Abstract
Lipid droplets (LDs) are found in most cells, where they play central roles in energy and membrane lipid metabolism. The de novo biogenesis of LDs is a fascinating, yet poorly understood process involving the formation of a monolayer bound organelle from a bilayer membrane. Additionally, large LDs can form either by growth of existing LDs or by the combination of smaller LDs through several distinct mechanisms. Here, we review recent insights into the molecular process governing LD biogenesis and highlight areas of incomplete knowledge.
Lipid droplets (LDs) are ubiquitous, dynamic cellular organelles that serve as important reservoirs of lipids. These lipids provide energy and serve as substrates for membrane synthesis, making LDs crucial metabolic hubs. Indeed, many of the enzymes that synthesize phospholipids (PLs), triacylglycerols (TGs), and their intermediates, as well as lipases and lipolytic regulators, localize to LD surfaces. In addition to their known role in lipid metabolism, increasing evidence suggests that LDs also participate in protein degradation [1,2], response to ER stress [3], protein glycosylation [4], and pathogen infection [5]. Further details about the general aspects of LD cell biology and physiology are discussed in numerous recent reviews [6–10]. However, despite recent focus and the application of new technologies to study LDs, a number of basic questions remain unanswered. Chief among these are the molecular processes governing how LDs form and grow. Here, we review recent advances in this area.
Lipid Droplet Composition
LDs span a wide range of sizes (tens of nm to several microns in diameter) and can grow and shrink in response to cellular signals. LD cores contain neutral lipids, predominantly sterol esters (SE) or TGs, and depending on cell type, may also include retinyl esters, waxes, and ether lipids. These lipids are surrounded by a phospholipid monolayer comprising mostly phosphatidylcholine (PC) and phosphatidylethanolamine (PE) [11]. The surface composition is highly relevant to regulating LD size and their ability to interact with other LDs or organelles, such as the endoplasmic reticulum (ER) ([12,13] and reviewed in [6,14,15]).
LD surfaces are decorated by specific proteins, and, not surprisingly, many of these function in lipid metabolism. LD proteins have been identified by microscopy analyses of individual proteins in yeast and mammalian cells [16,17] and through studies employing non-biased mass spectrometry analyses (reviewed in [18]). The latter approach is highly sensitive, but not always specific. From these data, it seems likely that most LDs have in the neighborhood of 50–200 different proteins at their surface (for example, see [4]). The composition of proteins can differ between LDs of different sizes [19–21] or different lipid compositions [22] within the same cell. Specific targeting signals for LD proteins are reviewed elsewhere [6,23].
LD Formation
LDs could either form de novo or could be derived from existing LDs by fission. Most evidence favors the former process as a major source, however, fission of LDs has been observed [24]. De novo formation of LDs in eukaryotes occurs from the ER [25,26], where neutral lipids are synthesized [27]. Precisely how LDs form, however, remains mostly unanswered. Here we present a model for LD formation in three stages (Figure 1): (1) neutral lipid synthesis, (2) lens formation (intra-membrane lipid accumulation), and (3) drop formation. We highlight recent advances in the understanding of each of these stages.
Figure 1.

A step-wise model of lipid droplet formation. Lipid droplets form in at least three discrete steps. (a) Neutral lipids are synthesized in the ER and accumulate within the bilayer. Neutral lipids are highly mobile in the bilayer and may spontaneously aggregate based on thermal fluctuations and electrostatic interactions with integral membrane proteins or other lipids. (b) Once the local concentration of neutral lipid reaches a critical threshold, a lens will form as the oil phase coalesces. (c) As the lens accumulates additional neutral lipids, the bilayer deforms and a nascent lipid droplet buds into the cytoplasm, possibly via a de-wetting mechanism. The nascent droplet might remains attached to the ER or separate completely.
Step 1: Neutral lipid synthesis
Neutral lipids are synthesized by enzymes of the membrane-bound O-acyltransferase (MBOAT) [i.e., acyl-CoA:cholesterol acyltransferase (ACAT)-1, ACAT2, and acyl-CoA:diacylglycerol acyltransferase (DGAT)-1] and DGAT2 gene families [28]. Generally, these enzymes localize to the ER, where they encounter their substrates. One common substrate is fatty acyl-CoA produced by acyl-CoA synthetase (ACSL) enzymes (reviewed in [29]), which activate fatty acids for use in metabolic pathways. Fatty acyl-CoAs join with lipid alcohols to form neutral lipids. For example, DGAT enzymes utilize fatty acyl-CoAs and diacylglycerol to form TGs. Similarly, cholesterol esters are produced by condensation of fatty acyl-CoA with cholesterol. Neutral lipid synthesis is essential for LD formation. Yeast lacking all enzymes of neutral lipid synthesis are viable but lack detectable LDs [30]. In mammals, knockout mouse studies show that ACAT1, ACAT2, and DGAT1 are not essential for life, whereas DGAT2 is [28]. DGAT2-deficient mice die shortly after birth due to lack of energy stores and skin defects related to essential fatty acid deficiency [31], Neutral lipid synthesis in the ER functions, in part, to maintain membrane lipid homeostasis, specifically by preventing the accumulation of excess lipids such as cholesterol or diacylglycerol.
Several different enzyme isoforms (for ACSL, glycerol-3-phosphate acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), phosphatidic acid phosphohydrolase (PAP), and DGAT) catalyze each step of the Kennedy pathway for TG synthesis. This raises the possibility that different isoforms prefer specific substrates (e.g., exogenous versus de novo–synthesized fatty acids). For example, knockout and inhibitor studies indicate that GPAT4 [32] and DGAT1 [33–35] appear to prefer exogenous or lipolysis-derived fatty acids, while GPAT1 [32] and DGAT2 [33–35] handle mostly endogenously synthesized fatty acids. Several isoforms, such as DGAT2, AGPAT3, and GPAT4, also localize to LDs under conditions of fatty acid excess (discussed below) [21,36–38]. Furthermore, recent studies show that DGAT2 prefers the substrate sn-1,3-diacylglycerol, one of the products of lipolysis by adipose triglyceride lipase (ATGL) on LDs [39]. This supports a role for DGAT2 in LD-localized re-esterification of fatty acids. In contrast, GPAT1, AGPAT1, AGPAT2, and DGAT1 localize to the ER [21,37,40]. Interestingly, the functions of these isoforms are not exclusively correlated with localization, as DGAT1 and DGAT2 can compensate for each other under certain conditions [41].
Step 2: Neutral lipid accumulation and lens formation
At relatively low concentrations, neutral lipids will accumulate between leaflets of the ER bilayer. Several groups have measured TGs in cellular membranes in the range of 3–7 (w/w)% [42,43], in agreement with biophysical [44] and in silico predictions [45] for the capacity of bilayer membranes to hold TG. As the concentration increases, lipid lenses may form in the ER (Figure 1), though this has not been clearly demonstrated. A simulation predicts that TGs form in “blisters” in the bilayer of at least 17 nm in diameter [45].
What determines lens formation sites within the ER is an open question. Recent data in yeast [26] and COS cells ([46] and J. H. and R.F., unpublished observations) indicate that they form in discrete foci dispersed throughout the ER. In the starvation-refeeding model in COS cells, LDs appear to form at pre-existing sites marked by an LD-targeted protein, suggesting that lenses form at sites of previous LDs [46]. Localization studies have found DGAT1 and DGAT2 are continuously distributed along the ER [37], but not perfectly overlapping [47], suggesting that neutral lipids accumulate in spots that are dissociated from the enzymes that synthesize them. Interestingly, the plant homologue of GPAT4 (called GPAT9) localizes to the same ER subdomains as DGAT2, suggesting that they might form sub-complexes for neutral lipid synthesis within the ER [47]. In yeast, the DGAT2 homolog Dga1p also distributes throughout the ER in the absence of LDs [25]. Thus, it is possible that TGs are synthesized throughout the ER and diffuse through the bilayer to LD formation sites. Alternatively, enzyme activity for TG synthesis may occur specifically at regions of the ER where LDs form. Several proteins have been implicated in organizing LD formation sites (e.g., seipin (BSCL2)[48], lipin (Pah1p) [49], and fat-storage inducing transmembrane protein (FITM)-2 [50]), although their precise roles remain undefined. Seipin is an ER protein whose deficiency dramatically alters LD numbers and size. Seipin is thought to localize at LD-ER junctions in yeast [51] and deficiency increases phosphatidic acid levels, which may contribute to LD fusion [12]. FIT2 is also an ER protein that binds TG and appears to be involved in organizing LDs [52]. Pah1 is required for normal LD formation in yeast [53].
Step 3: Droplet Formation
Above a certain size, depending on the oil and phospholipid composition, lipid lenses in the ER are predicted to be unstable and bud, by a mechanism similar to de-wetting, due to thermal fluctuations [6] (Figure 1). The smallest mature cytosolic LDs have diameters in the range of 250–500 nm [21,54], which establishes an upper limit for budding size. Reports of nascent LD size are quite broad, as simulations predict LD diameters of ~50–100 nm [45,55], and one study suggests that they might be even smaller [24]. Defining the lower limit of LD formation size is challenging as their formation events reach the temporal and spatial resolution limitations of current microscopy techniques. In yeast, some newly formed LDs appear to remain connected with the ER [25]. At least in some cells, LDs that have budded and separated from the ER have been observed [21,56].
Whether drop formation is protein-mediated is an open question. To date, no single protein has been identified that is required for this step. The ubiquity of LDs across organisms argues for a highly conserved mechanism. However, the heterogeneity of LDs in terms of size and protein composition may allow for multiple mechanisms that include a process that is facilitated by proteins. For example, perilipin (PLIN)-3 has been proposed to be a major regulator of LD formation [57]. However, LD formation also occurs in systems that do not express perilipins. Drop formation is predicted to occur spontaneously based on coarse-grained simulations [45,58]. However, there is some evidence that TG accumulation alone may not be sufficient to drive drop formation in vivo [53,59]. A recent study determined that incubating COS cells with increasing amounts of oleate had no effect on the number of LD nucleation sites, but did increase the size of forming LDs [46], suggesting these sites may be pre-determined. Given the current evidence, we speculate that proteins are not required for drop formation per se, but may act to facilitate and/or regulate this step.
Lipid Droplet Growth
Although nascent LDs are small relative to cell size, many cells possess very large LDs. Large droplets can arise from two general mechanisms: growth of a LD or by processes in which LDs combine to form a single, larger LD (Figure 2).
Figure 2.
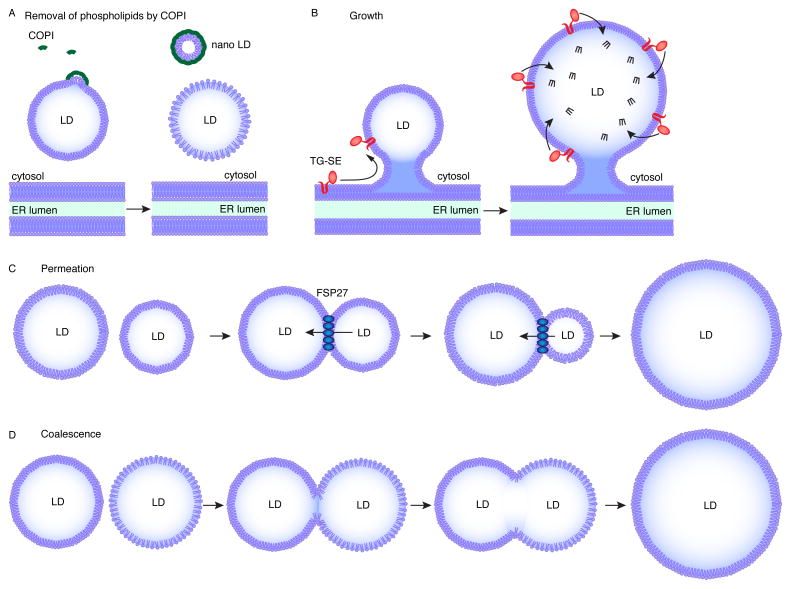
Features of lipid droplet growth (expansion) and fusion. Large LDs can form by at least two general mechanisms: growth of an LD or processes in which LDs combine to form a single, larger LD. Growth of LDs is triggered by relocalization of TG synthesis enzymes from the ER to the surface of LDs. (a) The COPI machinery buds small nano-LDs from a mature LD leading to a reduction of phospholipids on the LD surface. This leads to an increase in surface tension facilitating interactions of the LD with the ER. (b) Once connections are established, a subset of TG synthesis enzymes is able to relocalize to the LD surface to locally produce TG, which, in turn, leads to the growth of the LD. (c) Alternatively LDs can expand by a ripening process called permeation. Here neutral lipids are transferred from a smaller LD to a larger LD. In adipocytes, FSP27 is involved in this process. (d) Under certain conditions, for example when PC is limited and surface tension is relatively high, large LDs can form by fusion/coalescence of two or more LDs.
LD growth occurs by the local synthesis of TGs at the surface of LDs. Droplet growth thus requires a cellular trafficking pathway that delivers the enzymes necessary for TG synthesis to LDs. Both nascent and mature LDs can acquire enzymes for growth from the ER. Interestingly, LDs are often found in close proximity to the ER and, in some instances, have been found to be connected to the ER through ER–LD membrane bridges [56]. These ER–LD connections were first observed in plant cells by electron microscopy [60] and were more recently shown to participate in localization of TG enzymes to the LD surface [13,21]. This indicates that ER–LD connections are crucial for LD growth. Consistent with this, factors maintaining ER structure, such as atlastin, a GTPase that mediates membrane fusion to connect ER tubules, play a critical role in regulating LD size [61].
On the LD surface, coatamer protein (COP)-I machinery plays an important role in establishing connections to the ER [13,62]. COPI proteins act at LD surfaces by removing phospholipids, thereby increasing LD surface tension and favoring the fusion of LDs with other membranes [62]. Once ER–LD bridges are established, specific isoforms of TG synthesis enzymes (e.g., GPAT4, AGPAT3, DGAT2) use the connections to relocalize from the ER to LDs. The dual localization of these proteins is possible because of the special topology of these enzymes, which harbor a hairpin of two α-helices that extends into, without completely spanning, a bilayer membrane [21,37,63]. In support of the crucial role of localized TG synthesis for LD growth, depletion of DGAT2 or GPAT4 prevents the expansion of LDs [21]. It is unclear why, under conditions favoring LD expansion, the trafficking of enzymes to the LD is apparently unidirectional.
The expansion of the LD core by TG synthesis is tightly connected to the expansion of the phospholipid surface. Although the most abundant phospholipids in the LD monolayer are PC and PE, PC is key for coating LDs and preventing their coalescence [20]. Therefore, the expansion of droplets leads to an increased need of PC on the surface of LDs. Synthesis of PC consists of three enzymatic steps, the second of which is a rate-limiting step catalyzed by CTP:phosphocholine cytidylyltransferase (CCT). CCT uses phosphocholine and cytidine triphosphate (CTP) to form CDP-choline, which, in turn, is combined with diacylglyerol by cytidine diphosphate (CDP)-choline:1,2-diacylglycerol cholinephosphotransferase (CPT) in the ER to form PC. Under conditions of LD expansion, CCT is translocated from the cytosol to LD surfaces and becomes activated [20]. In this manner, PC synthesis increases in response to local demands. Since the last enzyme of the PC synthesis pathway, CPT, is exclusively localized at the ER, newly synthesized PC needs to be transferred to expanding LDs. How this is achieved is unknown.
Lipid Droplet Coalescence and Ripening
The generation of a large LD from two smaller LDs can occur either by direct coalescence/fusion or by ripening (diffusion-mediated transfer of core lipids; see [6]). Direct fusion of LDs in cells is rare under normal circumstances, but can be induced by modulating the LD surface (e.g., by limiting available PC [12,20] or by the addition of surfactants [64]).
In adipocytes, large LDs form by what appears to be a ripening process called permeation. Specifically, fat-specific protein of 27 kDa (FSP27), a member of the cell death-inducing DFF45-like effector (CIDE) family mainly expressed in adipocytes, is involved in transferring lipids between two adjacent LDs [65]. This process occurs over several minutes, with transfer of TG from the smaller LD to the larger LD [65]. Experimental observations, therefore, are most consistent with permeation, in which TG molecules diffuse to the larger LD at a contact site, driven by differences in Laplace pressures of the two LDs. This model is supported by the localization of FSP27 to LD–LD contact sites [65]. Additionally, overexpression of FSP27 in cells leads to increased LD size whereas depletion abolishes LDs with a diameter larger than ~12 μm in adipocytes [66]. Ripening-mediated transfer of TG by FSP27 is likely regulated by binding of FSP27 to PLIN1, which increases transfer by increasing pore size [66]. Whether other members of the CIDE family promote similar LD growth reactions in cell types other then adipocytes is unclear.
Conclusion
With renewed attention to LD organelles, various aspects of their biology are being uncovered. Recent advances have included new insights into their formation and subsequent growth. However, many questions remain. What drives lens formation in the ER? What determines or regulates the localization of the budding process? What are the functional roles of different enzymes in initial formation of LDs versus LD growth? Are ER-LD bridges stabilized and maintained, and if so, how? Is LD formation coupled with or distinct from ER-LD bridges? How are neutral lipid synthesis enzymes concentrated on LD surfaces? What happens to LD proteins during lipolysis and LD catabolism?
Since LDs touch many fields, insights into these questions are likely to arise from many avenues of investigation. With increasing knowledge, and model refinement of how each step occurs, a detailed insight into the membrane biology of this fascinating organelle will emerge, as well as new ideas on how to manipulate these hubs of metabolism for therapeutic or industrial benefits.
Acknowledgments
The authors thank members of the Farese and Walther laboratories for helpful discussions, Crystal Herron for editorial assistance, Daryl Jones for manuscript preparation, and NIH grants RO1GM099844 (R.V.F.) and R01GM097194 (T.C.W.), the G. Harold and Leila Y. Mathers Foundation (T.C.W.), a Boehringer Ingelheim Fellowship (F.W.), a NSF Graduate Research Fellowship (J.T.H.) and the Gladstone Institutes (R.V.F.) for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285:19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci U S A. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fei W, Wang H, Fu X, Bielby C, Yang H. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem J. 2009;424:61–67. doi: 10.1042/BJ20090785. [DOI] [PubMed] [Google Scholar]
- 4.Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herker E, Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol Metab. 2011;22:241–248. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nature Reviews in Molecular Cell Biology. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohsaki Y, Suzuki M, Fujimoto T. Open Questions in Lipid Droplet Biology. Chem Biol. 2013 doi: 10.1016/j.chembiol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 12.Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Brewersdorf J, Farese RV, et al. Arf1/COPI Machinery Acts Directly on Lipid Droplets and Enables their Connection to the ER for Protein Targeting. eLife. 2014 doi: 10.7554/eLife.01607. Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Galea A, Sytnyk V, Crossley M. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol. 2012;24:509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochim Biophys Acta. 2013;1831:589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 17.Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, et al. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, et al. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- **20.Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. Using in vitro and cellular models, the authors showed the crucial role of surface lipids in maintaining lipid droplet size. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. The authors identified a pathway by which TG synthesis enzymes, such as GPAT4, AGPAT3, and DGAT2, translocate to the lipid droplet surface for localized triglyceride synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *23.Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci. 2008;121:1852–1860. doi: 10.1242/jcs.012013. The authors defined a general targeting motif for LD proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Long AP, Manneschmidt AK, VerBrugge B, Dortch MR, Minkin SC, Prater KE, Biggerstaff JP, Dunlap JR, Dalhaimer P. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic. 2012;13:705–714. doi: 10.1111/j.1600-0854.2012.01339.x. This authors provide experimental evidence of LD fission. [DOI] [PubMed] [Google Scholar]
- **25.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. Using a yeast model, the authors found that Dga1p could move freely between the ER and newly formed LDs, suggesting physical connections exist between the ER and some LDs. [DOI] [PubMed] [Google Scholar]
- *26.Jacquier N, Mishra S, Choudhary V, Schneiter R. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J Cell Sci. 2013;126:5198–5209. doi: 10.1242/jcs.131896. This study shows that ectopic expression of PAT proteins and oleosins can induce LD formation in yeast, apparently in the absence of neutral lipid synthesis. [DOI] [PubMed] [Google Scholar]
- 27.Buhman KK, Chen HC, Farese RV., Jr The enzymes of neutral lipid synthesis. J Biol Chem. 2001;276:40369–40372. doi: 10.1074/jbc.R100050200. [DOI] [PubMed] [Google Scholar]
- 28.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis JM, Frahm JL, Li LO, Coleman RA. Acyl-coenzyme A synthetases in metabolic control. Curr Opin Lipidol. 2010;21:212–217. doi: 10.1097/mol.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- 31.Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV., Jr Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- 32.Wendel AA, Cooper DE, Ilkayeva OR, Muoio DM, Coleman RA. Glycerol-3-phosphate acyltransferase (GPAT)-1, but not GPAT4, incorporates newly synthesized fatty acids into triacylglycerol and diminishes fatty acid oxidation. J Biol Chem. 2013;288:27299–27306. doi: 10.1074/jbc.M113.485219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, Farese RV., Jr Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 2009;50:434–442. doi: 10.1002/hep.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurie HR, Buckett L, Zammit VA. Diacylglycerol acyltransferase 2 acts upstream of diacylglycerol acyltransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. Febs J. 2012;279:3033–3047. doi: 10.1111/j.1742-4658.2012.08684.x. [DOI] [PubMed] [Google Scholar]
- 35.Qi J, Lang W, Geisler JG, Wang P, Petrounia I, Mai S, Smith C, Askari H, Struble GT, Williams R, et al. The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and -2. J Lipid Res. 2012;53:1106–1116. doi: 10.1194/jlr.M020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 37.Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV., Jr The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, Jr, Mak HY. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichmann TO, Kumari M, Haas JT, Farese RV, Jr, Zimmermann R, Lass A, Zechner R. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J Biol Chem. 2012;287:41446–41457. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal AK, Sukumaran S, Cortes VA, Tunison K, Mizrachi D, Sankella S, Gerard RD, Horton JD, Garg A. Human 1-acylglycerol-3-phosphate O-acyltransferase isoforms 1 and 2: biochemical characterization and inability to rescue hepatic steatosis in Agpat2(-/-) gene lipodystrophic mice. J Biol Chem. 2011;286:37676–37691. doi: 10.1074/jbc.M111.250449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas JT, Winter HS, Lim E, Kirby A, Blumenstiel B, DeFelice M, Gabriel S, Jalas C, Branski D, Grueter CA, et al. DGAT1 mutation is linked to a congenital diarrheal disorder. J Clin Invest. 2012;122:4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackinnon WB, May GL, Mountford CE. Esterified cholesterol and triglyceride are present in plasma membranes of Chinese hamster ovary cells. Eur J Biochem. 1992;205:827–839. doi: 10.1111/j.1432-1033.1992.tb16847.x. [DOI] [PubMed] [Google Scholar]
- *43.King NJ, Delikatny EJ, Holmes KT. 1H magnetic resonance spectroscopy of primary human and murine cells of the myeloid lineage. Immunomethods. 1994;4:188–198. doi: 10.1006/immu.1994.1019. Using a simple biophysical approach, the authors defined a critical threshold for the solubility of TG in a model PC emulsion. [DOI] [PubMed] [Google Scholar]
- 44.Hamilton JA, Small DM. Solubilization and localization of triolein in phosphatidylcholine bilayers: a 13C NMR study. Proc Natl Acad Sci U S A. 1981;78:6878–6882. doi: 10.1073/pnas.78.11.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Khandelia H, Duelund L, Pakkanen KI, Ipsen JH. Triglyceride blisters in lipid bilayers: implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS One. 2010;5:e12811. doi: 10.1371/journal.pone.0012811. The authors provide evidence of a specialized subdomain for LD formation in the ER of mammalian cells in a starvation-refeeding model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kassan A, Herms A, Fernandez-Vidal A, Bosch M, Schieber NL, Reddy BJ, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol. 2013;203:985–1001. doi: 10.1083/jcb.201305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross DA, Zhan C, Silver DL. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci U S A. 2011;108:19581–19586. doi: 10.1073/pnas.1110817108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross DA, Zhan C, Silver DL. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci U S A. 2011;108:19581–19586. doi: 10.1073/pnas.1110817108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang CW, Miao YH, Chang YS. Size control of lipid droplets in budding yeast requires a collaboration of Fld1 and Ldb16. J Cell Sci. 2014 doi: 10.1242/jcs.137737. [DOI] [PubMed] [Google Scholar]
- 55.Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T, von Landenberg P, et al. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- 56.Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284:30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zanghellini J, Wodlei F, von Grunberg HH. Phospholipid demixing and the birth of a lipid droplet. J Theor Biol. 2010;264:952–961. doi: 10.1016/j.jtbi.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 59.Gubern A, Casas J, Barcelo-Torns M, Barneda D, de la Rosa X, Masgrau R, Picatoste F, Balsinde J, Balboa MA, Claro E. Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J Biol Chem. 2008;283:27369–27382. doi: 10.1074/jbc.M800696200. [DOI] [PubMed] [Google Scholar]
- 60.Wanner G, Theimer RR. Membranous appendices of spherosomes (oleosomes): Possible role in fat utilization in germinating oil seeds. Planta. 1978;140:163–169. doi: 10.1007/BF00384916. [DOI] [PubMed] [Google Scholar]
- 61.Klemm RW, Norton JP, Cole RA, Li CS, Park SH, Crane MM, Li L, Jin D, Boye-Doe A, Liu TY, et al. A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Rep. 2013;3:1465–1475. doi: 10.1016/j.celrep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiam AR, Antonny B, Wang J, Delacotte J, Wilfling F, Walther TC, Beck R, Rothman JE, Pincet F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc Natl Acad Sci U S A. 2013;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **63.Poppelreuther M, Rudolph B, Du C, Grossmann R, Becker M, Thiele C, Ehehalt R, Fullekrug J. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res. 2012;53:888–900. doi: 10.1194/jlr.M024562. The authors show that FSP27 and PLIN1 form a complex to maximize FSP27 lipid exchange activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy S, Martin S, Parton RG. Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One. 2010;5:e15030. doi: 10.1371/journal.pone.0015030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Z, Gong J, Wu H, Xu W, Wu L, Xu D, Gao J, Wu JW, Yang H, Yang M, et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun. 2013;4:1594. doi: 10.1038/ncomms2581. [DOI] [PMC free article] [PubMed] [Google Scholar]