Abstract
Despite exciting new possibilities for regenerative therapy posed by the ability to induce pluripotent stem cells, recapitulation of three-dimensional kidneys for repair or replacement has not been possible. ARID3a-deficient mouse tissues generated multipotent, developmentally plastic cells. Therefore, we assessed the adult mouse ARID3a−/− kidney cell line, KKPS5, which expresses renal progenitor surface markers as an alternative cell source for modeling kidney development. Remarkably, these cells spontaneously developed into multicellular nephron-like structures in vitro, and engrafted into immunocompromised medaka mesonephros, where they formed mouse nephron structures. These data implicate KKPS5 cells as a new model system for studying kidney development.
Keywords: ARID3a, Renal progenitor, Nephron structure, Medaka
1. Introduction
Damaged kidneys rarely recover function, and patients with chronic kidney diseases require dialysis or kidney transplantation for end-stage renal insufficiency. The renal transplantation approach suffers from donor shortages and complications of immune rejection [1]. The kidney is one of the most complicated organs in the human body, requiring multiple specialized cell types to maintain osmolality, excrete toxins, regulate blood pressure, promote erythropoiesis, and regulate bone density [2]. By birth, the human kidney has its full complement of about one million nephrons. To date, no one has been able to regenerate entire nephrons from normal or diseased kidneys. Regenerative medicine has therefore focused on the use of stem cell therapies to repair damaged nephrons, or to develop functional tissues that can perform some, but not all of the kidney’s functions. Producing kidney lineage cells from either mouse or human induced pluripotent stem cells (iPSCs) is a lengthy process that requires the timed addition of an expensive series of various growth factors to direct differentiation toward individual mature lineages and, disappointingly, results in insufficient kidney structures [3-7]. Thus, there is a clear need for additional approaches to generate kidney tissue.
ARID3a is one of fifteen proteins in the ARID family of DNA-binding proteins, many of which act in epigenetic regulation [8]. Tissues from transgenic mice expressing a dominant negative form of ARID3a and from the ARID3a knockout mouse formed developmentally plastic cells that expressed multiple pluripotency genes, suggesting they might be partially reprogrammed [9]. Although spontaneously renewing, these adult tissue-derived cell lines resembled primary cell lines rather than oncogenic lines, and were generated from a wide range of ARID3a-deficient tissues, including spleen, bone marrow, and kidney [9]. In addition, mouse embryonic fibroblasts from ARID3a knockout mice spontaneously formed induced iPSC-like colonies in vitro, and were reprogrammed more efficiently than wild-type cells using standard four-factor reprogramming [10]. ARID3a binds to the mouse OCT4 promoter and acts as a barrier to reprogramming by suppressing OCT4 transcription [10], but the mechanisms by which ARID3a inhibition allows generation of readily renewable, multipotent cell lines is currently unclear.
In this study, we explored the utility of an ARID3a−/− adult kidney cell line (KKPS5) for generating nephron structures in both in vitro and in vivo model systems. Surprisingly, these cells engrafted into immunocompromised medaka adult kidney (metanephros) and formed structures reminiscent of mouse kidney tubules. In addition, KKPS5 cells grown in semi-solid culture spontaneously generated complex structures composed of multiple mature kidney cell types within a few days. We are unaware of the existence of other cell lines that exhibit this unique multipotent property, and suggest that these cells provide a unique advantage over induced pluripotent stem cells for exploring kidney development. Moreover, we predict our findings will be relevant for future therapeutic manipulations in kidney disease.
2. Materials & methods
2.1. Cell culture
KKPS5 and iPSCs were cultured as previously described. Briefly, KKPS5 cells were maintained at 37 °C in RPMI-1640 (Life Technology, Grand Island, NY) with 5% fetal bovine serum (FBS; Life Technology), were washed in PBS with 2% FBS, and were resuspended at 3.5 × 104 cells/μl before implantation into medaka mesonephros, as described below. KKPS5 cells were resuspended at 105 cells/ml and mixed 1:4 with Matrigel (BD Biosciences, San Jose, CA) for 3-D cultures before we plated the suspension onto 1- or 4- well chamber slides (Nunc, Rochester, NY). Matrigel cultures were assessed daily using a Nikon Eclipse TS100 inverted microscope for up to 7 days, and were assessed for kidney structure formation by Olympus Provis AX-70 Epifluorescence Microscope (Olympus, Center Valley, PA).
2.2. FACS staining and analysis
Medaka mesonephros and adult C57Bl/6 mouse kidneys were mechanically dissociated to a single cell suspension. Whole kidneys and KKPS5 cells were stained for CD133-PE, CD34-PE, CD105-PE, CD90.2-FITC (eBiosciences, Inc., San Diego, CA), Sca-I–Pacific Blue, c-kit-PE-Cy7 (BioLegend, San Diego, CA), CD31-FITC, CD24-FITC, CD106-FITC, FLT-3-PE, CD9-Biotin, and Streptavidin-APC-Cy7 (BD Biosciences), and analyzed by flow cytometry using an LSRII (BD Biosciences) with FACSDiva (BD Biosciences) software version 4.1. Appropriate isotype controls (BD Biosciences, eBiosciences, Inc., BioLegend) were used for analyses. Doublet exclusion was used to ensure analyses of single cells prior to forward/side scatter gating. Data were analyzed using FlowJo (Tree Star, Inc., Ashland, OR) software version 10.
2.3. Fish maintenance
Medaka (Cab strain) were maintained and raised at 28.5 °C under a 14-h light/10-h dark cycle. Medaka embryos were kept at 28.5 °C in medaka embryo culture medium containing 17 mM NaCl, 0.4 mM KCl, 0.3 mM CaCl2, 0.65 mM MgSO4, and 0.01% methylene blue. All experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The medaka experiments were covered by protocols approved by the Institutional Animal Care and Use Committee of the University of Oklahoma Health Sciences Center (IACUC protocol No. 14-130-SSRCT to T.O.).
2.4. RT-PCR
RT-PCR of mouse and medaka transcripts was performed with total RNA isolated from mouse metanephros, medaka mesonephros, and KKPS5-engrafted medaka mesonephros using the RNAqueous®-4PCR Kit (Life Technology). Primers used to target mouse (mi) coding regions of WT-1, PAX2, OSR1, and GAPDH and medaka (me) GAPDH are listed in Table 1. RT-PCR was performed using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Life Technology), followed by nested PCR using Phusion High-Fidelity DNA Polymerase (Life Technology).
Table 1.
Oligonucleotide sequences, related to Fig. 3. Sequence of primers used in this study.
| Gene | Forward and reverse primer for RT-PCR | Forward and reverse primer for 2nd PCR |
|---|---|---|
| Mouse-WT1 | 5′-GAT GTG CGG CGT GTA TCT GG-3′ | 5′-ACT CTT GTC CGG TCA GCA TC-3′ |
| 5′-AGT GTG CTT CCG GCT ATG CAT CT-3′ | 5′-CTT CCG GCT ATG CAT CTG TAA-3′ | |
| Mouse-PAX2 | 5′-CTA CGA GAC TGG CAG CAT CA-3′ | 5′-GAG ACT GGC AGC ATC AA-3′ |
| 5′-GAT GGA TGA GAC ACT GGG AA-3′ | 5′-TGG ATG AGA CAC TGG GAA CTG-3′ | |
| Mouse-OSR1 | 5′-CAC CCA TCC CTG CAG CTT AC-3′ | 5′-CCA TCC CTG CAG CTT ACC AAT-3′ |
| 5′-TGG AGG TTT TGA GCT CCT TCA-3′ | 5′-TCT TGT GGA CAG CGA GAG TCC-3′ | |
| Mouse-GAPDH | 5′-GAC TCC ACT CAC GGC AAA TTC-3′ | 5′-CAA ATT CAA CGG CAC AGT CAA-3′ |
| 5′-CCT CAG TGT AGC CCA AGA TGC-3′ | 5′-GAC ACA TTG GGG GTA GGA ACA-3′ | |
| Medaka-GAPDH | 5′-CCG AGC TGC TTT CAC CTC TAA-3′ | 5′-GAC ACC AGA TCA CCG TCT TCC-3′ |
| 5′-CCA GCA TCA AAG ATG GAG GAG-3′ | 5′-GTG AGA TCA ACC ACC GAC ACA-3′ |
2.5. Immunohistochemistry
Medaka mesonephros was fixed with 4% PFA fixative (PBS containing 4% paraformaldehyde) overnight at 4 °C. Fixed samples were washed with PBS containing 0.3% Tween 20 (PBSTw), blocked with PBS-BB (PBS containing 0.5% Triton X-100, 0.2% (w/v) nonfat dry milk, and 1% Bovine Serum Albumin [BSA]) and incubated for 1 h with the primary antibodies diluted in PBS-BB. After washing three times with PBSTw for 30 min each time, samples were incubated for 1 h with Alexa-Fluor® 488-conjugated goat anti-rabbit IgG (H + L) (Life Technology) diluted with PBS-BB, washed three time with PBSTw for 30 min, dehydrated with a graded series of methanol, embedded in JB4 resin (Polysciences, Inc., Warrington, PA), and cut into 5-μm sections. Sections were stained with DAPI (KPL, Gaithersburg, MD), and mounted in Prolong® Gold Antifade Mountant (Life Technology). Matrigel slides were fixed with 4% PFA fixative for 10 min, washed twice with PBS +0.1 M glycine for 10 min each time, and cells were permeabilized with 0.1% Tween 20 in PBS for 1 h at room temperature (RT). Following permeabilization, wells were incubated with primary antibodies overnight at 4 °C and were washed three times with PBS for 5 min each time. Secondary antibodies were added for 2 h at RT. Cells were then stained with DAPI and mounted in ProLong® Gold Antifade mounting media (Life Technology). Primary antibodies were: Alexa Fluor® 488 anti-mouse Ly-6A/E (Sca-1) Antibody (BioLegend), Alexa Fluor® 488 anti-mouse CD24 Antibody (BioLegend), Anti-rabbit SIX2 (Proteintech, Chicago, IL), anti-WT1 (Abcam, Cambridge, MA), anti-NEPHRIN (US Biological, Salem, MA), anti-PAX2 (BioLegend), anti-JAGGED1 (Santa Cruz, California, CA), anti-thiazide-sensitive NaCl cotransport (NCC1, SLC12A3; EMD Milli- pore, Darmstadt, Germany), AQP2 (Santa Cruz), and anti-Na, K-ATPase (alpha6F; Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary antibodies were Alexa Fluor® 488-conjugated goat anti-rabbit IgG (H + L), Alexa Fluor® 568-conjugated goat anti-guinea pig IgG (H + L), Alexa Fluor® 568-conjugated goat anti-rabbit IgG (H + L), Alexa Fluor® 568-conjugated goat anti-mouse IgG (H + L), and Alexa Fluor® 568-conjugated donkey anti-goat IgG (H + L) from Life Technology. Samples were visualized using an Olympus FluoView 1000 confocal laser-scanning microscope and analyzed with FluoView imaging software (Olympus).
2.6. In situ hybridization and histological analyses
Mouse partial-length WT1 and OSR1 cDNA were obtained by RT-PCR from total RNA isolated from adult C57BL/6 mouse kidneys using the RNAqueous®-4PCR Kit (Life Technology). RT-PCR was performed using the SuperScript™ III One-Step RT-PCR System with Platinum® Taq High Fidelity, followed by a second PCR using Phusion® High-Fidelity DNA Polymerase (Life Technology). Table 1 lists all primers used. T7 RNA polymerase site (5′-GGT AAT ACG ACT CAC TAT AGG-3′ was added to WT1-R2 and OSR1-R2 3′ ends. The second PCR product was used as a template for the digoxigenin-labeled anti-sense RNA probe. All probes were synthesized using T7 RNA polymerase (New England BioLabs, Ipswich, MA) and DIG-RNA labeling (Roche, Indianapolis, IN) per the manufacturer’s instructions. Medaka mesonephros were fixed in 4% PFA, 0.1% Tween 20 in PBS for 2 h at RT and changed to 100% MeOH and stored at −20 °C. Whole mount in situ hybridization was performed as described previously [11]. Alkaline phosphatase-conjugated anti-digoxigenin (Roche) was used to localize the probes. NBT/BCIP (Roche) was used as the chromogenic substrate to produce the blue staining. After color development, samples were photographed on a Leica M165FC Z-stack microscope equipped with a Leica DFC295 camera (Leica, Buffalo Grove, IL). For histological analyses, medaka mesonephros were fixed, dehydrated and embedded in JB4 resin (Polysciences, Inc.) as described before [11,12]. We cut 4-μm sections using an RN2255 microtome (Leica, Buffalo Grove, IL) and stained them with Harris hematoxylin and special eosin II (BBC Biochemical, Mount Vernon, WA).
2.7. Engraftment to immunocompromised medaka mesonephros
Twenty-four hours before the implant, 8-week-old male medaka were treated with 10 Gy of gamma irradiation and were returned to the fish tank system without food. KKPS5 cells, as described above, were used within 30 min of harvest and were injected directly into one side of the mesonephros of medaka anesthetized in 0.02% ethyl 3-aminobenzoate methanesulfonate (MS222), using an insulin syringe. Implanted medaka were kept for 5 h in tank water containing 10 parts per million of MS222, before returning to the fish tank system with normal feedings.
3. Results
3.1. KKPS5 cells express surface markers used to identify renal progenitor cells in the adult mouse kidney
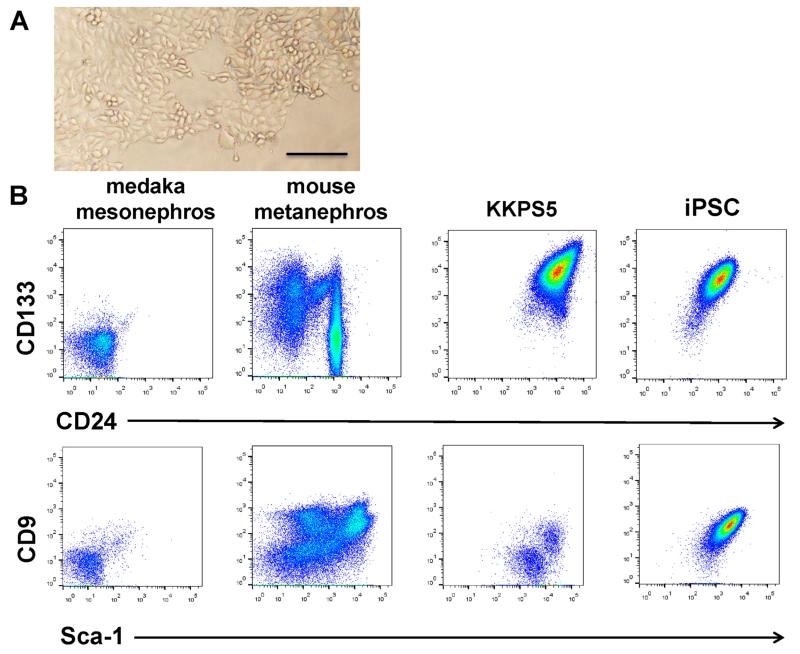
The KKPS5 adult ARID3a−/− kidney cell line was derived as a serially passaged bulk culture of kidney cells. In standard liquid culture media, KKPS5 cells are semi-adherent, heterogeneous cells that exhibit clusters of multiple cell types (Fig. 1A). Flow cytometric analyses revealed expression of high levels of CD133 and CD24 (Fig. 1B), markers previously used to identify kidney progenitor cells [13]. In contrast, adult (3-month-old) C57Bl/6 mouse kidney cells contained large numbers of Sca-I- and CD9-expressing cells, but fewer cells that expressed CD133 and CD24 (Fig. 1B). Although CD9 is typically expressed on iPSCs [14,15], KKPS5 cells expressed variable levels of CD9 and Sca-I (Fig. 1B). Flow cytometry revealed that KKPS5 cells also expressed varying levels of other surface markers characteristic of progenitor cells, including CD34 (57%), CD90 (18%), and c-kit (36%) (data not shown). These markers are expressed on some hematopoietic progenitors and mesenchymal stem cells [16]. The cells were negative for expression of other progenitor markers, including CD31, CD105, CD106, and FLT3. Therefore, KKPS5 cells express a combination of progenitor markers that are not consistent with predefined populations of precursors.
Fig. 1.
KKPS5 cells express a number of early lineage surface markers. (A) Phase contrast images of KKPS5 cells maintained in liquid culture. Scale bars: 200 μm. (B) Flow cytometric analyses of medaka mesonephros, cells from mouse metanephros, KKPS5 cells, and an ARID3a−/− iPSC line revealed expression levels of the progenitor markers CD133 and CD24, CD9, and Sca-I.
3.2. Mature kidney cell types and structures spontaneously develop from KKPS5 cells grown in matrigel
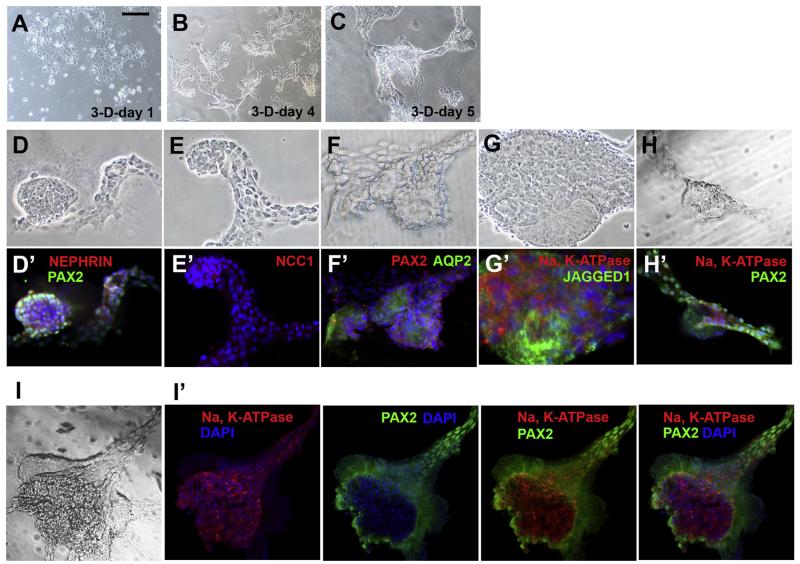
Several studies showed that kidney progenitors form some types of nephron structures in 3-D matrix cultures with the addition of specific growth factors [17–19]. To determine the differentiation potential of KKPS5 cells, we seeded cells into standard culture media with 25% Matrigel. After 1 day, cells were not visibly different than cells grown in liquid culture (Fig. 2A). However, by day 4 of culture aggregates formed (Fig. 2B), and these continued to develop into multicellular, organized structures at day 5 (Fig. 2C). More complex structures were observed after a week (Fig. 2D–H, brightfield panels). These complex structures contained cells that expressed immunofluorescent markers for mouse proximal tubule proteins (PAX2 and JAGGED1), glomerulus podocytes (NEPHRIN), the thick ascending limb of the loop of Henle (NCC1), and the collecting duct (AQP2, Fig. 2D′–H′) [4,7,19]. Interestingly, matrigel cultures of KKPS5 cells grown for more than a week began to further form polarized tubule-like epithelial structures, and Na K-ATPase was localized in the basolateral membranes of both PAX2-positive and -negative epithelial structures (Fig. 2I–I′). Together, these data indicate that the KKPS5 cell line is a unique source of progenitors that spontaneously develop into multicellular structures that resemble structural components of the mature kidney and express appropriate mature kidney markers.
Fig. 2.
KKPS5 cells grown in matrigel spontaneously form complex multicellular structures that express mature mouse kidney cell markers. (A–C) Brightfield images of KKPS5 cells plated in 3-D matrigel cultures at 1, 4, and 5 days are shown. (D–H) Representative brightfield images and (D′–H′) the corresponding immunostaining for mature kidney markers (red and green labels) of 3-dimensional structures spontaneously formed within 7 days after KKPS5 cells were plated in semisolid matrigel cultures. Nuclei were stained with DAPI. Complex structure shown as brightfield (I) and co-stained with multiple markers (I′). Scale bars: 200μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. KKPS5 cells engrafted into immunocompromised medaka better than conventional iPSCs
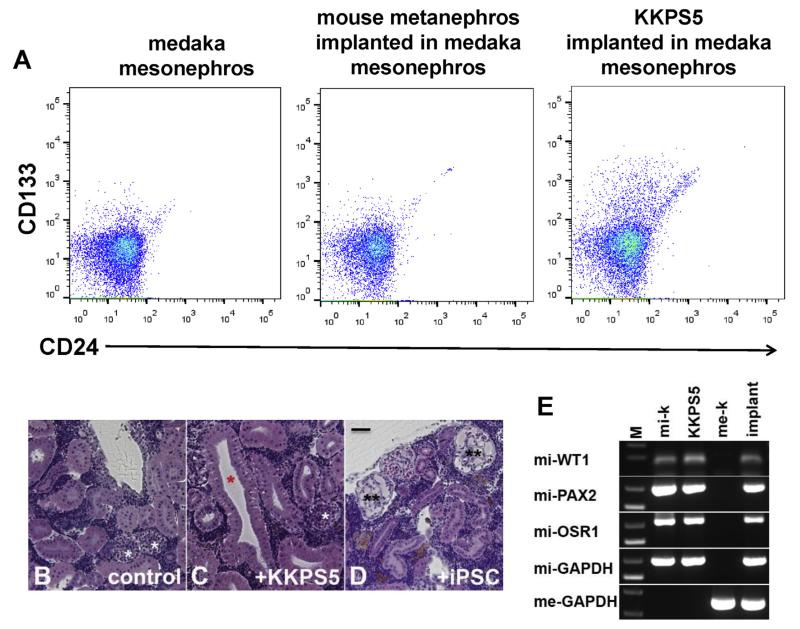
Next, we asked whether KKPS5 cells could form kidney structures in vivo. Because this cell line was developed from mouse kidney, which would make tracking implanted cells in a mouse kidney difficult, we assessed a xenograft model using medaka as recipients. We implanted 35,000 KKPS5 cells or conventionally generated ARID3a−/− iPSCs into irradiated, immunocompromised 8-week-old male medaka mesonephros and assessed them for implantation by flow cytometric analyses (Fig. 3A) on day 34 after implantation (34 dai). Neither control nor mouse metanephros-implanted medaka showed expression of mouse surface markers CD133 and CD24 (<1%), but KKPS5-implanted medaka contained small numbers of cells (7%) that expressed both markers (Fig. 3A). Histological examination of control medaka mesonephros revealed closed nephron structures typical of fish mesonephros (white asterisks, Fig. 3B), while KKPS5-implanted mesonephros showed distinct open tubule structures more typical of mouse kidney structures (red asterisk) incorporated into the medaka mesonephros (white asterisk, Fig. 3C). The glomeruli diameters in control and KKPS5-implanted medaka mesonephros were 45–65 μm (Fig. 3B–C), while mesonephros implanted with ARID3a−/− iPSCs had enlarged glomeruli (90–120 μm), with mesangial expansion and glomerular capillary dilation (two black asterisks, Fig. 3D) more typical of diseased kidneys [12]. Mouse-specific RNA transcripts were detected as early as 7 dai (Fig. 3E). Mature mouse kidney transcripts mi-WT1 (glomerulus podocytes), mi-PAX2 (proximal tubule), and the renal progenitor marker mi-OSR1 [4,7,19], were observed in adult mouse kidney (mi-k), KKPS5 cells, and medaka mesonephros implanted with KKPS5 cells, but were not evident in total RNA prepared from medaka mesonephros (Fig. 3E). As expected, mouse-specific GAPDH (mi-GAPDH) was only detected in RNA prepared from mouse mesonephros, KKPS5 cells, and medaka mesonephros implanted with KKPS5 cells. Medaka GAPDH (me-GAPDH) was specifically expressed in RNA prepared from medaka mesonephros and medaka mesonephros implanted with KKPS5 cells. Together, these data indicate that KKPS5 cells engraft into the medaka kidney and can be distinguished, genetically and morphologically, from the medaka mesonephros.
Fig. 3.
KKPS5 cells implant into medaka mesonephros and differentiate into mature renal cell types. (A) Flow cytometric analyses of medaka mesonephros at 34 dai from control, mouse metanephros-implanted, and KKPS5-implanted medaka revealed expression of mouse CD133 and CD24. (B–D) H&E-stained histology sections from representative medaka mesonephros of control, KKPS5-implanted, and iPSC-implanted medaka are shown. Colored asterisks indicate: medaka glomerulus (white), mouse-like tubules (red), and dilated glomerulus (black). Scale bars: 50 μm. (E) RT-PCR for mi-WT1 (glomerulus podocytes), mi-PAX2 (proximal tubules), mi-OSR1 (renal progenitors), mi-GAPDH (internal marker), and me-GAPDH (internal marker) are shown for representative mouse metanephros (mi-k), KKPS5, medaka mesonephros (me-k), and KKPS5-implanted medaka mesonephros (implant). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. KKPS5 cells generated multiple types of mouse kidney cells and structures in the immunocompromised medaka kidney
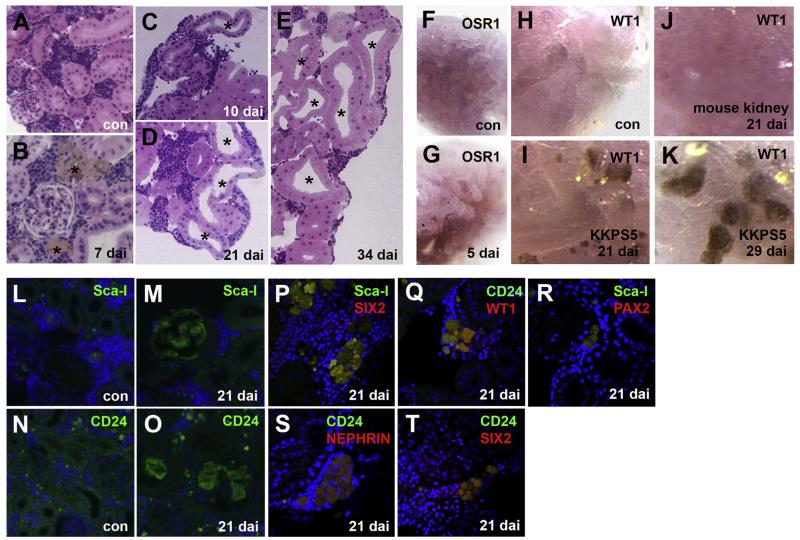
We further assessed the capability of KKPS5 cells to generate mature mouse kidney structures in a developmentally progressive fashion from 5 to 34 dai (Fig. 4). Medaka tubules characteristically form a long brush border in the apical region of cells (Fig. 4A–E), while mouse tubules show a shorter brush border (black asterisk shows apical open region of the mouse kidney tubule) and a wider apical region (Fig. 4C–E). Mouse tubule structures in medaka mesonephros were confirmed in histology sections (Fig. 4B–E). Tubules were detected at 10 dai (Fig. 4C), and numbers of tubules increased by 21 and 34 dai (Fig. 4D, E). At 5 dai, mi-OSR1 was detected in implanted medaka mesonephros (Fig. 4G), but was not present in the control medaka mesonephros (Fig. 4F). The earliest we observed KKPS5-derived cells, marked by Sca-I in the medaka mesonephros, was at 7 dai, (Fig. 4B). At 21 dai, mi-WT1, a mouse podocyte marker was detected in some cells within the medaka mesonephros (Fig. 4I). By 29 dai, mi-WT1 was also detected in structures reminiscent of newly formed mouse glomerulus podocytes intermingled in the medaka mesonephros (Fig. 4K). mi-WT1 was undetected in control medaka mesonephros without KKPS5 implantation (Fig. 4H). Furthermore, implantation of adult mouse kidney cells failed to generate mi-WT1 expressing cells or structures (Fig. 4J). We also examined a number of mouse kidney cell surface markers in KKPS5-implanted medaka at 21 dai (Fig. 4M, O–T). At that time point, the progenitor markers CD24 and Sca-I were found with co-expression of SIX2 (progenitor and proximal marker), PAX2 (proximal tubules), and podocytes markers WT1 and NEPHRIN (Fig. 4M, O–T) [4,7,19]. Sca-I and CD24 were undetected in the un-implanted medaka mesonephros (Fig. 4L, N). Therefore, the KKPS5 cell line exhibits unique properties compared with freshly isolated adult mouse kidney cells that allow the cell line to engraft, survive, and form structures within the medaka mesonephros.
Fig. 4.
KKPS5 cells implanted in the medaka mesonephros express mouse-specific kidney protein markers. (A–E) Representative H&E-stained histology sections show overall features of the medaka nephron structure at the indicated time points post KKPS5 implantation. Scale bars: 50 μm *indicates mouse tubule structures in medaka mesonephros. (F–K) Representative in situ hybridization of sections show expression of mouse OSR1 and WT1 in control (con), mouse kidney-implanted, and KKPS5-implanted mesonephros. (L–T) Representative immunostaining of KKPS5-implanted medaka mesonephros for Sca-I (green), SIX2 (red), CD24 (green), WT1 (red), and PAX2 (green) are shown. DAPI was used to counterstain nuclei (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The present study demonstrated that an ARID3a-deficient adult mouse kidney cell line that expresses multiple kidney progenitor surface antigens spontaneously develops into mouse nephron-like structures in 3D culture that express mature kidney markers. In addition, using a xenograft model system to help distinguish KKPS5-derived cells from host nephrons, we showed that KKPS5 cells can be successfully engrafted into medaka mesonophros, where they develop into structures reminiscent of mouse kidney tubules. These data implicate this multipotent, and readily renewable, cell line as a unique source of tissue-specific progenitors.
The metanephric kidney is composed of multiple kidney-specific cell types derived from the ureteric bud and metanephric mesenchyme, two distinct progenitor populations. This cell line is the first reported to allow generation of nephron structures derived from both ureteric bud and metanephric nephron-derived cells. Therefore, the KKPS5 cell line provides several advantages over current technology for studying kidney organogenesis. KKPS5 cells are easily grown in culture and do not require isolation of embryonic renal progenitors, such as metanephric mesenchyme and ureteric bud cells, from tissues. Our system allows spontaneous generation of different types of kidney structures within 5 days in 3-D culture, and polarized epithelial structures within a week. While others succeeded in generating intermediate mesoderm, producing 90% OSR1+ cells from human iPSCs, differentiation of multiple mature cell types required 10–22 days [3]. While Kitamura et al. (2015) showed reconstitution of a 3-D kidney-like structure from the S3 segment of adult rat kidney nephrons using an extracellular matrix gel and the presence of several growth factors, our model system did not require the addition of exogenous growth factors. Furthermore, KKPS5 cells were amenable to growth in vivo, and when implanted into medaka kidney, allowed production of OSR1+ renal progenitors at 5 dai, with mature kidney cell markers following soon afterward. Our data suggest that the KKPS5 cell line is a readily renewable source of tissue-specific progenitors, which can be experimentally manipulated.
Although KKPS5 cells form structures reminiscent of glomeruli, additional studies will be required to explore their functional attributes. Nevertheless, KKPS5 cells represent a new model system that fills a major gap in our ability to study the development of multicellular kidney structures in an efficient and cost-effective fashion. Whether human cell lines similar to KKPS5 can be developed remains to be seen.
Acknowledgments
We thank Dr. Anupam Agarwal, Dr. Troy D. Templeton, Ms. Meghna D. Mehta, and Mr. James Griffith for technical support. The authors thank Drs. Deborah Hyink and Kathy Kyler for helpful criticism and comments on the manuscript. T.O. acknowledges financial support from the University of Oklahoma Health Sciences Center (OUHSC). C.F.W. was supported by the Oklahoma Center for Adult Stem Cell Research. T.O. was supported by OCAST (HR14-082), a PHF grant, and a VPR grant. This work was also supported in part by the Diabetes Histology and Image Acquisition and Analysis Core Facility at OUHSC (NIH: COBRE-1P20RR024215).
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.06.130.
References
- [1].Coresch J, Selvin E, Steven LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- [2].Saxen L. Organogenesis of the Kidney. Cambridge University Press; New York: 1987. [Google Scholar]
- [3].Mae S, Shono A, Shiota F, et al. Monitoring and robust induction of nephrogenic intermediated mesoderm from human pluripotent stem cells. Nat. Commun. 2013;4 doi: 10.1038/ncomms2378. http://dx.doi.org/10.1038/ncomms2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Taguchi A, Kaku Y, Ohmori T, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell. Stem Cell. 2013;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- [5].Araoka T, Mae S, Kurose Y, Uesugi M, et al. Efficient and rapid induction of human iPSCs/ESCs into nephrogenic intermediate mesoderm using small molecule-based differentiation methods. PLoS One. 2014;9:e84881. doi: 10.1371/journal.pone.0084881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Harari-Steinberg O, Metsuyanim S, Omer D, Gnatek Y, et al. Identification of human nephron progenitors capable of generation of kidney structures and functional repair of chronic renal disease. EMBO Mol. Med. 2013;5:1556–1568. doi: 10.1002/emmm.201201584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takasato M, Er PX, Becroft M, Vanslambrouck JM, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- [8].Wilsker D, Patsialou A, Dallas PB, Moran E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002;13:95–106. [PubMed] [Google Scholar]
- [9].An G, Miner CA, Nixon JC, Kincade PW, et al. Loss of Bright/ARID3a function promotes developmental plasticity. Stem Cells. 2010;28:1560–1567. doi: 10.1002/stem.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Popowski M, Templeton TD, Lee BK, Rhee C, et al. Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell. Rep. 2014;2:26–35. doi: 10.1016/j.stemcr.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ichimura K, Powell R, Nakamura T, Kurihara H, et al. Podocalyxin regulates pronephric glomerular development in zebrafish. Physiol. Rep. 2013;1:e00074. doi: 10.1002/phy2.74. http://dx.doi.org/10.1002/phy2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ichimura K, Kawashima Y, Nakamura T, Powell R, et al. Medaka fish, Oryzias latipes, as a model for human obesity-related glomerulopathy. Biochem. Biophys. Res. Commun. 2013;341:712–717. doi: 10.1016/j.bbrc.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lazzeri E, Crescioli C, Ronconi E, Mazzinghi B, et al. Regenerative potential of embryonic renal multipotent progenitors in acute renal failure. J. Am. Soc. Nephrol. 2007;18:3128–3138. doi: 10.1681/ASN.2007020210. [DOI] [PubMed] [Google Scholar]
- [14].Mugford JW, Sipilä P, McMahon JA, McMahon AP. Osr1 expression demarcates a multiple-potent population of intermediated mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dekel B, Zangi L, Shezen E, Reich-Zeliger S, Eventov-Friedman S, et al. Isolation and characterization of nontubular sca1+lin− multipotent stem/progenitor cells from adult mouse kidney. J. Am. Soc. Nephrol. 2006;17:3300–3314. doi: 10.1681/ASN.2005020195. [DOI] [PubMed] [Google Scholar]
- [16].Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, et al. Isolation of a candidate human hematopoietic stem-cell population. Proc. Natl. Acad. Sci. U. S. A. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Joraku A, Stem AK, Atala A, Yoo JJ. In vitro generation of three-dimensional renal structures. Methods. 2009;47:129–133. doi: 10.1016/j.ymeth.2008.09.005. [DOI] [PubMed] [Google Scholar]
- [18].Lindgren D, Bostrom AK, Nilsson K, Hansson J, et al. Isolation and characterization of progenitor-like cells from human renal proximal tubules. Am. J. Pathol. 2011;178:828–837. doi: 10.1016/j.ajpath.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kitamura S, Sakurai H, Makino H. Single adult kidney stem/progenitor cells reconstitute 3-dimensional nephron structure in vitro. Stem Cells. 2014;33:774–784. doi: 10.1002/stem.1891. [DOI] [PubMed] [Google Scholar]