•GMPPNP ternary complex resulted in a shift of the toe-print signal by +4 nt from +14–15 to +18–19 (Fig. 1B, lanes 8 and 10) indicating that the position of the ribosome on the CrPV-STOP RNA has changed. Importantly, the change in the toe-print signal was dependent on the presence of eEF2, in excellent agreement with previous studies (Jan et al., 2003; Yamamoto et al., 2007).
CrPV-STOP IRES is fully translocated by eEF2
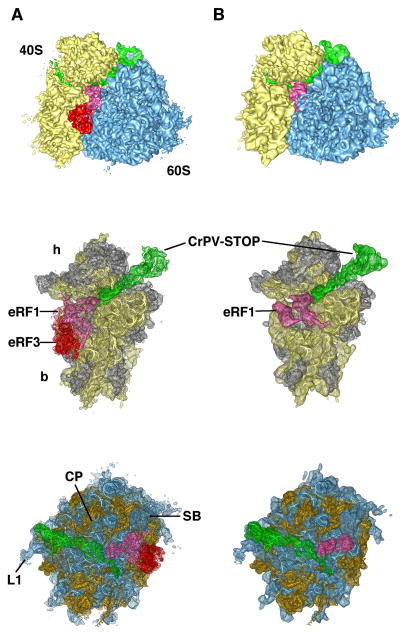
The resulting 80S•CrPV-STOP complexes were analyzed by multiparticle cryo-EM (Loerke et al., 2010) in order to overcome the expected substoichiometric occupancy of the factors and the overall heterogeneity of the samples (Fig. S1). The final reconstructions of the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP and 80S•CrPV-STOP•eRF1 complexes were obtained from 64,902 (10% of total number) and 109,596 particle images (22% of total number), respectively. The resolution was determined to 8.9 Å for the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex and 8.7 Å for the 80S•CrPV-STOP•eRF1, according to the FSC curve using the 0.5 cutoff criterion (Fig. S1). Both maps exhibit strong densities for the factors as well as the CrPV-STOP IRES (Fig. 2). The conformation of the present 80S complexes does not show intersubunit rotation or 40S subunit rolling and is overall similar to the POST state of the mammalian 80S ribosome (Budkevich et al., 2014).
Figure 2. Cryo-EM maps of the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP (A) and 80S•CrPV-STOP•eRF1 (B) termination complexes.
In vitro termination complexes were prepared on CrPV-STOP mRNA (green). Upper panel displays the 80S•CrPV-STOP•eRF1 eRF3•GMPPNP and the 80S•CrPV-STOP•eRF1 complexes with 40S subunit shown in yellow, 60S subunit in blue, eRF1 in hot pink and eRF3 in red. Middle and bottom panels display the corresponding 40S and 60S subunits with docked ligand models, respectively. Ribosomal RNA is shown in yellow or blue and ribosomal proteins in gray or orange for the 40S and 60S subunit, respectively. Landmarks of the 40S subunit are the head (h) and the body (b) domains. Landmarks of the 60S subunit: central protuberance (CP), the L1 stalk (L1) and the stalk base (SB).
The subnanometer resolution of our maps facilitated the analysis of interactions between release factors, CrPV IRES and the 80S ribosome. For an interpretation in molecular terms, we docked our previous cryo-EM based homology model of the human 80S ribosome (Budkevich et al., 2014), which was generated on the basis of S. cerevisiae 80S (Ben-Shem et al., 2011) and T. thermophila 40S and 60S X-ray structures (Klinge et al., 2011; Rabl et al., 2011). eRF3 was modeled based on the homology with its archaeal eEF1A ortholog (aEF1α) from the aEF1α/aRF1 complex (Kobayashi et al., 2012) and eRF1 by docking individual domains of the human factor (Mantsyzov et al., 2010; Song et al., 2000) into the cryo-EM density. The CrPV-STOP IRES has been modeled based on the previously published X-ray structure for domain 3 (Zhu et al., 2011) and cryo-EM model for domains 1 and 2 (Schüler et al., 2006). During preparation of our manuscript a cryo-EM structure of a 80S•CrPV IRES at high resolution has been published (Fernández et al., 2014) presenting a refined CrPV IRES model. However, the RMSD calculation for domains 1 and 2 between the recent structure (Fernández et al., 2014) and the previous model (Schüler et al., 2006) resulted in a value of only 2.9 Å indicating a high degree of similarity between both models. Only the SL2.1 of domain 2 has been modeled in a slightly different conformation in both studies (Fig. S2).
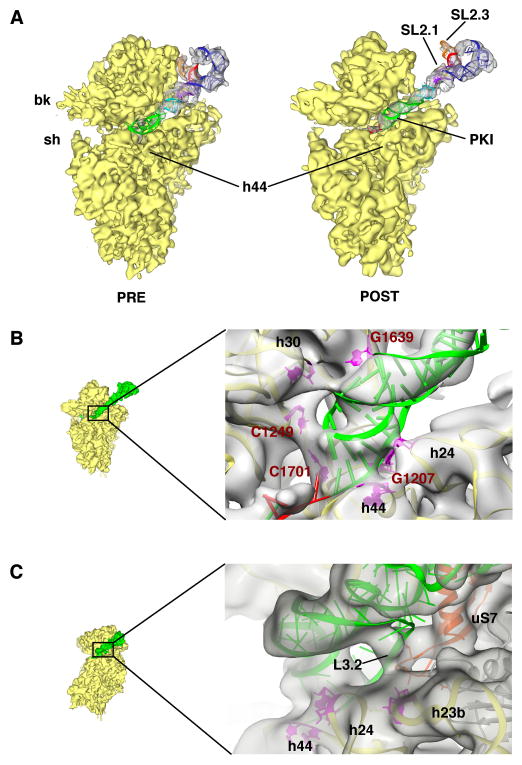
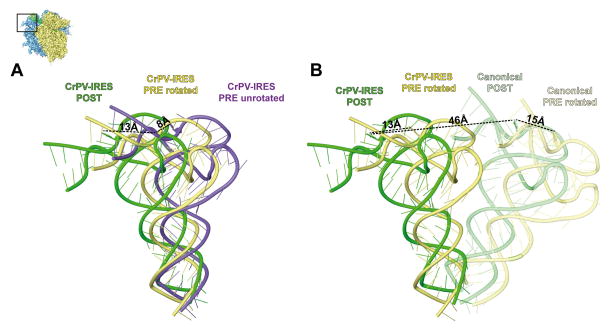
Comparison of the present complexes with the previous binary 80S•IRES structures (Fernández et al., 2014; Schüler et al., 2006) reveals a previously unseen binding state of the IRES in the presence of release factors (Fig. 3A). The CrPV-STOP IRES density in both of our complexes is indistinguishable in terms of location and conformation. Therefore the following interpretations concerning the IRES are based on the reconstruction of the 80S•CrPV-STOP•eRF1 complex but are also valid for the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex. As predicted from the toe-printing analysis the PK I element has been translocated by eEF2 to the P-site leading to a POST state (Fig. 4A). The 60S alignment of the PRE (Fernández et al., 2014) and POST state IRES complexes indicates a shift of the PK I by 22 Å, which is comparable to the distance between ASL of the A- and P-site bound tRNAs (Budkevich et al., 2011) (Fig. 4B). Moreover, like a tRNA ASL being translocated from the A to the P site, the PK I undergoes a rotational movement. Accordingly, the apical part of the PK I mimics an ASL-codon duplex not only in the A site of the PRE state but also in the present P site of the POST state like in the crystal structure of the PK I element bound to the bacterial 70S ribosome (Zhu et al., 2011).
Figure 3. Post-translocational state of the CrPV IGR IRES and its interactions with the 40S subunit.
(A) Extracted densities for CrPV IRES and ribosomal 40S subunit are shown for the PRE- (left) (Schüler et al., 2006) and POST-translocational state from the 80S•CrPV-STOP•eRF1 complex (right). (B) Close-up view of the apical tip of PK I (green). Interactions to conserved nucleotides G1639, C1249, C1701 and G1207 are depicted (red) as well as interactions to h24 and h44. (C) Interactions of loop L3.2 of CrPV IRES (green) with the ribosomal protein uS7 and helix 23b.
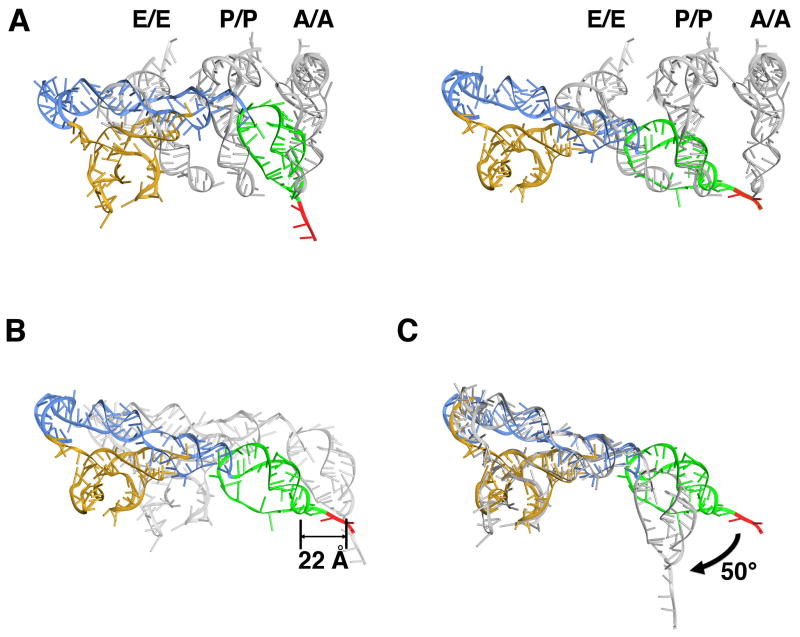
Figure 4. Comparison of the POST state IRES with tRNA positions and with PRE state IRES.
(A) Superposition of the PRE state IRES (left) (Fernández et al., 2014) and POST state IRES in the 80S•CrPV-STOP•eRF1 complex (right) with positions of canonical bound tRNAs (Budkevich et al., 2011) after 60S alignment. The CrPV IRES is colored according to the domain organization: domain 1 blue, domain 2 gold, domain 3 green. The first codon of the open reading frame is depicted in red. (B) Superposition of the PRE state IRES (gray) (Fernández et al., 2014) with the POST state IRES after corresponding 60S alignment. (C) Superposition of the PRE (gray) and POST state IRES after alignment of the corresponding ribosome-binding domains (domains 1 and 2).
However, our cryo-EM analysis clearly shows that not only the PK I element has been translocated but also that the ribosome-binding domain (domains 1 and 2) moves as a rigid body by ~25 Å towards the E-site region (Fig. 4A, B). While the lateral movement of the ribosome binding domain and PK I is comparable, the ribosome binding domain cannot undergo the same rotational movement as PK I because this would lead to a steric clash with the 40S subunit. As a consequence, the IRES changes its conformation and PK I undergoes a rotation relative to domains 1 and 2 of ~50° around the linker region (U6171-C6172) between domains 1 and 3 (Fig. 4C). Overall, the IRES undergoes a transformation from a bent conformation in the PRE state to a more stretched conformation in the POST state.
Interactions of the translocated CrPV IRES with the 80S ribosome
In the POST state, contacts between the PK I element and the 18S rRNA at positions around nucleotides C1249, G1639, C1701 and G1207 can be observed (Fig. 3B). Interestingly, all these interactions have been found in the X-ray structure of the isolated PK I element bound to the bacterial ribosome (Zhu et al., 2011) indicating that the P-site occupation in the bacterial complex corresponds to our present POST state of the IRES and not to the initial PRE state. In addition, we see contacts of PK I to h30, h24 as well as h44 (nt 1827–1829, (1495–1497 in E. coli)) of the 18S rRNA. As a consequence of the translocation event, the IRES loses the PRE state contacts (Fernández et al., 2014; Schüler et al., 2006) to the central protuberance of the 60S subunit and to the head of the 40S subunit. Also, the stem loops SL2.1 and 2.3 of CrPV IRES domain 2 have lost their contacts with uS7 and eS25, respectively, at the head of the 40S subunit and are solvent exposed. Both elements have been shown to play a crucial role for IRES-driven translation initiation and their interactions with the 40S head provide binding affinity (Jan and Sarnow, 2002; Nishiyama et al., 2003).
Instead with SL2.1 of the PRE state IRES, protein uS7 interacts with the POST state IRES via loop L3.2 of domain 3 (Fig. 3C). The contact seems to exist in the context of an interaction with helix 23b at the 40S body. The same site of the ribosome is occupied by the anticodon-stem loop (ASL) of E-site bound tRNA in the bacterial termination (Jin et al., 2010) as well as in the mammalian POST complex (Budkevich et al., 2014). In the PRE state 80S•CrPV IRES structures (Fernández et al., 2014; Schüler et al., 2006), the density assigned to L3.2 was fragmented and also in the crystal structures of domain 3 bound to the P-site of the bacterial ribosome (Zhu et al., 2011) the loop was disordered, revealing a dynamic, flexible state of this IRES part. However, in our present structures the solid density at the contact site between L3.2 and the 40S subunit indicates a potential ordering of the loop caused by interactions with the ribosome (Fig. 3C).
The interactions of the IRES and the 60S subunit are confined to interactions of IRES domain 1 with the L1 stalk. Like in the binary 80S•IRES structure (Fernández et al., 2014; Schüler et al., 2006) the internal loop L1.1 interacts with rpL1 and 28S rRNA helices 76–78 of the L1 stalk. This highly mobile element of the 60S subunit is part of the E-site. The L1 stalk can swing in and out to facilitate interactions with tRNA in P/E or E/E states and is known to exist in at least three conformational states, i.e. closed, open and an intermediate half-open state (Cornish et al., 2009). In both POST state IRES complexes, we find the L1 stalk in a more open position than in PRE state IRES complexes (Fernández et al., 2014; Schüler et al., 2006) (Fig. 5A ) or in the canonical POST state with P/P- and E/E-tRNAs (Budkevich et al., 2014) (Fig. 5B ), and even in a more outward position than in the ribosome with a vacant E-site (Becker et al., 2011). This extended outward position is apparently required to adapt the POST state IRES. During IRES translocation the L1 stalk moves by ~13 Å. Remarkably, comparison of the canonical 80S complex carrying classical P/P and E/E tRNAs (canonical POST) (Budkevich et al., 2014) with canonical PRE complex carrying hybrid A/P and P/E tRNAs (rotated PRE) (Budkevich et al., 2011) reveals a similar shift of ~15 Å for the L1 position (Fig. 5B). Accordingly, despite the dramatic overall difference in the L1 stalk position caused by IRES binding (46 Å between canonical POST and IRES bound POST), the relative movement of the L1 stalk during the translocation step is preserved.
Figure 5. Interaction of the POST state IRES with the L1 stalk and comparison of L1 positions in different ribosomal states.
(A) Depiction of three distinct L1 positions during translocation of the IRES, distance measurements were made between axes of mass-centers. From the unrotated PRE state (purple) to the rotated PRE state (yellow) (both from Fernández et al., 2014) L1 moves 14 Å towards the E-site. Upon translocation to the POST state (green) a movement of another 13 Å can be observed. (B) Comparison of the L1 stalk position in rotated 80S•CrPV IGR IRES complex (yellow) (Fernández et al., 2014) and 80S•CrPV-STOP (POST) complex (green) with corresponding canonical complexes (Budkevich et al., 2014).
The 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex is structurally related to the mammalian decoding complex
The POST state 80S•CrPV-STOP complex allows stable binding of eRF1•eRF3•GMPPNP or eRF1. The structure of the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex is overall similar to a previous structure of an equivalent canonical termination complex (des Georges et al., 2014) in terms of ribosome conformation and the binding sites of eRF1 and eRF3. Accordingly, the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex faithfully represents a canonical termination complex. The POST state CrPV-STOP IRES with a P-site ASL mimic and the stop codon in the ribosomal decoding center are sufficient for stop codon recognition by eRF1. Thus, the CrPV-STOP construct can be used as a tool to study termination factor binding in a minimal system.
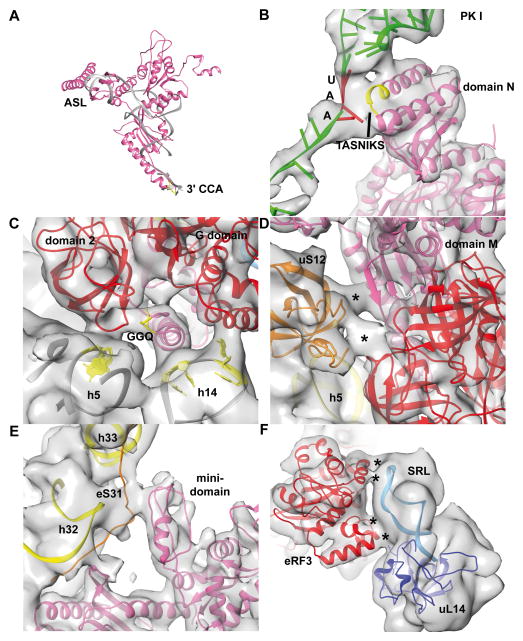
Comparison between the present 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex and our recent cryo-EM map of the mammalian decoding complex (Budkevich et al., 2014) reveals striking similarities. Domains N and M of eRF1 mimic the tRNA in the A/T state and eRF3 resembles eEF1A (Fig. S3). The M domain of eRF1 overlaps with the acceptor stem of the A/T tRNA and even appears to mimic the 3′-CCA-end of the tRNA (Fig. 6A). The GGQ motif (aa 183–185) of eRF1’s M-domain is in a similar position to C75 of the A/T tRNA (Budkevich et al., 2014) and both elements appear to interact with a homologous region of domain 2 (aa 441–442) of eRF3 or eEF1A, respectively (Fig. 6C).
Figure 6. The 80S•eRF1•eRF3•GMPPNP complex strongly resembles the eukaryotic decoding complex.
(A) Superposition of the A/T tRNA model (gray) (Budkevich et al., 2014) with the eRF1 model (pink) from the 80S•eRF1•eRF3•GMPPNP complex. The GGQ motif at the tip of the long α5 helix is colored in yellow (B) Interactions of the N domain of eRF1 with the stop codon of CrPV IRES mRNA (green) via the TASNIKS motif (yellow). The UAA stop codon is shown in red. (C) GGQ motif of eRF1 contacts domain 2 of eRF3 (red) in context with h5 of the 18S rRNA. Furthermore, eRF1 contacts h14 of the 18S rRNA via its α5 helix, interaction not seen in the decoding complex. (D) Ribosomal protein uS12 positioned at the shoulder of the 40S forms a network of interactions including domains 2 and 3 of eRF3 as well as M domain of eRF1 at the position which would overlap with nt 68 of the A/T tRNA. (E) eRF1 specific contact of the minidomain of eRF1 with eukaryotic specific ribosomal protein eS31 at the head of the 40S. (F) eRF3 contacts the SRL via its helical expansion of SW I region as well as via a eukaryotic insertion around Glu324-Lys325.
Furthermore, the M domain of eRF1 interacts with ribosomal protein uS12, localized at the shoulder of the 40S (Fig. 6D). An analogous contact with nucleotide 68 in the acceptor stem of the A/T tRNA is present in bacterial (Schmeing et al., 2009) and mammalian decoding complexes (Fig. 6D). Moreover, a mammalian specific interaction between the C-terminal end of uS12 and the translational GTPase factor can be observed in both systems. Domain C of eRF1 is interacting with the stalk base (H43/H44) of the 28S rRNA at the 60S subunit and with eS31 at the head of the 40S via the characteristic mini-domain of eRF1 (Fig. 6E). The eS31 interaction seems to be unique for eRF1, as it could not be observed for Dom34 in the Dom34/Hbs1-complex (Becker et al., 2011) or A/T tRNA in the decoding complex (Budkevich et al., 2014) because both are lacking a similar domain. Mutational analysis indicated that the mini-domain influences the specificity of eRF1 for certain stop codons during stop-codon recognition (Mantsyzov et al., 2010), which may explain its exclusive presence in eRF1.
The mammalian decoding complex with a GMPPNP stalled ternary complex has been visualized in two substates, i.e. a codon sampling state and a codon recognition state (Budkevich et al., 2014). A smaller rotational movement of the ternary complex was found to have an influence on the exact position of eEF1A leading to full interactions with the functionally important sarcin-ricin loop (SRL, H95 of 28S rRNA) of the ribosome’s GTPase associated center only in the codon-recognition state. eRF3 seems to be closer to the codon recognition state and we observe interactions to the 60S subunit, in particular the SRL, via the G-domain (Fig. 6F) as well as to h5, h14 and uS12 of the 40S subunit incorporating the helical insertion and domain 2.
Structure of the 80S•CrPV-STOP•eRF1 complex
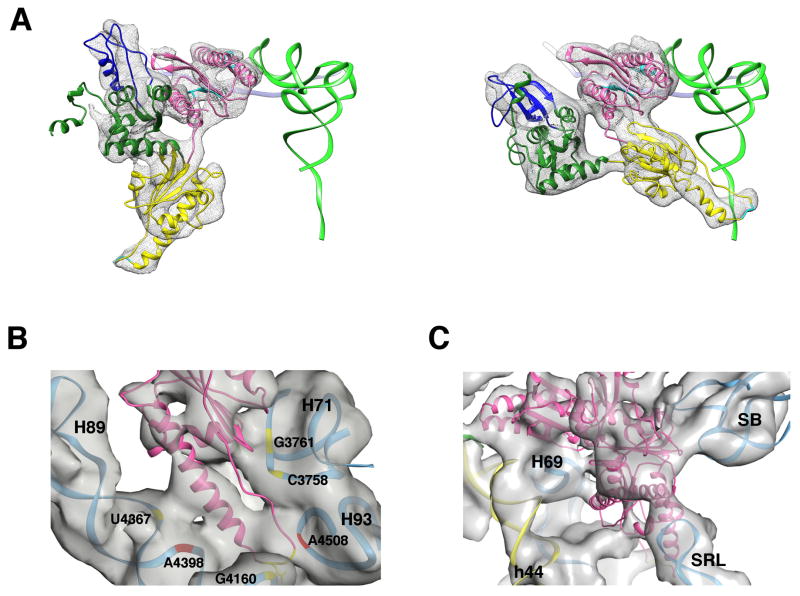
In the context of our minimal system we were able to directly visualize eRF1 within the 80S•CrPV-STOP•eRF1 complex in the absence of eRF3. This state of eRF1 resembles an important stage between eRF1 delivery to the ribosome by eRF3 and the recycling step mediated by ABCE1. In current models of termination the structure of this intermediate is unclear (Becker et al., 2012; Preis et al., 2014). When compared with the eRF3 bound state, the orientation of the M domain of eRF1 is dramatically changed and rotates by nearly 150°, orienting the GGQ motif at the tip of the long α5 helix towards the peptidyl transferase center on the 60S subunit (Fig. 7A). eRF1 in its catalytically active state has been proposed to adopt a similar conformation as its homolog Dom34 within the Dom34/Rli1 complex involved in mRNA decay (Franckenberg et al., 2012). Indeed, both factors adopt a similar but not identical orientation. Domain C of eRF1 overlaps nicely with domain C of Dom34, whereas domains N and M of eRF1 are rotated around domain C by additional ~18°. As a result, the GGQ motif of eRF1 is positioned more closely to the peptidyl-transferase center (PTC). It appears that the presence of a stop codon in the A-site and an ASL mimic in the P-site is requisite and sufficient for the binding and accommodation of eRF1 to the ribosome. Thus, eRF1 requires neither eRF3 nor ABCE1 to adopt its catalytically active conformation on the ribosome, bringing the GGQ motif close to its substrate in the PTC.
Figure 7. Ribosomal contacts of eRF1 in the 80S•CrPV-STOP•eRF1 complex.
(A) eRF1 conformation in the 80S•CrPV-STOP•eRF3•eRF1•GMPPNP (left) and the 80S•CrPV-STOP•eRF1 complex (right). As a marker, the canonical P-site bound tRNA is shown in green. eRF1 model is colored according to its domain organization: N domain in pink, M domain in yellow, C domain in green with the minidomain in blue. (B) The M domain of eRF1 is positioned in the PTC on the 60S subunit and forms conserved (red) as well as unique (yellow) ribosomal contacts. (C) In the 80S•eRF1 complex, the tip of the SRL interacts with the C domain of eRF1, which is analogous to the eukaryote-specific interaction involving the T-loop of A/T tRNA seen in the decoding complex (Budkevich et al., 2014).
Despite the lack of sequence homology, in bacterial termination complexes a similar orientation of domain 3 comprising the extended α7 helix with GGQ motif of RF1 or RF2 has been observed (Fig. S4) (Jin et al., 2010; Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). Consistent with this observation, several ribosomal contacts of the M domain of eRF1 are preserved between bacteria and eukaryotes. The M domain appears to make extensive interactions in the PTC with conserved nucleotides A4508, U4398 and C4465 of the 28S rRNA (human numbering, corresponding residues in E. coliare A2602, U2492 and C2558, respectively) ( Fig. 7B). The presence of these evolutionary preserved interactions corroborate that eRF1 bound to the present 80S•IRES complexis in a biologically relevant intermediate state of the pathway of eukaryotic translation termination/recycling.
In addition, we see several interactions between domain M of eRF1 and the ribosome, which have not been described for bacterial termination complexes (Fig. 7B). Two of these eukaryotic specific contacts appear to involve H71 of the 28S rRNA, around G3761 and C3758 and Ser253 and Ala226 of the M domain, respectively. Further eukaryotic specific contacts seem to exist around G4367 of H89 and Tyr197 of the long α5-helix of domain M and, less obviously, between the G4158 region of H80 and the GGQ motif of eRF1.
In the bacterial system, residues 1913–1915 in the loop of H69 of 23S rRNA have been shown to play a crucial role in stabilizing the rearrangements of the switch loop of RF1/2 required for proper positioning of the extended α7 helix and docking of the GGQ motif in the PTC (Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). However, these events are strongly interdependent with the recognition of the stop codon and subsequent molecular rearrangements in A1492–1493 of h44 of the 16S rRNA. As far as the stop codon is recognized, A1492 flips out from its position whereas A1493 remains stacked within h44. The gap caused by flipping-out of A1492 is filled by A1913 of 23S rRNA, which stacks on A1493 (Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). We do not observe such an interaction in our maps, suggesting a different mode of stop codon recognition in eukaryotes. However, higher resolution will be required to interpret the changes in the decoding centre in detail.
Due to a rotation of the C domain of eRF1 by ~39° the contact formed by the minidomain of the factor and eS31 at the head of the 40S, as seen in the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP structure, is disrupted. Furthermore, a contact not present in the 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex is formed between the tip of the SRL and Ser295 of eRF1 (Fig. 7C). Interestingly, a similar interaction was described for the mammalian decoding complex between the T-loop of the A/T tRNA and the SRL (Budkevich et al., 2014) (Fig. S3) but not in the homologous bacterial system.
Discussion
The mechanism of IGR IRES facilitated internal initiation
IRES-containing viral RNAs are capable of initiating translation with a strongly reduced set of initiation factors (Wilson et al., 2000b). In the case of the CrPV IGR IRES family neither cellular initiation factors nor initiator Met-tRNA are needed for synthesis of viral gene products by the host. IGR IRESs start translation from the A-site and only need elongation factor eEF2 to assemble elongation competent 80S ribosomes from 40S and 60S subunits. Initial biochemical studies suggested that after subunit association the IRES occupies the ribosomal P site and sets the translation frame so that the first codon is positioned in the A site. In contrast, we present here cryo-EM maps of 80S•CrPV-STOP•eRF1•eRF3•GMPPNP and 80S•CrPV-STOP•eRF1 complexes directly showing that an eEF2-dependent translocation resulting in a POST state of the IRES is required to allow the first A-site occupation.
Our finding is in excellent agreement with previous structural studies (Schüler et al., 2006; Fernández et al., 2014; Koh et al., 2014) demonstrating, that in the binary 80S•CrPV IRES complex domain 3 of the CrPV IRES is placed between the ribosomal A- and P-sites with the apical part of the PK I element mimicking an ASL of an A-site bound tRNA. Consequently, domain 3 has to move completely out of the A-site to allow the decoding of the first codon at the 3′ edge of the IRES structure (Schüler et al., 2006) and in line with previous experiments showing that binding of the first aa-tRNA•eEF1A•GTP ternary complex requires the action of eEF2 (Yamamoto et al., 2007) an initial eEF2-dependent translocation of the IRES on binary 80S•IRES complex has been suggested (Fernández et al., 2014; Koh et al., 2014). Our present cryo-EM maps show that the action of eEF2 leads to the movement of the entire CrPV IRES molecule towards the ribosomal E-site resulting in the previously unseen POST state with the apical part of the PK I structure mimicking the ASL of a tRNA in the P/P-state (Fig. 4A).
It appears that during the translocation reaction the IRES becomes more loosely bound. In particular, the two important stem loops SL2.1 and 2.3 of CrPV IRES domain 2 no longer interact with the ribosome in the POST state. However, it has been shown previously by mutagenesis studies that a major part of the binding energy of the IRES for the ribosome is derived by the SL2.1 and 2.3 interactions with uS7 and eS25, respectively (Jan and Sarnow, 2002). Therefore, the PRE state may be energetically preferred over the POST complex and it is therefore very likely that after translocation and dissociation of eEF2•GDP the A-site has to be occupied by a ligand in order to stabilize the POST complex and to prevent spontaneous back-translocation of the IRES. A short life time of the translocated binary 80S•CrPV IRES complex could be the reason, why it has been difficult to detect this state and why the presence of release factors is needed to shift the toe-print signal (Fig. 1B, lane 4 versus lanes 8–10). We note that during our multiparticle processing we obtained a sub-population of the binary 80S•IGR-IRES complex in the PRE but not in the POST state.
The question arises, why the IGR IRES family employs not the more simple mechanism with direct P-site binding of the PK I structure and only one unusual translocation (Pestova and Hellen, 2003; Wilson et al., 2000b) but a more complicated one with two unusual translocation events. A possible reason may be that during internal initiation the need for a high binding affinity to efficiently recruit the translational apparatus has to be balanced with the need to dissociate the IRES again during downstream events. It may be more efficient to partition the energetic penalty for IRES dissociation into two translocation steps or even three, when we consider the translocation of the first elongation complex with two tRNAs in A and P sites and the IRES possibly still interacting in the ribosomal E site.
A second factor may be the general task of translation initiation to thread the open reading frame into the mRNA entry tunnel of the 40S subunit. The CrPV IRES (Spahn et al., 2004) and also the unrelated HCV IRES (Spahn et al., 2001) as well as the canonical initiation factors eIF1 and eIF1A (Passmore et al., 2007) all induce a similar conformational change in the ribosomal 40S subunit to open the latch of the mRNA entry tunnel for inserting the mRNA. In this context initial positioning of the PK I element into the A site places the neighboring first sense codon directly into the opened mRNA entry tunnel and after closing the latch, the upstream part of the mRNA will be stably bound. In the case of P site location however, the coupling to mRNA insertion into the entry tunnel would be less direct due to the flexible character of the single stranded RNA. Thus, the initial PK I positioning into the A site rather than the P site appears advantageous in the context of mRNA loading.
Eukaryotic translation termination
Eukaryotic translation termination is mediated by two release factors: eRF1 and eRF3. The position of the release factors in our 80S•CrPV-STOP•eRF1•eRF3•GMPPNP complex as well as the overall conformation of the complex is essentially the same compared to the canonical termination complex (des Georges et al., 2014; Taylor et al., 2012). We can therefore conclude that using the CrPV-STOP RNA as a tool to study termination factor binding in a simplified minimal system is a valid approach. It follows that domain 3 of the CrPV IRES resembles the ASL of a P-site tRNA sufficiently well and that the presence of the IRES does not alter the termination complex in a major way.
The minimal system allowed us to analyze eRF1 binding in the absence of eRF3 and to visualize the factor in a previously unseen state in the 80S•CrPV-STOP•eRF1 complex. This complex mimics the intermediate state of the eukaryotic termination process that arises after binding of eRF1•eRF3•GTP, decoding of the stop codon, GTP hydrolysis and subsequent dissociation of eRF3•GDP, leaving eRF1 bound to the ribosome alone. In the next step, the hydrolysis of peptidyl tRNA is induced by eRF1 followed by release of the peptide chain and recycling of the post-termination complex. Our complex reveals major rearrangements in eRF1 providing structural insights into the “active” conformation of eRF1. Accordingly, the M-domain of the factor is reoriented compared to its position when bound to eRF3 positioning the GGQ motif into the peptidyl transferase centre. The adopted conformation of the factor and its ribosomal contact sites suggest an idea where eRF1 combines features of a eukaryotic A/T and A/A tRNA in one molecule by mimicking the A/T elbow region and the A/A acceptor stem, respectively.
Taking into account that the 80S•CrPV-STOP•eRF1 complex has been prepared without addition of eRF3, the presence of a stop codon in the ribosomal A-site and the ASL in the P-site are sufficient for eRF1 to adopt the “active” conformation regardless of the presence and activity of eRF3. The results are in good agreement with biochemical studies showing that eRF1 can mediate peptide hydrolysis in vitro alone (Alkalaeva et al., 2006). The established role of eRF3 is to facilitate efficient binding of eRF1 to ribosomal pre-termination complexes. However, the finding, that eRF1 can adopt the active conformation on the ribosome without additional factors suggests that eRF3 may have an additional function to prevent eRF1 from initiating premature translation termination and recycling. Regarding the cellular levels (Ghaemmaghami et al., 2003) of eRF1, eRF3, GDP and GTP it is reasonable to presume that eRF1 within the cell is found predominantly as ternary complex with eRF3 and GTP. It is tempting to speculate that eRF3, like the homologous elongation factor EF-Tu/eEF1A, may be involved in enhancing the accuracy of termination by separating codon recognition from peptidyl-hydrolysis via interposed GTPase activation and eRF1 accommodation steps.
Experimental Procedures
Cryo-EM of mammalian termination complexes
Mammalian termination complexes were prepared with isolated ribosomal subunits from RRL and in vitro transcribed CrPV-STOP RNA. Translocation was achieved by incubation with eEF2•GTP. In case of 80S•CrPV-STOP•eRF1, the release factor was added directly to the translocation reaction, for the 80S•CrPV-STOP•eRF1•eRF3 sample a preformed eRF1•eRF3•GMPPNP complex was used. Micrographs were recorded with a FEI Tecnai G2 Polara electron microscope and multiparticle refinement was carried out with SPIDER as described previously (Loerke et al., 2010; Penczek et al., 2006a, 2006b). Further details can be found in the Supplemental Experimental Procedures.
Modeling
Modeling of the translocated CrPV IRES was done based on the pre-translocated CrPV IRES (Schüler et al., 2006) and a crystal structure of PK I (Zhu et al., 2011) by MDfit (Ratje et al., 2010), COOT (Emsley and Cowtan, 2004) and ERRASER (Chou et al., 2013). The initial structural model for eRF1 was built by domain-wise rigid-body fitting (Mantsyzov et al., 2010; Song et al., 2000) accompanied by MDfit (Ratje et al., 2010; Whitford et al., 2011), COOT and CNS (Brünger et al., 1998). eRF3 was modeled based on homology to aEF1a (Kelley and Sternberg, 2009; Kobayashi et al., 2012; Tung and Sanbonmatsu, 2004) and yeast eRF3 (Kong et al., 2004).
Supplementary Material
See also Figure S1. Heterogeneity in the 80S termination complexes and resolution curves.
See also Figure S2. Comparison of the CrPV IRES structure modeled based on the cryo-EM map from Schüler et al. (Schüler et al., 2006) with the structure based on the high resolution cryo-EM map from Fernández et al. (Fernández et al., 2014)
See also Figure S3. Comparison of the eEF1A•A/T complex with the eRF1•eRF3 complex and the interaction with the SRL.
See also Figure S4. Overlap of the bacterial RF1 structure with eRF1 model.
Acknowledgments
We thank L. Yu. Frolova for eRF1 and eRF3 expression vectors, and E. Alkalaeva for constructing the transcription vector for CrPV-STOP mRNA. This work was supported by Senatsverwaltung für Wissenschaft, Forschungund Kultur Berlin (UltraStructureNetwork, Anwenderzentrum), HFSP grant RGP0062/2012 to C.M.T.S, T.V.P and K.S., and by NIH grant GM80623 to T.V.P.
Footnotes
Accession numbers
The electron density maps of mammalian termination complexes were deposited in the Electron Microscopy Data Bank (European Molecular Biology Laboratory-European Bioinformatics Institute, Cambridge, UK) with the accession numbers EMD-2810 and EMD-2813. Models for the mammalian release factors and the post-translocated CrPV-IRES have been deposited in the Protein Data Bank database with PDB IDs 4d5n, 4d5l, 4d5y, 4d5z (eRF1 complex) and 4d61, 4d66, 4d67, 4d68 (eRF1/eRF3 complex).
AUTHOR CONTRIBUTIONS
M.M. carried out sample preparation, data acquisition, image processing, structure determination and interpretation of the 80S•CrPV-STOP•eRF1 complex. T.H. carried out sample preparation, image processing, structure determination and interpretation of the 80S•CrPV-STOP•eRF1•eRF3 complex. T.V.P. and M.A.S. purified factors and ribosomal subunits and performed toeprint analysis. T.M. was responsible for electron. K.Y.S. made MDfit simulations for the CrPV IRES and the eRF1 structure. T.H. refined molecular models for eRF1, eRF3 and the CrPV IRES. T.V.P. and C.M.T.S. designed the study. T.V.P., M.M. and T.H. prepared the figures. M.M., T.H., T.V.P., K.Y.S. and C.M.T.S. interpreted results and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24C:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat Methods. 2013;10:74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson R, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernández IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Bai X, Murshudov G, Scheres SHW, Ramakrishnan V. Initiation of Translation by Cricket Paralysis Virus IRES Requires Its Translocation in the Ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckenberg S, Becker T, Beckmann R. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr Opin Struct Biol. 2012;22:786–796. doi: 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Des Georges A, Hashem Y, Unbehaun A, Grassucci Ra, Taylor D, Hellen CUT, Pestova TV, Frank J. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic Acids Res. 2014;42:3409–3418. doi: 10.1093/nar/gkt1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hertz MI, Thompson SR. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011;411:355–361. doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci U S A. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci U S A. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley La, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Saito K, Ishitani R, Ito K, Nureki O. Structural basis for translation termination by archaeal RF1 and GTP-bound EF1α complex. Nucleic Acids Res. 2012;40:9319–9328. doi: 10.1093/nar/gks660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc Natl Acad Sci U S A. 2014;111:9139–9144. doi: 10.1073/pnas.1406335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C, Ito K, Walsh Ma, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Loerke J, Giesebrecht J, Spahn CMT. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- Mantsyzov AB, Ivanova EV, Birdsall B, Alkalaeva EZ, Kryuchkova PN, Kelly G, Frolova LY, Polshakov VI. NMR solution structure and function of the C-terminal domain of eukaryotic class 1 polypeptide chain release factor. FEBS J. 2010;277:2611–2627. doi: 10.1111/j.1742-4658.2010.07672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Penczek Pa, Yang C, Frank J, Spahn CMT. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006a;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Penczek Pa, Frank J, Spahn CMT. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol. 2006b;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Conservation and Diversity Among the Three-dimensional Folds of the Dicistroviridae Intergenic Region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CUT, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin Ma, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CUT, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio-garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014:1–7. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci U S A. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek Pa, Westhof E, Spahn CMT. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci Ra, Penczek Pa, Zhou K, Doudna Ja, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Jan E, Mulder A, Grassucci Ra, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci Ra, Pestova TV, Frank J. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci U S A. 2012;109:18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012;20:558–566. doi: 10.1016/j.tim.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CS, Sanbonmatsu KY. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys J. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CMT, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000a;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of Protein Synthesis from the A Site of the Ribosome. Cell. 2000b;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Korostelev A, Costantino DA, Donohue JP, Noller HF, Kieft JS. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci U S A. 2011;108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See also Figure S1. Heterogeneity in the 80S termination complexes and resolution curves.
See also Figure S2. Comparison of the CrPV IRES structure modeled based on the cryo-EM map from Schüler et al. (Schüler et al., 2006) with the structure based on the high resolution cryo-EM map from Fernández et al. (Fernández et al., 2014)
See also Figure S3. Comparison of the eEF1A•A/T complex with the eRF1•eRF3 complex and the interaction with the SRL.
See also Figure S4. Overlap of the bacterial RF1 structure with eRF1 model.