Abstract
Carcinogenesis is a multistep process attributable to both gain-of-function mutations in oncogenes and loss-of-function mutations in tumor suppressor genes. Currently, most molecular targeted therapies are inhibitors of oncogenes, because inactivated tumor suppressor genes have proven harder to “drug.” Nevertheless, in cancers, tumor suppressor genes undergo alteration more frequently than do oncogenes. In recent years, several promising strategies directed at tumor suppressor genes, or the pathways controlled by these genes, have emerged. Here, we describe advances in a number of different methodologies aimed at therapeutically targeting tumors driven by inactivated tumor suppressor genes.
Keywords: cancer, tumor suppressor gene, oncogene, therapeutic targeting
INTRODUCTION
Cancer is a genetic disease. Historically, the idea that cancer could be etiologically attributed to genetic alterations was first recognized when cancer-causing viruses were found to be able to transform normal cells. The responsible genes within these viruses were identified and called “oncogenes.” The existence of “antioncogenes” had also been posited, but definitive evidence was lacking for many years. In 1969, Knudson first predicted the existence of tumor suppressor genes (TSGs), based on the kinetics of the development of sporadic and inherited retinoblastomas.1 He proposed a “2-hit” model for carcinogenesis that was ultimately supported in 1986 with the successful cloning of the retinoblastoma 1 (RB1) gene.2 Classically, inactivation of tumor suppressor genes will only lead to a phenotype when both copies of the gene have been lost. In cancer, the inactivation of one copy of a TSG will generally need to be followed by loss of the remaining copy of the gene, followed then by emergence of the tumor phenotype.
One of the early logical arguments against the existence of TSGs was that it was difficult to reconcile Knudson’s 2-hit model with the model of clonal evolution of cancer as put forth by Nowell, in which cancer is the result of cells progressing through successive waves of clonal selection, with a growth advantage at each step along the way.3 The Knudson 2-hit model assumed that TSGs are recessive and that a precancerous cell would only enjoy an advantage once it loses both functional copies of a TSG that had been suppressing growth. Given the very low spontaneous mutation rate in normal cells (estimated at between 1×10−6 and 1× 10−7 mutations per gene, per cell division), the requirement that both alleles be lost before a cell developed a growth advantage would make such events too rare to account for the observed incidence of human cancer.4,5 Quon and Berns proposed that a tumor requiring 4 mutations would arise at an approximate frequency of 1× 10−21 cells, orders of magnitude below the 1014 cells comprising the human body, a fact that appeared to be inconsistent with the statistic that 1 of 3 individuals will develop a cancer during their lifetime.6 This led to the conclusion that there could be a phenotype associated with loss of a single copy of certain TSGs; this ultimately was shown to be the case with some. Indeed, it is now more or less accepted that for many TSGs, heterozygous loss of function can be associated with reduced gene dosage and tumorigenesis via haploinsufficiency.6,7 Additional explanations for this question also appeal to alternate methods by which TSGs can be silenced, such as epigenetic mechanisms, or changes in mutation frequency, such as those that occur in hypermutator phenotypes. Together, these mechanisms begin to provide insight into potential therapeutic approaches.
Kinzler and Vogelstein proposed that TSGs fall into 2 categories: “gatekeeper” genes and “caretaker” genes.8 Gatekeeper genes control how cells progress through cycles of growth or division, whereas caretaker genes maintain the integrity of the genome. The distinction between these 2 classes of genes is critical to developing approaches to therapy. Currently, nearly all molecular targeted therapies are inhibitors of oncogenes such as kinases. Kinase inhibitors have been one of the most successful classes of cancer drugs developed to date. Intuitively, it appears more straightforward to inhibit a hyperactivated oncogene than to restore the function of an inactivated TSG. Despite their being harder to “drug,” loss-of-function alterations in TSGs make equally important contributions to tumorigenesis. In recent years, several promising strategies for targeting TSGs therapeutically have emerged. Here, we describe advances in several different methodologies aimed at targeting inactivated TSGs. Although attempts to restore TSG function have demonstrated some potential, the most promising approaches are those that focus on molecules that regulate, inhibit, or epigenetically silence TSGs; shut down signaling pathways that have been abnormally activated by loss of the TSG; or exploit vulnerabilities in cancer cells lacking certain TSGs.
Overview of Cancer Genome Data
In cancer genomes, alterations that lead to the development of cancer tend to more commonly affect TSGs rather than oncogenes. Early on, this was evident in exome sequencing studies performed across multiple types of human cancer, revealing a set of cancer “driver” genes, the majority of which were TSGs.9,10 More recently, pan-cancer analyses of data from The Cancer Genome Atlas have supported this initial finding on a broader scale, with the majority of copy number alterations in these cancer genome studies comprising deletions of putative TSGs.11 Zack et al have demonstrated that approximately 60% of peak regions of copy number alteration in cancer are deletions, and the majority of genes within these peaks are either known TSGs or appear to be novel TSGs.12
Of the TSGs mutated in cancer that are deemed most likely to be driver genes, several pathways and processes are implicated. Well-described TSGs include genes in pathways such as Wnt/APC (adenomatous polyposis coli gene [APC], AXIN1, and CDH1); apoptosis/cell cycle (cyclin-dependent kinase inhibitor 2A [CDKN2A], tumor protein 53 [TP53], RB1, TRAF7,and CASP8); chromatin modification (ARID1A/B/2, ASXL1, ATRX, CREBBP, KDM5C, KDM6A, MEN1, MLL2/3, SETD2, ten-eleven translocation-2 [TET2], WT1, and BAP1); DNA damage repair (ataxia telangiectasia mutated [ATM], ataxia telangiectasia and Rad3 related [ATR], BRCA1/2, mutL homo-log 1 [MLH1], and MSH2/6); hedgehog (PTCH1); Notch (FBXW7 and NOTCH1); phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) (PIK3R1, phosphatase and tensin homolog [PTEN], and TSC1); Ras (CEBPA, von Hippel-Lindau [VHL], and NF1); transforming growth factor-β (SMAD2/4); and transcriptional regulation (GATA3 and RUNX1).
Targeting p53
Mutations in p53, encoded by the TP53 gene, are the most frequent genetic alterations in cancer.13–15 TP53 is mutated in 30% to 50% of human cancers, with a particularly high prevalence (>50%) of mutation in types of ovarian, lung, colorectal, head and neck, pancreatic, uterine, breast, and bladder cancer.16 p53 was initially believed to be an oncogene, based on experiments demonstrating its transforming ability. However, these early data were ultimately attributed to the finding that the experimental TP53 cDNA had been cloned from a tumor cell and harbored a mutation.17 Subsequently, wild-type p53 was confirmed to suppress growth; ultimately, TP53 was correctly classified as a TSG. It is interesting to note that p53 exerts dominant-negative activity when mutated. In contrast to the Knudson 2-hit model, in which mutations are thought of as creating inactive alleles, mutated TP53 is associated with an altered gain-of-function phenotype.18 This finding is consistent with cancer genomics data, in which the vast majority of mutations in TP53 are missense, rather than nonsense.16
In recent years, targeting mutated p53 has been a field characterized by intense research that is beginning to bear fruit. TP53 is a frequently inactivated gene that represents a highly tumor-specific target. However, “drugging” mutant p53 via the standard mechanisms used for anticancer therapies is not straightforward because p53 is not a cell surface protein or an enzyme, and therefore not targetable with antibodies or enzyme inhibitors. In vivo studies have supported the desirability of reactivating p53 activity in p53-null or p53-mutant tumors, indicating that doing so is sufficient to cause tumor stability or regression.19–22 In many cases, the transformed tumor cells were observed to be highly responsive to restoration of p53 activity, which often turns on an apoptosis or senescence pathway. These findings have led to interest in finding a way to reactivate wild-type p53 in tumor cells. In addition, radiotherapy and the majority of chemotherapy agents are more effective in the presence of a functional p53 pathway, implying that biologic approaches to reactivating p53 could also sensitize cancer cells to chemotherapy or radiotherapy.
The main approaches to targeting p53 in cancer include targeting molecules that posttranslationally regulate, inhibit, or mediate the downstream effects of p53; reintroducing wild-type p53; or selectively killing p53-mutant cancer cells.
p53: Targeting its Regulators
Several compounds have been identified that affect p53 posttranslational modification. These agents include tenovin-1 and tenovin-6, which are now known to inhibit the protein deacetylation activities of sirtuins. Inhibiting these processes leads to acetylated, and thereby stabilized, p53.23 Another class of molecules are nuclear export inhibitors such as leptomycin B, an inhibitor of the nuclear export protein CRM1, which is able to increase local p53 protein levels.24
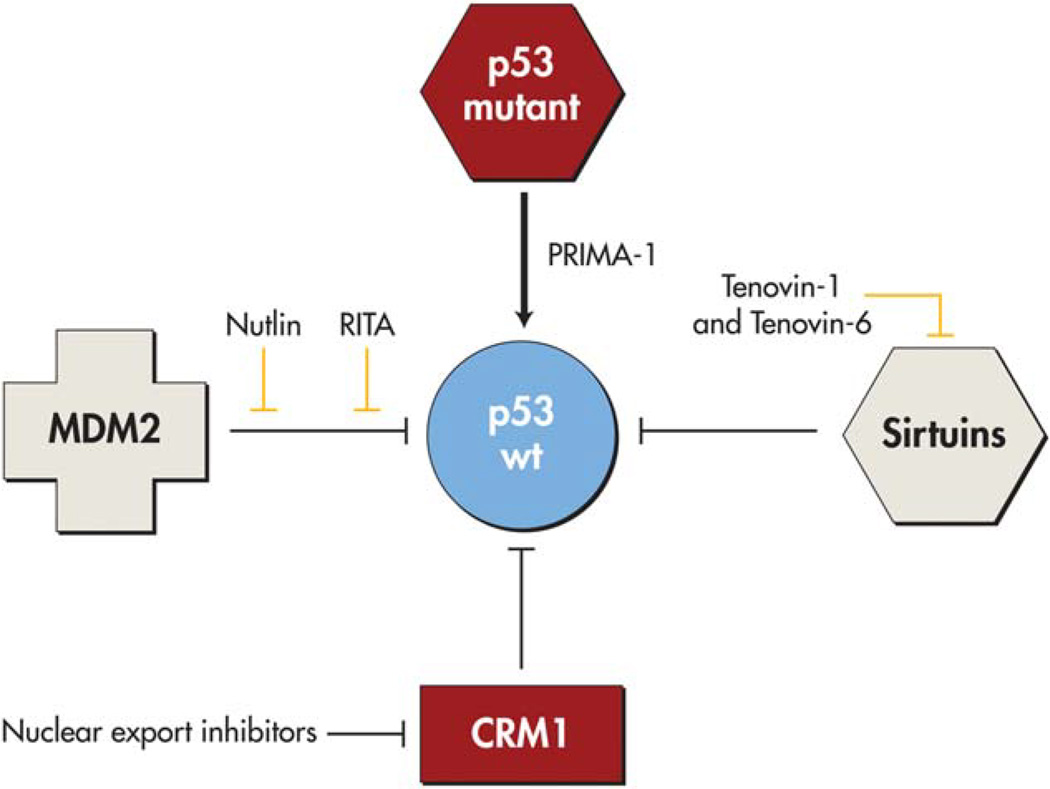
The protein MDM2 (mouse double minute 2 homolog) is a negative regulator of p53. Several compounds have been developed to specifically target protein-protein interactions between p53 and MDM2. The first molecule in this category is nutlin, which has been identified in several large in vitro biochemical screens as a molecule that inhibits interaction between p53 and MDM2 by occupying the p53-binding pocket.25 Early preclinical data have demonstrated that these agents have activity against tumors in vivo, and research has now advanced to several clinical trials in which nutlin in combination with cytotoxic chemotherapy or targeted therapies.26 Several additional molecules have been developed to target the N-terminal interaction between p53 and MDM2orMDMX. Thereisnowknown to be a deep hydrophobic cleft in MDM2 into which p53 is embedded, a potentially “druggable” pocket.27,28 There are now orally bioavailable molecules with submolar affinity for MDM2 in phase 1 studies. These compounds increase p53 levels, as well as the levels of the p53 target genes cyclin-dependent kinase inhibitor 1 (CDKN1A), MDM2, and PUMA (BBC3), in cancer cells.29 Complete tumor regression has been achieved with several of these compounds in preclinical studies. Of particular interest, these compounds appear to have very little toxicity in normal cells. As used, many of these compounds are only present in the cells for a brief time but long enough to induce a pulse of p53 activity. It is important to note that only a subset of tumors have upregulated MDM2; although up to 30% of sarcomas harbor MDM2 gene amplification, this is far less prevalent in other cancer types. In addition, nutlin has been demonstrated to select for certain p53-mutated cells, which can accumulate and lead to drug resistance.30 Several newer, second-generation compounds may be able to overcome this limitation, and are currently in phase 1 trials.31 Another recent approach is restoring p53 function using molecules that stabilize the protein as chaperones. For example, the molecule PRIMA-1 is a small molecule that restores the wild-type conformation of certain mutant p53 proteins by incompletely understood mechanisms, and has recently completed phase 1 clinical trials.32 Several of these approaches to targeting p53 are depicted in Figure 1.
Figure 1.
Restoring wild-type p53 function by targeting its regulators. The interaction between MDM2 and p53 is targeted by nutlin (which binds MDM2) or RITA (which binds p53). Nuclear export inhibitors such as leptomycin B target the nuclear export protein CRM1. Sirtuins are protein deacetylases that are inhibited by tenovin-1 and tenovin-6. PRIMA-1 restores the wild-type conformation of mutant p53. Adapted with modifications from Chen F, Wang W, El-Deiry WS. Current strategies to target p53 in cancer. Biochem Pharmacol. 2010;80:724–730.
p53: Gene Therapy Approaches
Gene therapy approaches to p53 functional restoration have been an area of investigation for years. The rationale behind gene replacement therapy strategies is to use a viral vector, such as a replication-deficient adenovirus, to introduce wild-type p53 into cancer cells. These viral vectors can be administered intratumorally or into body cavities (eg, intraperitoneally or intravesically). The toxicity to normal cells of such an approach would theoretically be minimal, because the introduction of a TSG into a normal cell at physiologic levels would not be expected to have any significant effect. In early-phase clinical trials, this therapy has been well tolerated by patients with minimal toxicity. Unfortunately, the major limitation to this approach has been efficacy. The viral vectors used for gene therapy have not been able to achieve the necessary efficiency of transduction of p53 within tumors to be curative.33,34 Furthermore, repeat administration is hampered by host immune reactions to the virus vectors.
Therefore, an alternative approach has been to use tumor-specific replication-competent oncolytic viruses. An example of this is an adenovirus in which a 55-kilodalton gene in the E1B region of the virus, which normally binds and inactivates p53, has been attenuated. As a result, the virus can only replicate within (and kill) cells lacking functional p53, and cannot survive in normal cells with functional p53.35,36 This approach has also been explored in numerous clinical trials and, although generally safe, has had varying levels of efficacy, most likely due to low efficiency of delivery and nonspecific expression.37,38 This agent did not advance through phase 3 trials in the United States, but has been approved in China for use in combination with chemotherapy for certain types of head and neck cancer.
p53: Moving Downstream
Several downstream mediators of the mutant p53 tumorigenic phenotype have been identified, including C-X-C chemokine receptor type 4 (CXCR4), cyclin G2, MYC, TERT, p63/p73 transcription factors, and the mevalonate pathway.13,39 This opens the possibility of targeting downstream targets of mutated p53. For example, mutant p53 (but not p53 loss) has been recognized to facilitate a prometastatic phenotype in a pancreatic adenocarcinoma model. Mutant p53 induces expression of platelet-derived growth factor receptor-β (PDGFRβ), which in turn mediates invasion and metastasis. Pharmacologic inhibition of PDGFRβ with agents such as crenolanib or imatinib was able to significantly reduce the invasive potential of models of pancreatic adenocarcinoma.40
p53: Vaccine Approaches
The frequent mutations in p53, together with its high levels of expression, make it likely to be identified by the immune system as a target antigen. Indeed, patients with cancer are known to produce anti-p53 antibodies and p53-reactive T cells. This has led to several nascent but promising approaches using vaccines. For example, vaccines containing multiple p53 peptides are able to generate a T-helper type I response in patients, although the responses have not yet been potent enough to be clinically beneficial.41 More recently, vaccines derived from dendritic cells transfected with the TP53 gene have been noted to generate stronger immune responses.42 Related approaches use dendritic cells loaded with human leukocyte antigen class I p53 peptides, which appear to induce changes in immune regulatory mechanisms, although a continuing challenge is overcoming strong immune suppressive mechanisms in patients with cancer.43
Inhibiting Hyperactivated Pathways Resulting From TSG Inactivation
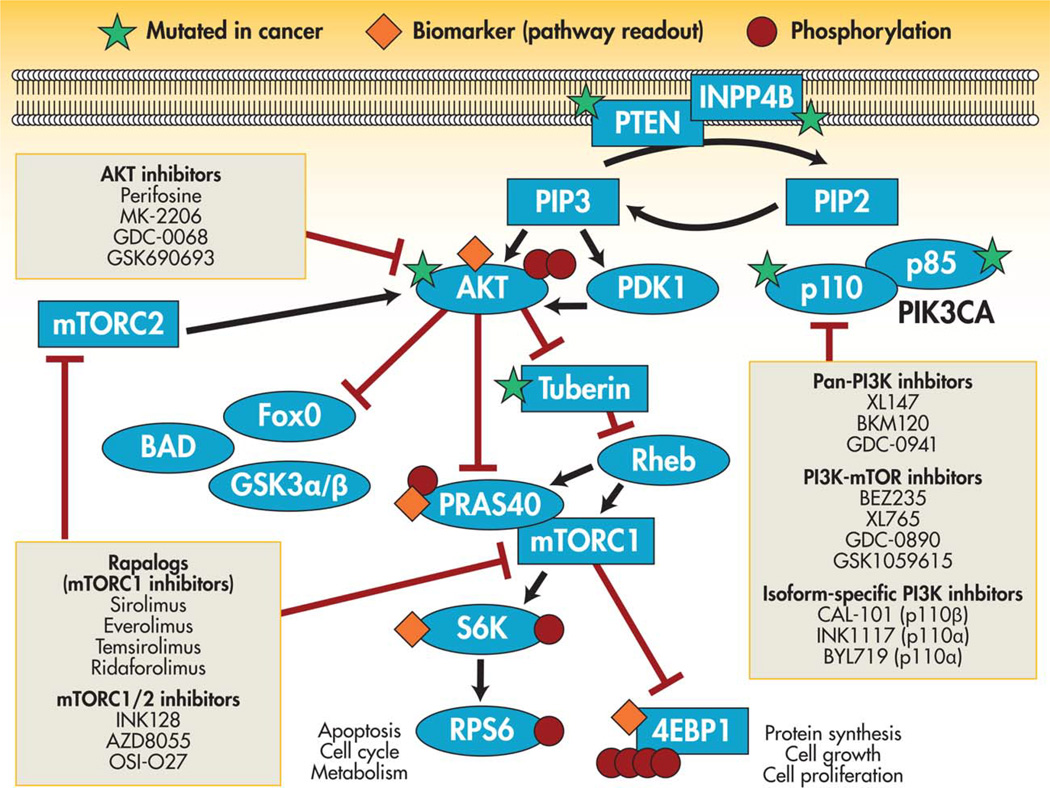
In cases such as the example provided above of p53 and PDGFRβ, the TSG undergoing loss of function is part of a broader signaling pathway, and the cancer phenotype is mediated by hyperactivation of that pathway. In this scenario, the inactivated TSG can be therapeutically targeted by inhibiting the relevant pathway further downstream. An example of this paradigm is provided by PTEN, which is one of the most commonly altered TSGs across human malignancies. PTEN is inactivated by mutation or deletion in a significant percentage of diverse cancer types including glioblastoma; endometrial, prostate, uterine, and breast cancer; and melanoma.16,44,45 PTEN is a well-described TSG, functioning as a phosphatase that removes the D3 phosphate from phosphatidyl (3,4,5) tri-phosphate (PIP3). In their resting state, cells have low levels of PIP3, which are then increased at the time of growth factor stimulation or activation of PI3K. Loss of PTEN leads to constitutively high levels of PIP3, which mimics the state of growth factor stimulation of hyperactivated PI3K.46,47 Ultimately, the second messenger PIP3 goes on to activate target proteins including the kinases phosphoinositide-dependent kinase-1 (PDK1) and AKT1/2/3. AKT then phosphorylates as many as 20 pro-growth targets relevant to cancer, including those activating the cell cycle, preventing apoptosis, and promoting cell growth via the kinase mTOR (Fig. 2).48–51
Figure 2.
Mutation in the tumor suppressor gene PTEN leads to hyperactivation of the PI3K/AKT/MTOR pathway, which can potentially be targeted at various sites downstream of PTEN. PIP indicates phosphatidyl phosphate. Adapted with modifications from Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10:143–153.
It is believed that hyperactivation of the PI3K/AKT/ mTOR pathway resulting from inactivation of PTEN is, at least in part, similar to the sequelae of oncogenic alterations elsewhere in the pathway such as epidermal growth factor receptor amplification or mutation, human epidermal growth factor receptor-2 (HER2) amplification, PIK3CA (the gene encoding the catalytic subunit of PI3K) mutation, or AKT1/2 mutation.44,52 Accordingly, downstream inhibition of signaling is an attractive approach to targeting inactivated PTEN. PTEN-mutated tumors, similar to PIK3CA-mutated tumors, appear to be dependent on this signaling pathway for maintenance of the transformed phenotype, and as a result are vulnerable to inhibition of the pathway.53,54 Unfortunately, these approaches have been limited by the complexity of feedback networks in this pathway. For example, inhibition of mTOR with agents such as rapamycin is effective in attenuating signaling but also relieves feedback inhibition of other upstream components such as insulin, insulinlike growth factor receptor, HER3, and HER4, which can then signal through other branches of the pathway such as forkhead box O (FOXO)-dependent transcription.55,56 In vivo data have demonstrated that combined inhibition of AKT, together with agents inhibiting HER kinases or with inhibitors of receptor tyrosine kinase stabilization by heat shock protein 90, is necessary to truly shut down signaling. These combination approaches can be effective in promoting tumor regression, which AKT inhibition alone is not able to achieve.56,57 The approach of targeting the hyperactivated pathway affected by an inactivated TSG is likely to be more complex than the simple linear models of signal transduction pathways. Effective targeting of PTEN mutation-driven cancers will require comprehensive dissection of the feedback networks activated by inhibition of AKT/mTOR signaling, and durable therapy will undoubtedly require combination approaches.
Synthetic Lethality: Vulnerabilities in DNA Damage Repair
Cancer cells are reliant on intact DNA repair mechanisms to be able to continuously divide. At the same time, many cancers result from genetic aberrations that lead to impaired DNA damage repair, such as mutations in the caretaker TSGs ATM, BRCA1, BRCA2, and the Fanconi anemia genes. Mutations in these TSGs are most commonly found in breast, ovarian, colorectal, and hematologic malignancies. In cancer cells with an impaired DNA damage repair pathway, the cell becomes addicted to another DNA damage repair pathway. Synthetic lethality refers to the principle by which secondary addictions such as these can be exploited therapeutically, either by inhibiting the remaining vital pathway or by enhancing DNA damage from chemotherapy.
There are 2 major pathways used for the repair of DNA double-strand breaks: nonhomologous end joining (NHEJ) and homologous recombination (HR); and 3 major pathways used to repair single-strand breaks: base excision repair (BER), nucleotide excision repair, and mismatch repair. “ Several important mediators of these pathways play a role in more than one pathway; for example, the gene poly(ADP-ribose) polymerase 1 (PARP1) is involved in NHEJ, HR, and BER.
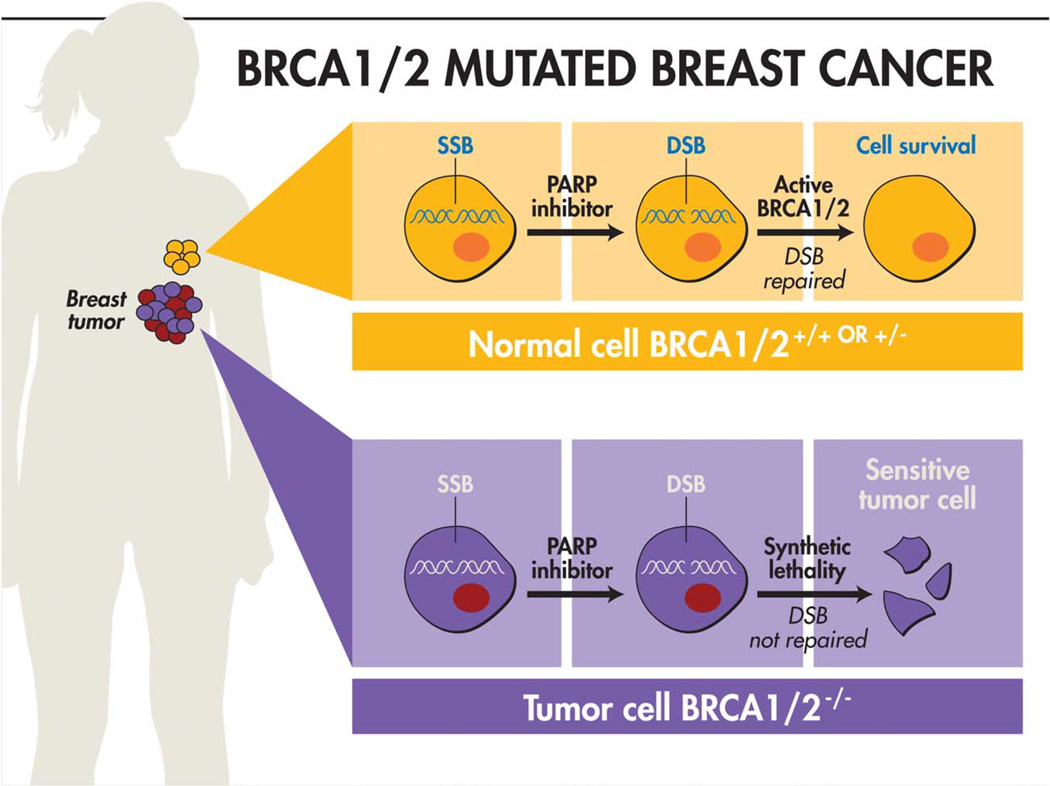
BRCA1 or BRCA2 loss leads to cells being defective in HR repair of double-strand breaks. This leads to a reliance on potentially mutagenic mechanisms such as NHEJ or single-strand annealing. Ultimately, this results in significant genomic instability, causing a cancer predisposition. This knowledge has led to the emergence of a synthetic lethal approach to targeting BRCA-mutated cancers. The best known example of this approach is targeting PARP1 (and other PARPs) in cancers deficient in BRCA1 or BRCA2. In these cells, if PARP1 is inhibited, the cell loses its ability to repair single-strand DNA breaks via BER. When the replication fork reaches an area with a single-strand break, a double-strand break results. These double-strand breaks are then unable to be repaired because of the BRCA1I2 mutation impairing HR As a result, the BRCA-mutated cells undergo apoptosis, whereas normal cells with intact BRCA are able to repair the double-strand DNA lesions and survive (Fig. 3). Accordingly, PARP1 inhibition is theoretically specific for BRCA-mutated cells. Preclinical data with PARP inhibitors demonstrate as high as 1000- fold relative sensitivity of BRCA-mutated cells compared to normal cells.62,63 In recent years, several clinical trials of PARP inhibitors have been undertaken in patients with breast and ovarian cancer and several of these are in middle-stage to late-stage trials. An initial disappointing phase 3 trial of the PARP1 inhibitor iniparib has been attributed to the relatively low on-target potency of this particular agent, and there remains considerable interest in other PARP inhibitors.64,65 Because radiotherapy causes double-strand DNA breaks, there is also considerable interest in combining radiation with PARP inhibitors for patients with tumors with BRCA1/2 mutations.66
Figure 3.
The paradigm of synthetic lethality as exemplified by the use of PARP inhibitors in BRCA1 or BRCA2 mutant breast cancers. PARP inhibitors impair the cell’s ability to repair single-strand DNA breaks (SSBs), which then progress to become double-strand breaks (DSBs). DSBs are normally repaired via homologous recombination. However, cancer cells lacking BRCA1 or BRCA2 (BRCA1/2−/−) lose the ability to repair DSBs, leading to specific death of these cancer cells undergoing PARP inhibition. Adapted with modifications from Polyak K, Garber J. Targeting the missing links for cancer therapy. Nat Med. 2011;17:283–284.
The list of potential candidates for a synthetic lethal approach most likely extends beyond patients with familial BRCA syndromes. Some sporadic (nonfamilial) breast and ovarian cancers are now recognized to harbor de novo mutations or promoter hypermethylation in BRCA1/2. Some types of cancers harbor alterations in other HR-associated genes such as RAD50, RAD51, ATR, ATM, or FANC family genes, conferring sensitivity to PARP1 inhibition. It is interesting to note that mutations in PTEN have also been found to sensitize cells to PARP1 inhibition, most likely due to downregulation of RAD51, a critical HR gene, although the precise mechanism remains unclear.67
Synthetic Lethality: “BRCAness”
This constellation of findings in BRCA-related pathways has led to the recognition of a broader concept of “BRCAness” in cancer. This term refers to the evidence that other genetic lesions in sporadically occurring cancers can phenocopy familial BRCA1/2 mutation-driven cancers. These cancers harbor impaired DNA repair mechanisms, potentially rendering them more sensitive to therapies with DNA-damaging agents such as crosslinking drugs such as cisplatin or mitomycin, and potentially establishing vulnerability to synthetic lethal therapies. There are great similarities in gene expression profiles among BRCA1/2-mutated, basal-like, and triple-negative breast cancers, as well as serous ovarian cancers.68 For example, there are molecular fingerprints such as expression signatures that identify BRCA1-like breast cancers.69,70 There are implications of BRCAness outside of breast cancer as well; for example, there are lung cancers with mutations in BRCA1, XRCC5, XRCC3, ERCC1, or RRM1, in which PARP inhibitors appear to have activity in combination with radiotherapy,71 and also chronic lymphocytic leukemias with ATM mutations that are sensitive to cytotoxic agents.72
Synthetic Lethality: Beyond PARP
Although PARP inhibition has received the most attention, several other synthetic lethal strategies targeting DNA repair proteins also have demonstrated significant promise. For example, the TSGs MLH1 or MSH2, implicated in hereditary nonpolyposis colon cancer, mediate the mismatch repair pathway. These alterations are synthetically lethal with inhibition of DNA polymerases, specifically, MLH1 with POLG and MSH2 with POLB.73,74 Therefore, inhibitors of these DNA polymerases may have activity in hereditary nonpolyposis colon cancer. Another example is inhibition of the enzyme DNA-dependent protein kinase, catalytic subunit (DNA-PKcs), which cooperates with ATM and ATR to phosphorylate proteins involved in NHEJ. ATM-deficient cancer cells are addicted to DNA-PKcs activity to survive DNA damage, and DNA-PKcs agents may therefore have activity in ATM-deficient or ATR-deficient cancers.75,76 WEE1 inhibitors capitalize on the finding that cancer cells with dysfunctional p53 rely on the G2 checkpoint to repair DNA damage. WEE1 is a nuclear serine/threonine kinase that drives G2/M-phase checkpoint progression. After DNA damage, WEE1 is phosphorylated and stabilized, leading to cell cycle arrest. In combination with DNA damaging agents, WEE1 kinase inhibition selectively induce death of p53-mutant cancer cells.77 This results in mitotic catastrophe.78 A WEE1 inhibitor is currently in early-phase clinical trials for the treatment of patients with p53-deficient ovarian cancer.
More recently, the field has moved toward using high-throughput unbiased screening methodologies to identify novel synthetic lethal interactions. These techniques are generally based on modeling in yeast79 or human cancer cell lines,80 and use RNA interference or chemical compounds, or both.81 Bioinformatic approaches can also predict synthetic lethal interactions. For example, the DAISY pipeline leverages pan-cancer sequencing data to identify coinactivated genes that occur less frequently than expected, suggesting that they have been eliminated from surviving cancer cell populations. As a proof of principle, this approach was able to identify numerous genes that were then experimentally validated as synthetically lethal partners of VHL in renal cancer cells.82
“Collateral Lethality”
An intriguing concept related to synthetic lethality was recently proposed and named “collateral lethality.” When tumor suppressor genes undergo homozygous deletion, the region of deletion can be quite broad, and usually encompasses several (or many) neighboring genes. Because these broad genetic alterations are not selected against during the development of cancer, the collateral effect on other genes must not be ultimately harmful to the cell. However, these passenger deletions of nearby genes may generate vulnerabilities if the nearby deleted gene is one of several redundant genes that perform a housekeeping function critical to cell survival. As a proof of principle, Muller et al examined genes in the 1p36 locus, which is deleted in a small percent (1%–5%) of glioblastomas.83 The ENO1 gene is located in this region and is one of 3 mammalian genes encoding enolase, an enzyme that mediates glycolysis. In glioma cells with a passenger deletion affecting ENO1, knockdown of ENO2 abrogated tumorigenic potential. Similarly, an enolase inhibitor was selectively toxic for ENO1-deleted cells. There are numerous other candidates for such a collateral lethality approach. There are several similarly situated housekeeping genes within functionally redundant families that can be identified within the deletion peaks targeting chromosome 9p21 (deleting CDKN2A) and 10q23 (deleting PTEN).83 It has been estimated that 11% of protein-coding genes undergo homozygous deletion in patients with cancer,84 making such a personalized therapeutic approach potentially promising in many malignancies.
Taking Aim at Epigenetic Mechanisms
It is now well understood that many epigenetic processes are altered during the development of cancer. There are several complex mechanisms by which TSGs are silenced in cancer as a result of dysregulated epigenetic mechanisms. The important processes altered in cancer center around modulation of the chromatin landscape, and include DNA methylation, histone modifications, and nucleosome remodeling. During the process of tumor initiation and progression, the cancer epigenome is remodeled in several complex ways, including global hypomethylation, increased promoter methylation at CpG islands, and alterations in nucleosome occupancy.85Ultimately, it is the balance between transcriptionally permissive and transcriptionally repressive chromatin modifications that affects gene expression, and it is an imbalance in these modifications that is observed in cancer.86
At its simplest, the role of epigenetic alterations in cancer is best exemplified by promoter hypermethylation of TSGs, leading to gene silencing. Many canonical TSGs are hypermethylated in cancer, including BRCA1/2, RB, and PTEN.87 In contrast, there are several important TSGs that are silenced in cancer but rarely undergo mutation or deletion, including the DNA repair gene MGMT88; the cell cycle regulator CDKN2B; the RAS-binding protein RASSF1A; MLH1, which plays an important role in genomic stability; and secreted frizzled-related proteins, which negatively regulate WNT signal-ing.89 In certain types of cancer, a genome-wide CpG island methylator phenotype has been reported, and these tumors demonstrate unique genetic and epigenetic char-acteristics.90–92
DNA methylation in mammalian cells is regulated by DNA methyl transferases (DNMTs). Mutations in several members of this family have been identified in cancer, including DNMT1 mutations in colorectal cancer93and DNMT3A mutations in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).94,95 Inhibitors of DNMTs such as 5-azacytidine and 5-aza-2-deoxycytidine reverse hypermethylation of DNA and have now been approved by the US Food and Drug Administration for use in patients with MDS and selected patients with AML. Patients with MDS experienced improved survival with 5-azacytidine in a randomized controlled trial. Response appears to be modulated, in part, by the status of the demethylating genes DNMT3A and TET2.96,97
Mutation of epigenetic regulators can cause profound changes in gene expression and cellular behavior. For example, mutation of the noncanonical oncogene iso-citrate dehydrogenase 1 (IDH1) results in fulminant DNA hypermethylation, silencing of differentiation mediators, and a block in differentiation.98,99 Although a specific inhibitor of mutant IDH is quite effective in reversing this block in differentiation in AML, it has been less active in solid tumors with extensive mutant IDH-induced hypermethylation.100 In IDH-mutant glioma cells and mutant IDH-transformed mesenchymal cells, DNMT inhibitors have been quite effective, acting by demethylating and reactivating the tumor suppressors silenced by mutant IDH.101–103
Histones are critical regulators of the chromatin landscape, and modification of histones is able to alter chromatin dynamics and gene expression. Histone methylation can be described as either activating (eg, trimethylation of histone H3 at lysine 4, H3K4me3) or repressing (eg, H3K27me3, H3K9me3) transcription. Histones can also undergo modification via acetylation, which is associated with active transcription. Histone deacetylases (HDACs) are “eraser” genes that remove acetyl groups from histone tails and thereby repress transcription. Aberrant activity of HDACs has been implicated in cancer and plays a role in silencing TSGs. For example, the promoter of CDKN1A is hypoacetylated in cancer; this can be reversed by inhibiting HDAC activity.104 Overexpression of HDACs has also been implicated in the silencing of BRCA1 and ATR.105 Mutations in HDAC genes have also been identified in several types of cancer.106 There are currently 2 HDAC inhibitors (vorinostat and romidepsin) that are approved by the US Food and Drug Administration for patients with refractory cutaneous T-cell lymphoma.107,108 These and several other agents currently are in clinical trials for solid and hematologic malignancies. Despite responses in patients with hematologic malignancies, durable responses in patients with solid tumors have been uncommon with current agents, and significant toxicity has been observed in some trials. Additional trials with combination therapies are planned.105
The Polycomb group of repressor proteins are chromatin remodelers that control transcription by regulating the accessibility of gene regulatory elements to transcrip-tional machinery (eg, by occupying the transcription start site and compacting chromatin).109 These proteins have been observed to undergo alteration in cancer. For example, the Polycomb Repressive Complex 2 is formed by enhancer of zeste homolog 2 (EZH2), SUZ12, and embryonic ectoderm development (EED), which together can silence gene expression by trimethylating H3K27. EZH2 is the catalytic subunit of this complex and is over-expressed or mutated, and associated with H3K27me3, in several cancer types, including breast, prostate, and lung cancer.110 Among other TSGs, EZH2 has been shown to repress the expression of the CDKN2A (p14/p16) locus.111 This has led to great interest in targeting EZH2 in cancer as a means of disabling the Polycomb Repressive Complex 2, with several emerging agents including 3-deazaneplanocin A and novel agents that disrupt the interaction between EZH2 and EED.112,113
Immunotherapy
The field of cancer immunotherapy has recently experienced remarkable advances, with immune checkpoint inhibitors demonstrating strong promise in several tumor types.114,115 Clinical responses can be dramatic and durable. Although the molecular determinants of response are ill-defined, these immunotherapy approaches likely target neoantigens formed by somatic mutations that can cause tumors to present “non-self” peptides on major histocompatibility complex molecules. This is in contrast to the more focused tactic of vaccines targeting specific, nonforeign proteins such as mutant p53, as discussed earlier. Bioinformatic approaches are now able to examine exome sequencing data across multiple types of cancer, to identify tumor-specific mutated peptides, combined with predicted binding to major histocompatibility complex class I proteins, inferring neoantigens that would be expected to be presented to CD8-positive T cells. Many of these mutations represent TSGs. In fact, the number of neoantigens generated from missense and frameshift mutations was proportional to the mutational rate in the tumor.116
Conclusions
Progress in recent years has fortunately belied the concerns of some researchers that TSGs were going to be a “neglected” area of therapeutics.117 Nevertheless, the translation of basic cancer research findings into therapy is a long journey. The steady progress being made in targeting p53 and other TSGs is a decades-long effort, not dissimilar to the history of the development of molecular targeted therapies directed at oncogenes such as BRAF and PIK3CA. Continued progress in basic research into TSG biology, and the allied fields of DNA damage repair, p53 biology, oncolytic viruses, signaling pathways, the cancer epigenome, and the immune system, will be essential to informing translational laboratory work in functional genomics and pharmacology, and ultimately bringing novel compounds to the clinic.
Acknowledgments
FUNDING SUPPORT
Supported by the Department of Defense (to Dr. Chan), the Sontag Foundation (to Dr. Chan), the Adenoid Cystic Carcinoma Research Foundation (to Dr. Chan and Dr. Morris), and the Damon Runyon Cancer Research Foundation (to Dr. Morris).
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
REFERENCES
- 1.Knudson AG., Jr Mutation and cancer: statistical study of retinoblas-toma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.DeMars R, Held KR. The spontaneous azaguanine-resistant mutants of diploid human fibroblasts. Humangenetik. 1972;16:87–110. doi: 10.1007/BF00393992. [DOI] [PubMed] [Google Scholar]
- 5.Seshadri R, Kutlaca RJ, Trainor K, Matthews C, Morley AA. Mutation rate of normal and malignant human lymphocytes. Cancer Res. 1987;47:407–409. [PubMed] [Google Scholar]
- 6.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 7.Cook WD, McCaw BJ. Accommodating haploinsufficient tumor suppressor genes in Knudson’s model. Oncogene. 2000;19:3434–3438. doi: 10.1038/sj.onc.1203653. [DOI] [PubMed] [Google Scholar]
- 8.Kinzler KW, Vogelstein BC. ancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 18.Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 20.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcino-genesis. 2010;31:1311–1318. doi: 10.1093/carcin/bgp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Suh YA, Fuller MY, et al. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest. 2011;121:893–904. doi: 10.1172/JCI44504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutka SC, Yang WQ, Dong SD, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69:510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 26.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 27.Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 28.Chene P, Fuchs J, Bohn J, Garcia-Echeverria C, Furet P, Fabbro D. A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines. J Mol Biol. 2000;299:245–253. doi: 10.1006/jmbi.2000.3738. [DOI] [PubMed] [Google Scholar]
- 29.Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaelis M, Rothweiler F, Barth S, et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2011;2:e243. doi: 10.1038/cddis.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Sun W, Zhao Y, et al. SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann S, Bykov VJ, Ali D, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 33.Schuler M, Rochlitz C, Horowitz JA, et al. A phase I study of adenovirus-mediated wild-type p53 gene transfer in patients with advanced non-small cell lung cancer. Hum Gene Ther. 1998;9:2075–2082. doi: 10.1089/hum.1998.9.14-2075. [DOI] [PubMed] [Google Scholar]
- 34.Swisher SG, Roth JA, Nemunaitis J, et al. Adenovirus-mediated p53 gene transfer in advanced non-small-cell lung cancer. J Natl Cancer Inst. 1999;91:763–771. doi: 10.1093/jnci/91.9.763. [DOI] [PubMed] [Google Scholar]
- 35.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff DD, Kirn DH. ONYX-015, an E1B gene-attenuated adenovi-rus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 36.Lamfers ML, Grill J, Dirven CM, et al. Potential of the conditionally replicative adenovirus Ad5-Delta24RGD in the treatment of malignant gliomas and its enhanced effect with radiotherapy. Cancer Res. 2002;62:5736–5742. [PubMed] [Google Scholar]
- 37.Khuri FR, Nemunaitis J, Ganly I, et al. A controlled trial of intratu-moral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6:879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 38.Nemunaitis J, Ganly I, Khuri F, et al. Selective replication and on-colysis in p53 mutant tumors with ONYX-015, an E1B-55kD gene-deleted adenovirus, in patients with advanced head and neck cancer: a phase II trial. Cancer Res. 2000;60:6359–6366. [PubMed] [Google Scholar]
- 39.Mehta SA, Christopherson KW, Bhat-Nakshatri P, et al. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007;26:3329–3337. doi: 10.1038/sj.onc.1210120. [DOI] [PubMed] [Google Scholar]
- 40.Weissmueller S, Manchado E, Saborowski M, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leffers N, Lambeck AJ, Gooden MJ, et al. Immunization with a P53 synthetic long peptide vaccine induces P53-specific immune responses in ovarian cancer patients, a phase II trial. Int J Cancer. 2009;125:2104–2113. doi: 10.1002/ijc.24597. [DOI] [PubMed] [Google Scholar]
- 42.Chiappori AA, Soliman H, Janssen WE, Antonia SJ, Gabrilovich DI. INGN-225: a dendritic cell-based p53 vaccine (Ad.p53-DC) in small cell lung cancer: observed association between immune response and enhanced chemotherapy effect. Expert Opin Biol Ther. 2010;10:983–991. doi: 10.1517/14712598.2010.484801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuler PJ, Harasymczuk M, Visus C, et al. Phase I dendritic cell p53 peptide vaccine for head and neck cancer. Clin Cancer Res. 2014;20:2433–2444. doi: 10.1158/1078-0432.CCR-13-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 45.Teng DH, Hu R, Lin H, et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- 46.Myers MP, Pass I, Batty IH, et al. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci U S A. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 48.Datta SR, Dudek H, Tao X, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 49.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phos-phorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 51.Puc J, Keniry M, Li HS, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 53.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haruta T, Uno T, Kawahara J, et al. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteaso-mal degradation of insulin receptor substrate-1. Mol Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 56.Chandarlapaty S, Sawai A, Scaltriti M, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao JJ, Castel P, Radosevic-Robin N, et al. Antagonism of EGFR and HER3 enhances the response to inhibitors of the PI3K–Akt pathway in triple-negative breast cancer. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005125. ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 60.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 62.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) poly-merase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 63.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mateo J, Ong M, Tan DS, Gonzalez MA, de Bono JS. Appraising iniparib, the PARP inhibitor that never was-what must we learn? Nat Rev Clin Oncol. 2013;10:688–696. doi: 10.1038/nrclinonc.2013.177. [DOI] [PubMed] [Google Scholar]
- 65.O’Shaughnessy J, Osborne C, Pippen JE, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 66.Shall S, Gaymes T, Farzaneh F, Curtin N, Mufti GJ. The use of PARP inhibitors in cancer therapy: use as adjuvant with chemotherapy or radiotherapy; use as a single agent in susceptible patients; techniques used to identify susceptible patients. Methods Mol Biol. 2011;780:239–266. doi: 10.1007/978-1-61779-270-0_15. [DOI] [PubMed] [Google Scholar]
- 67.Gupta A, Yang Q, Pandita RK, et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8:2198–2210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- 68.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez AA, Makris A, Wu MF, et al. DNA repair signature is associated with anthracycline response in triple negative breast cancer patients. Breast Cancer Res Treat. 2010;123:189–196. doi: 10.1007/s10549-010-0983-z. [DOI] [PubMed] [Google Scholar]
- 70.Turner N, Tutt A, Ashworth A. Hallmarks of ’BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 71.Lee MN, Tseng RC, Hsu HS, et al. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res. 2007;13:832–838. doi: 10.1158/1078-0432.CCR-05-2694. [DOI] [PubMed] [Google Scholar]
- 72.Weston VJ, Oldreive CE, Skowronska A, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 73.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin SA, McCabe N, Mullarkey M, et al. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang H, Reinhardt HC, Bartkova J, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, Thomas HD, Batey MA, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 77.Mizuarai S, Yamanaka K, Itadani H, et al. Discovery of gene expression-based pharmacodynamic biomarker for a p53 context-specific anti-tumor drug Wee1 inhibitor. Mol Cancer. 2009;8:34. doi: 10.1186/1476-4598-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Wurdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 79.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multipara-metric phenotyping. Nat Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 81.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011;21:34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Jerby-Arnon L, Pfetzer N, Waldman YY, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. cell. 2014;158:1199–1209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 83.Muller FL, Colla S, Aquilanti E, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 89.Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol. 2012;4:a007989. doi: 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 92.Fang F, Turcan S, Rimner A, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3001875. 75ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanai Y, Ushijima S, Nakanishi Y, Sakamoto M, Hirohashi S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003;192:75–82. doi: 10.1016/s0304-3835(02)00689-4. [DOI] [PubMed] [Google Scholar]
- 94.Yamashita Y, Yuan J, Suetake I, et al. Array-based genomic rese-quencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 95.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. International Vidaza High-Risk MDS Survival Study Group. Efficacy of azaciti-dine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bejar R, Lord A, Stevenson K, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu C, Venneti S, Akalin A, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27:1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Borodovsky A, Salmasi V, Turcan S, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–1747. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turcan S, Fabius AW, Borodovsky A, et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget. 2013;4:1729–1736. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 105.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16(spec no 1):R28–R49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- 107.Duvic M, Talpur R, Ni X, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Piekarz RL, Frye R, Turner M, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 111.Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim W, Bird GH, Neff T, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolchok JD, Kluger H, Callahan MK, et al. Nivolumab plus ipili-mumab in advanced melanoma. N. Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rajasagi M, Shukla SA, Fritsch EF, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H, Han H, Mousses S, Von Hoff DD. Targeting loss-of-function mutations in tumor-suppressor genes as a strategy for development of cancer therapeutic agents. Semin Oncol. 2006;33:513–520. doi: 10.1053/j.seminoncol.2006.04.013. [DOI] [PubMed] [Google Scholar]