Abstract
Ankyloblepharon-ectodermal defects-cleft lip/palate (AEC) syndrome is a rare monogenetic disorder that is characterized by severe abnormalities in ectoderm-derived tissues, such as skin and its appendages. A major cause of morbidity among affected infants is severe and chronic skin erosions. Currently, supportive care is the only available treatment option for AEC patients. Mutations in TP63, a gene that encodes key regulators of epidermal development, are the genetic cause of AEC. However, it is currently not clear how mutations in TP63 lead to the various defects seen in the patients’ skin. In this review, we will discuss current knowledge of the AEC disease mechanism obtained by studying patient tissue and genetically engineered mouse models designed to mimic aspects of the disorder. We will then focus on new approaches to model AEC, including the use of patient cells and stem cell technology to replicate the disease in a human tissue culture model. The latter approach will advance our understanding of the disease and will allow for the development of new in vitro systems to identify drugs for the treatment of skin erosions in AEC patients. Further, the use of stem cell technology, in particular induced pluripotent stem cells (iPSC), will enable researchers to develop new therapeutic approaches to treat the disease using the patient’s own cells (autologous keratinocyte transplantation) after correction of the disease-causing mutations.
Keywords: ectodermal dysplasia, ankyloblepharon-ectodermal defects-cleft lip/palate syndrome (AEC), TP63, induced pluripotent stem cells, iPS cells, iPSC-derived keratinocytes, iPSC-based cell therapy, skin equivalents, genodermatoses, inherited skin disorders, in vitro disease models
INTRODUCTION
Ectodermal dysplasias (ED) represent a group of inherited disorders that are characterized by developmental abnormalities in ectoderm-derived tissues and organs, including the epidermis and ectodermal appendages, such as hair follicles. Approximately 200 distinct ED have been described, however, the underlying genetic mutations have been identified only in approximately 30% of these disorders [Priolo, 2009; Visinoni et al., 2009]. Mutations in the TP63 gene have been found to underlie several different ED. ED caused by TP63 mutations include Ectrodactyly, ectodermal dysplasia, and cleft lip/palate syndrome (EEC; OMIM# 604292) [Celli et al., 1999], ADULT syndrome (OMIM# 103285) [Duijf et al., 2002], Limb-mammary syndrome (LMS; OMIM# 603543) [van Bokhoven et al., 2001], and Ankyloblepharon-ectodermal defects-cleft lip/palate syndrome (AEC or Hay Wells syndrome; OMIM# 106260) [McGrath et al., 2001]. A fifth condition caused by TP63 mutations is Rapp-Hodgkin syndrome (OMIM# 129400) [Kantaputra et al., 2003]; however, this syndrome is now considered to represent the same clinical entity as AEC [Bertola et al., 2004; Clements et al., 2010]. Thus, in this manuscript, we will use the inclusive term “AEC” to refer to Hay Wells syndrome, Rapp-Hodgkin syndrome, or AEC syndrome.
ANKYLOBLEPHARON-ECTODERMAL DEFECTS-CLEFT LIP/PALATE (AEC) SYNDROME
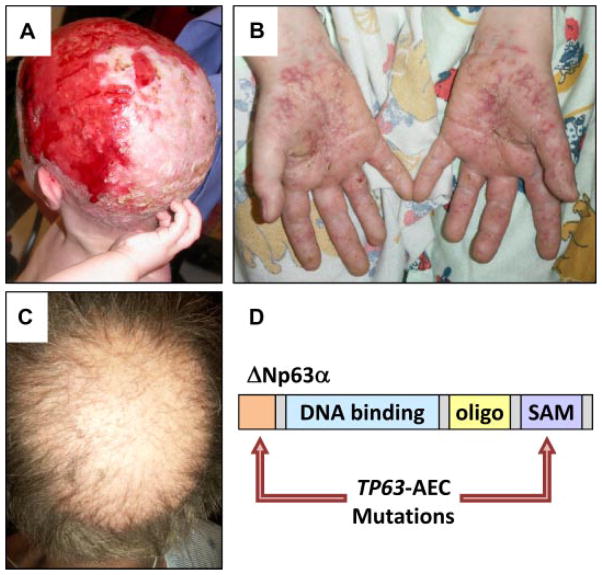
A hallmark of the clinical AEC phenotype is the presence of severe scalp erosions (Fig. 1A). However, other skin sites may also be affected by erosions, including palms and soles (Fig. 1B). Skin erosions are a major cause of morbidity in infants with AEC, often complicated by local and systemic infections, which may be life-threatening [Vanderhooft et al., 1993; Siegfried et al., 2005; Julapalli et al., 2009]. Currently, there is no cure for the skin erosions, and symptomatic wound care is only partially effective [Julapalli et al., 2009].
FIG. 1.
Mutations in TP63 underlie AEC, characterized by skin fragility and hair abnormalities. A: Scalp erosions on an AEC patient. B: Palmar epidermal erosions. C: Hair abnormalities, including partial alopecia, reduced hair shaft density, and hair shaft abnormalities. D: Schematic of ΔNp63α indicating the most common mutation sites identified in AEC patients.
In addition to the extensive skin erosions, the AEC phenotype often includes cleft lip, cleft palate, and abnormalities in several other ectoderm-derived tissues, including sweat glands, teeth, nails, limbs, and hair [Bree, 2009; Cole et al., 2009; Koster, 2010]. The observation that different appendages are affected in AEC patients reflects the crucial role for TP63 in the initial steps of forming all of these structures during development [Koster and Roop, 2004; Mikkola, 2007]. We chose to study the role of TP63 in the hair follicle in part because the regulatory pathways controlling normal hair follicle development and homeostasis have been relatively well-characterized [Schmidt-Ullrich and Paus, 2005; Duverger and Morasso, 2009], thus allowing us to link the effects of TP63-AEC mutations to specific signaling pathway abnormalities. Hair abnormalities in AEC patients include patchy or complete hair loss (alopecia), diminished hair density in areas where hair is present, and structural hair shaft abnormalities [Salinas and Montes, 1988; Hicks et al., 2001; Dishop et al., 2009] (Fig. 1C). Although sparse hair is a feature of most ED, the extensive and sometimes complete alopecia in AEC is likely a consequence of scarring as a result of scalp erosions. Systematic evaluation of AEC scalp biopsies and trichograms (i.e., microscopic analysis of plucked hair) would be valuable tools to help define the nature and causes of the alopecia, including the identification of the hair follicle cell types affected by TP63-AEC proteins. Research in our laboratories and others is currently addressing this question, and is expected to lead to a better understanding of the role of TP63 in hair follicle biology.
TP63 Mutations in AEC
The TP63 gene encodes at least 10, and possibly more, transcription factors that differ only in their N- (TA and ΔN) and C-termini (α–ε) [Yang et al., 1998; McGrath et al., 2001; Koster, 2010]. Further, all known TP63 isoforms contain identical DNA binding and oligomerization domains. ΔNp63α is the predominantly expressed TP63 isoform in the epidermis and in skin appendages [Yang et al., 1998; Liefer et al., 2000; Koster et al., 2004; Laurikkala et al., 2006]. This isoform was also found to be critical for normal development and homeostasis of the skin in mice and humans [Mills et al., 1999; Yang et al., 1999; Koster and Roop, 2004; Koster and Roop, 2008].
TP63 mutations in AEC patients (TP63-AEC) primarily cluster in the α C-terminus of TP63, a domain that is encoded by exons 13 and 14 of the TP63 gene and that contains a SAM domain (sterile α motif; a predicted protein-protein or protein-RNA interaction motif) [McGrath et al., 2001; Rinne et al., 2007, 2009]. In addition, mutations in the N-terminus of ΔNp63 isoforms have been described [Rinne et al., 2008], collectively suggesting a central role for mutant ΔNp63α isoforms (ΔNp63α-AEC) in the pathogenesis of AEC (Fig. 1D).
Cellular and Molecular Abnormalities in AEC Patient Skin
To gain insight into the pathological roles of TP63-AEC proteins, a first prerequisite is the identification of the cellular abnormalities that occur in the skin of AEC patients. To this end, we and others have analyzed skin biopsies from AEC patients. The overall consensus is that epidermal differentiation fails to occur normally in the skin of AEC patients. Importantly, these abnormalities were observed even when biopsies were obtained from non-lesional skin. At this time, the underlying reason for the focal nature of the skin erosions is not known; however, one possibility is that erosions are triggered by mechanical trauma.
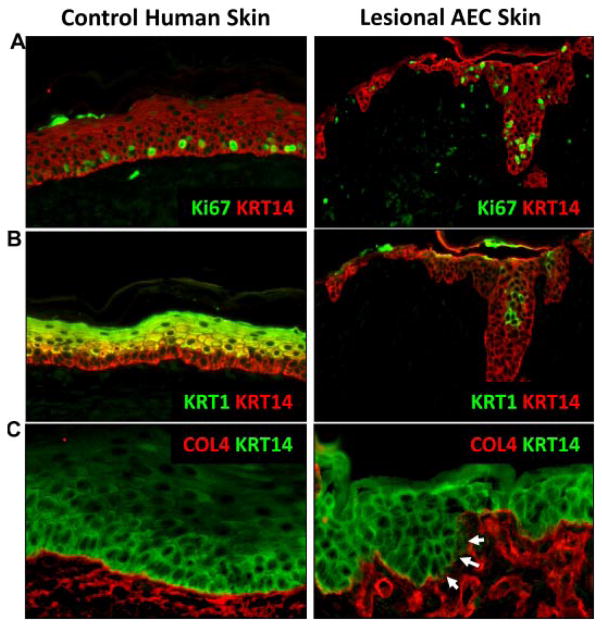
In normal epidermis, proliferating cells are restricted to the basal layer and cells permanently withdraw from the cell cycle as they move suprabasally and embark on the terminal differentiation program [Koster and Roop, 2007]. This withdrawal from the cell cycle is, in part, mediated by the induction of IKKα and IRF6, two ΔNp63α target genes [Hu et al., 1999; Li et al., 1999; Takeda et al., 1999; Ingraham et al., 2006; Richardson et al., 2006; Koster et al., 2007; Marinari et al., 2008; Moretti et al., 2010]. Concomitant with the cell cycle withdrawal, markers of epidermal differentiation are induced, including keratins 1 and 10 (KRT1 and KRT10) and involucrin. Analysis of skin biopsies from AEC patients demonstrated abnormalities in epidermal differentiation as indicated by the persistence of proliferating cells in the suprabasal cell layers and the failure to induce KRT1, either completely or focally [McGrath et al., 2001; Marinari et al., 2008; Koster et al., 2009; Browne et al., 2011; Clements et al., 2012] (Fig. 2A,B).
FIG. 2.
Cellular abnormalities observed in the skin of AEC patients. A: Proliferation, as shown here by Ki67 immunofluorescent staining (green) is restricted to the basal layer in normal human skin, but extends suprabasally in AEC patient skin. B: The marker for terminal differentiation, KRT1 (green/yellow), is induced in the first suprabasal layer in control human skin, but is delayed in its expression or absent from AEC patient skin. C: Immunofluorescent staining for the basement membrane component Collagen IV (COL4; red) shows uninterrupted staining along the basement membrane in control human skin, but is disrupted in AEC patient skin. Immunofluorescent staining for keratin 14 (KRT14) was used to highlight the epidermis in A (red), B (red), and C (green). Image reproduced from [Koster et al., 2009].
In addition to impaired terminal differentiation, AEC patient skin shows abnormalities in the basement membrane zone [Koster et al., 2009; Clements et al., 2012]. These defects include aberrant and/or reduced synthesis of the basement membrane components collagen IV (COL4) [Koster et al., 2009] and collagen VII (COL7A1) [Clements et al., 2012] (Fig. 2C). Currently, it is not known whether these collagen genes are direct transcriptional targets of TP63. Further, expression of FRAS1, a basement membrane component that is under the direct transcriptional control of ΔNp63α, was reduced in AEC patients [Koster et al., 2007; Clements et al., 2012]. Collectively, these data indicate that expression of TP63-AEC proteins impairs epidermal differentiation and basement membrane integrity, two defects that likely contribute to the skin fragility observed in AEC patients.
INSIGHTS FROM CELL CULTURE STUDIES
The analysis of skin biopsies from AEC patients has led to important insights into the pathogenesis of AEC. However, a shortcoming of skin biopsies is that they represent a single point in time at which the disease phenotype has developed. Thus, these tissues do not allow one to distinguish between direct and indirect effects of TP63-AEC protein expression and how the disease develops over time. In order to obtain mechanistic insights into the disease mechanism, cell culture systems are a valuable tool.
For example, although AEC patient skin demonstrated increased TP63 protein expression, the underlying reason for this was not clear [Browne et al., 2011; McGrath et al., 2001; Clements et al., 2012]. Cell culture studies demonstrated that the stability of ΔNp63α-AEC proteins is increased compared to ΔNp63α-wt (wild type ΔNp63α) proteins, at least in part due to their inability to be appropriately targeted for ubiquitin- or phosphorylation-dependent degradation [Di Costanzo et al., 2009; Bellomaria et al., 2010; Browne et al., 2011]. In addition, an altered ability of TP63-AEC proteins to interact with other proteins in the cell may affect its ability to induce or repress TP63 target genes. Few proteins that differentially interact with TP63-AEC and TP63-wt proteins have been identified so far. Of these, SATB2 (special AT-rich binding protein-2) displays a stronger interaction with TP63-AEC than with TP63-wt proteins [Chung et al., 2011]. Due to this interaction, TP63-AEC proteins fail to induce PERP, a TP63 target gene that is required for epidermal cell adhesion and that is downregulated in a subset of AEC patients [Ihrie et al., 2005; Beaudry et al., 2009; Chung et al., 2011], thus potentially contributing to the tissue fragility observed in patient skin. Conversely, unlike TP63-wt proteins, TP63-AEC proteins cannot interact with the CCAAT-binding factor NF-Y [Testoni and Mantovani, 2006]. This in turn leads to a failure to repress Cyclin B2 (CCNB2) expression, a mechanism that may contribute to the suprabasal keratinocyte proliferation observed in AEC patient epidermis. Consistent with the findings that TP63-AEC proteins affect the expression of known TP63 target genes, reporter gene assays have demonstrated that TP63-AEC proteins have a dominant-negative effect towards TP63-wt proteins on the promoters of several target genes including IKKα [Marinari et al., 2008; Koster et al., 2009], Claudin 1 [Lopardo et al., 2008], KRT14 [Browne et al., 2011], and BPAG1 [Browne et al., 2011]. The latter observation raises the interesting possibility that structural or functional defects in hemidesmosomes (of which BPAG1 is a component) might contribute to the epidermal fragility observed in AEC patient skin. Collectively, data obtained from cell culture studies indicate that expression of TP63-AEC proteins leads to diminished expression of critical TP63 target genes involved in keratinocyte differentiation and tissue resistance to mechanical stress. However, whether, analogous to mutant TP53 proteins [Brosh and Rotter, 2009], TP63-AEC proteins also have gain-of-function effects could not be addressed in these studies. Unbiased transcriptome analysis of AEC cells is necessary to gain a more precise understanding of the molecular role of TP63-AEC proteins.
MODELING OF AEC
Mouse Models
To gain a better understanding of the pathological mechanism underlying AEC and the role of TP63-AEC proteins in the disease process, it is important to generate models that replicate the disease. Although skin biopsies from patients represent a biologically ideal model, they are difficult to obtain for rare disorders, yield patient cells with a limited life-span in vitro, and have a diverse genetic background. Mouse models offer an attractive alternative due to the availability of inbred stains in which all mice have an identical genetic background. Further, mice that are genetically engineered to carry Trp63 mutations can be utilized to study the role of this gene in the development and maintenance of organs and tissues such as the skin.
To model human genetic disorders in mice, two main approaches have been undertaken. In the first model, a TP63 mutation originally identified in AEC patients (L514F) was genetically engineered into the mouse genome. Mice heterozygous for this mutant allele developed clefting of the secondary palate, hypoplasia and blistering of the epidermis, and a delay in hair follicle morphogenesis [Ferone et al., 2012; Ferone et al., 2013]. Thus, some important features of AEC, such as cleft palate and hair follicle abnormalities, are replicated in this model. Importantly, however, the defective epidermal proliferation and hypoplasia are inconsistent with data obtained using skin biopsies from AEC patients [Marinari et al., 2008; Koster et al., 2009; Clements et al., 2012].
A second model generated by the Koster laboratory took advantage of data indicating that ΔNp63α-AEC proteins function, at least in part, as dominant-negative molecules. This would suggest that AEC keratinocytes, that is, keratinocytes carrying one TP63-AEC allele and one TP63-wt allele, exhibit impaired TP63 function. To test this hypothesis, we generated a mouse model that enables us to downregulate ΔNp63 expression in the epidermis of mice in a spatially and temporally controlled manner [Koster et al., 2007]. We observed that downregulating ΔNp63 in postnatal mouse epidermis led to skin erosions with histological characteristics similar to those observed in AEC patients [Koster et al., 2009]. Skin erosions in AEC patients and in our mouse model displayed impaired terminal differentiation, as demonstrated by the presence of proliferating cells in the suprabasal epidermis and the failure to induce markers of terminal differentiation such as Krt1 [Koster et al., 2007, 2009]. In addition, we observed basement membrane abnormalities including a discontinuous pattern of collagen IV synthesis, also observed in AEC patient skin [Koster et al., 2007, 2009]. Finally, we detected a reduced expression of those Trp63/TP63 target genes that are downregulated in AEC patient skin, including Fras1 and IKKα [Koster et al., 2007; Marinari et al., 2008; Clements et al., 2012]. Thus, in addition to the skin erosions, we have been able to successfully mimic critical cellular and molecular features of AEC patient skin in our mouse model, suggesting that this model will be a useful tool to understand AEC pathology.
Human Models
As outlined above, genetically engineered mouse models have provided important insights into the function of TP63 during skin development and homeostasis. Further, at least some aspects of AEC have been recapitulated in these model organisms. However, none of the mouse lines developed to date completely mimics the AEC skin phenotype at both the genetic and the phenotypic level. This observation might, at least in part, be due to intrinsic differences in the biology of mouse and human skin [Godin and Touitou, 2007; Wong et al., 2011]. Human cell-based disease models are thus required to fill the gap in our understanding of the molecular and cellular mechanisms that lead to skin erosions in AEC patients.
Cell culture-based systems have a lower complexity than animals, thus facilitating theidentification of molecular defects specific to a particular cell type, such as skin keratinocytes. For AEC, one approach to generate AEC tissue models has been to introduce TP63-AEC expression constructs into normal human keratinocytes via retroviral vectors [Zarnegar et al., 2012]. These keratinocytes were then used to generate skin equivalents in cell culture. A transcriptome analysis of these cells led to the identification of signaling pathways that link TP63-AEC to the aberrant expression of epidermal differentiation genes. Consistent with AEC patient skin, a reduction of KRT1 expression was observed in this model. A disadvantage of this system is that ratio of TP63-AEC and TP63-wt expression is likely different from that found in patients. Consequently, transcriptome changes caused by TP63-AEC expression in these cultures might not accurately reflect transcriptome changes in AEC patient skin. Another concern is that the type of cell culture system described above relies on cells isolated from discarded surgical material, that is, the genetic background of these cells is not known and may not be identical in each experiment. Importantly, such differences in genetic background can significantly affect disease phenotypes. For example, it is well-known that the same TP63 mutation can lead to different clinical manifestations, even within individuals from the same family [Dianzani et al., 2003; Bertola et al., 2004; Clements et al., 2010].
As described in more detail below, technology developed in recent years now enables us to introduce or correct TP63-AEC mutations in the mammalian genome. Thus, we can now generate genetically matched pairs of cells that differ only with respect to the TP63 gene (TP63-wt vs. TP63-AEC). Because gene expression variability as a result of differences in genetic background are effectively eliminated in these matched pairs, they are ideally suited to conduct transcriptome and signaling pathway analyses, crucial steps in elucidating the molecular pathology underlying AEC.
INDUCED PLURIPOTENT STEM CELLS (IPSC) AS TOOLS TO STUDY AEC
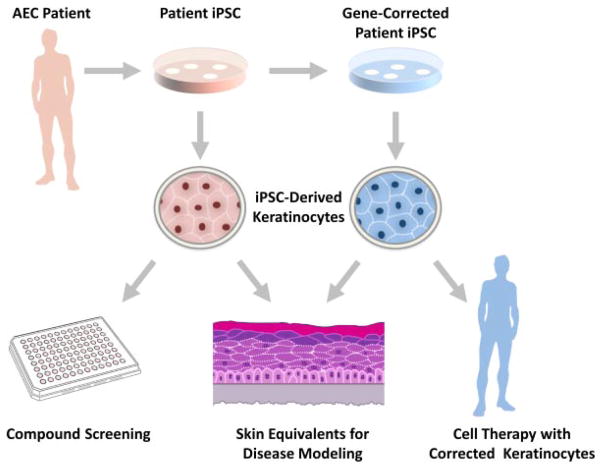
Patient-derived cells are not only essential for elucidating disease mechanisms; they are also invaluable tools for screening and testing drugs aimed at correcting cellular defects (Fig. 3). However, AEC research has been hampered by the limited availability of patient cells. First, epidermal keratinocytes, the cell type affected in AEC skin, have a limited life span in vitro. Second, obtaining multiple skin biopsies of pediatric patients is not a desirable approach to obtain cells for large scale experiments.
FIG. 3.
Disease models and new approaches to develop effective AEC therapies. Cells derived from patient skin biopsies are converted into iPSC. Using genome editing tools such as TALEN, it is possible to correct the TP63-AEC mutation, leading to the generation of conisogenic cell lines that differ only with respect to the status of the TP63 gene (TP63-AEC, TP63-wt). These cells can be differentiated into keratinocytes which will be useful tools to model the disease in 3D cultures, to test compounds for therapeutic effects on mutant keratinocytes, and to develop cell-based therapies (transplantation of gene-corrected keratinocytes onto the patient).
Fortunately, the recent development of induced pluripotent stem cell (iPSC) technology has provided an elegant approach to circumvent these difficulties [Schambach et al., 2010; Selvaraj et al., 2010; Yamanaka and Blau, 2010; Hockemeyer et al., 2011] (Fig. 3). iPSC have properties similar to embryonic stem cells. They can be propagated indefinitely, they can be genetically modified, and they can be induced to differentiate into keratinocytes [Bilousova et al., 2011; Itoh et al., 2011; Tolar et al., 2011]. Consequently, iPSC technology can provide us with the unlimited source of genetically defined material necessary for disease studies and the development for new cell-based therapies for AEC (see below).
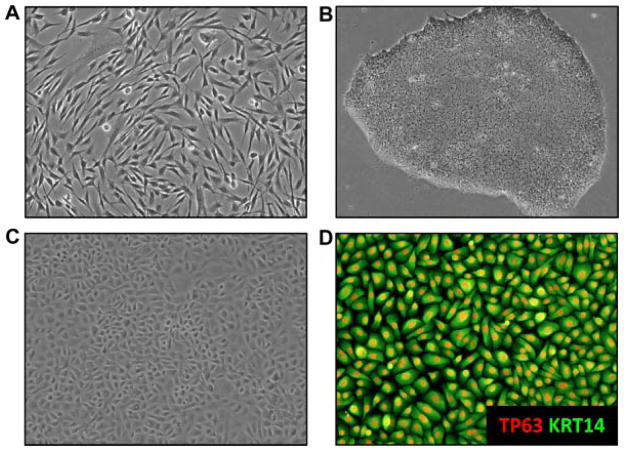
The first step in the generation of iPSC is the isolation of skin cells from AEC patients, either fibroblasts (the most common approach) or keratinocytes [Aasen et al., 2008; Polo et al., 2010; Tolar et al., 2011] (Fig. 5A). Amazingly, iPSC have also been generated using plucked human hair [Aasen and Izpisua Belmonte, 2010]. Primary patient cells are then transduced with genes encoding so-called reprogramming factors, which convert these cells into iPSC [Yamanaka, 2008]. A critical issue at this step is the choice of a vector system to introduce these factors. Retroviral and lentiviral vector systems have been widely used due to their efficiency in transducing cells. However, because these vectors integrate into the genome, they induce mutations, an undesired side effect. Other non DNA-integrating vector systems, such as plasmids, in vitro transcribed RNA, or proteins [Qin et al., 2009; Carey et al., 2009; Yakubov et al., 2010; Lai et al., 2011] have successfully been used, but are either very inefficient in generating iPSC or are technically difficult to use. New vector systems are thus needed, especially if iPSC are to be used for human therapy. For research purposes, the non-integrating but highly effective Sendai virus vectors that we routinely use to derive iPSC from AEC patient samples, appear to be ideal [Fusaki et al., 2009] (Fig. 5B).
FIG. 5.
Generation of iPSC-derived keratinocytes. A: Fibroblasts isolated from a skin biopsy of a pediatric AEC patient were transduced with reprogramming factors. B: Example of an iPSC colony derived from patient fibroblasts. Exposure of the iPSC to differentiation factors such as BMP4, retinoic acid, or ascorbic acid induces the development of ectodermal precursor cells. C: Further differentiation of these cells leads to the development of cells with a typical keratinocyte morphology. D: FACS sorting using antibodies against keratinocyte markers such as α6 integrin and β4 integrin leads to the isolation of a pure population of cells homogeneously expressing the keratinocyte markers TP63 (red) and keratin 14 (KRT14; green).
Editing the iPSC Genome
As outlined above, correcting TP63-AEC mutations in iPSC will provide us with the opportunity to generate matched pairs of iPSC-derived keratinocytes that differ only in their TP63 status. Recent advances in mammalian genome editing make it possible to accomplish this gene correction in an efficient way. Central to these approaches are sequence-specific DNA nucleases that target the TP63 gene locus. Both zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) have been shown to be effective in genetically manipulating the iPSC genome [Zou et al., 2009; Hargus et al., 2010; Hockemeyer et al., 2011; Miller et al., 2011].
The TALEN shown in Figure 4 were designed to target the TP63 gene close to a cluster of TP63-AEC mutations that occur in our cohort of AEC patients. Two “half” (enzymatically inactive) TALEN are designed which bind adjacent DNA segments. Because correct binding of two TALEN components is required to generate an active nuclease, the specificity of gene targeting using this system is very high. Upon correct dimerization, the active nuclease generates a double strand (ds) DNA break, which facilitates homologous recombination between the TP63 locus and gene targeting vectors (also referred to as exchange matrices), which are introduced into the cells together with the TALEN (Fig. 4). A gene targeting vector carrying wild type TP63 sequences can thus be used to correct TP63-AEC mutations. Because TP63 mutations in AEC cluster in the same region of the gene, a single pair of TALEN can be used to repair several different TP63-AEC mutations. This represents a tremendous advantage of this technology over other methods, and is important for the development of effective cell therapies to treat skin erosions in AEC patients (see below).
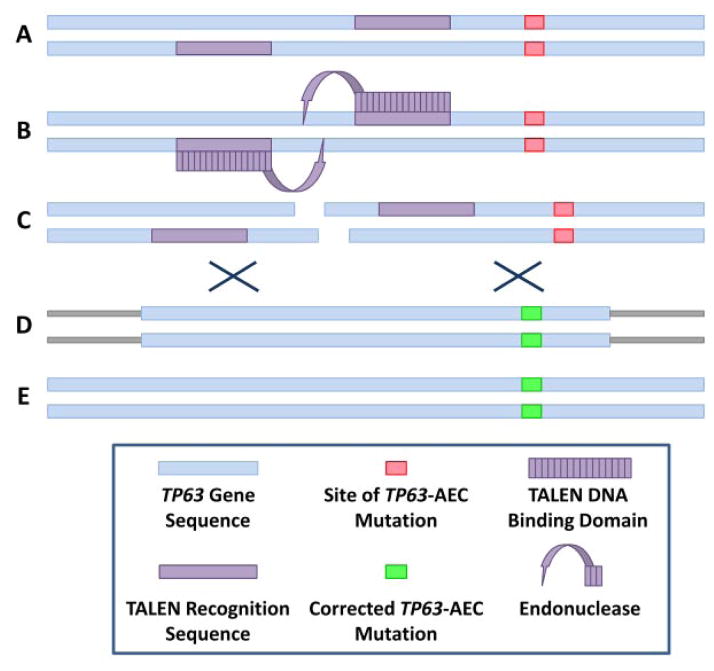
FIG. 4.
TALEN as tools to correct TP63-AEC mutations. A: Schematic representation of the TP63-AEC allele carrying a mutation (pink box). B: Two half TALEN bind to target sequences in the gene (purple boxes). Binding of these proteins leads to dimerization and activation of a DNA endonuclease activity. C: Double-stranded break in the TP63 DNA sequence. D: Homologous recombination of the TP63-AEC locus with a targeting vector carrying TP63 wild type sequences (green box). E: TP63 locus after correction of the AEC mutation.
BUILDING PATIENT-DERIVED SKIN IN VITRO
Human iPSC can be induced to differentiate into the ectodermal lineage. The required cell culture protocols were originally developed for embryonic stem cells [Aberdam et al., 2008; Metallo et al., 2008] and later adapted for iPSC [Bilousova et al., 2011; Itoh et al., 2011; Tolar et al., 2011]. Essentially, iPSC are exposed to combinations of factors such as retinoic acid, BMP4, or ascorbic acid. As a result, the cells begin to express epithelial markers such as keratins 8 and 18 (KRT8, KRT18) before synthesizing TP63, KRT14, and other keratinocyte markers. After 30 days of in vitro differentiation, a pure population of iPSC-derived keratinocytes can then be isolated via fluorescence activated cell sorting (FACS; Fig. 5C,D). Keratinocyte surface markers such as α6 integrin and β4 integrin are useful markers for FACS isolation of keratinocytes.
Besides conventional 2D cell culture (culture of keratinocytes on collagen IV-coated plastic dishes), different methods to generate in vitro skin equivalents can be applied to such FACS-sorted keratinocytes. For example, these cells can form an epidermis-like structure on artificial extracellular matrix (ECM) substitutes (Fig. 6A). Alternatively, fibroblasts embedded into a collagen matrix can be used as a substrate for these cells to form epidermis-like 3D cultures [Simpson et al., 2010] (Fig. 6B). The ultimate approach to generate skin equivalents is a xeno-transplant onto immunodeficient mice, for example, by grafting a mixture of fibroblasts and keratinocytes onto mice using a silicone grafting chamber [Lichti et al., 2008]. The latter approach connects the newly formed tissue to the blood circulation of a mammal, a step closer to mimicking true skin biology.
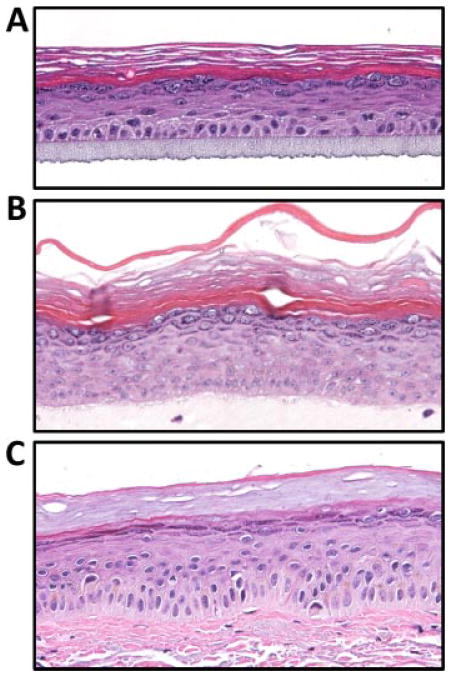
FIG. 6.
Examples of in vitro culture systems used to generate AEC models. Monolayer cultures of keratinocytes are transferred to an extracellular matrix allowing for the development of a 3D epidermal structure. A: Three dimensional culture generated on an artificial extracellular matrix. B: Three dimensional culture generated on a collagen plug with embedded fibroblasts. C: Example of normal human skin. Note the histological similarity of in vitro generated skin equivalents and normal human skin.
Each of the systems described above has distinct advantages and disadvantages. Simple 2D cell cultures consist of a rather homogeneous population of keratinocytes under culture conditions that suppress differentiation. These cultures are considered a model for undifferentiated keratinocytes, such as those found in the basal layer of the epidermis. Since basal keratinocytes express high levels of TP63, such 2D cultures are well-suited for characterizing cell autonomous functions of TP63. Three dimensional culture systems (skin equivalents) are ideally suited to test the effects of TP63-AEC mutations on keratinocyte differentiation. Given that skin of AEC patients is characterized by severe epidermal differentiation defects, these in vitro skin equivalents are important tools to replicate and thus analyze the AEC disease phenotype in vitro. Lastly, the recent development of xeno-transplant technology that allows for the generation of hair follicles using iPSC-derived human keratinocytes and mouse dermal papilla cells [Veraitch et al., 2013] opens up the possibility to replicate the hair defects observed in AEC in an animal transplant model using human keratinocytes, thus providing us with new tools to understand the underlying pathology and to develop new treatment options.
INVESTIGATING THE DISEASE MECHANISM
A few genes affected in their expression by TP63-AEC mutations have been identified [Marinari et al., 2008; Koster et al., 2009; Moretti et al., 2010; Clements et al., 2012; Sen et al., 2012; Zarnegar et al., 2012]. However, we currently do not have a comprehensive understanding of the molecular and cellular pathways affected by TP63-AEC proteins. Identifying molecular mechanisms will not only advance our understanding of TP63-AEC function, it might also lead to the identification of potential therapeutic targets for the treatment of skin erosions in AEC patients. A first step towards this goal will be a comparative transcriptome analysis of cells expressing TP63-AEC and TP63-wt. Conisogenic cell systems (i.e., genetically matched pairs of cells that differ only in the TP63 gene), will be of particular importance since their use will minimize genetic background effects on gene expression. To this end, patient iPSC in which the TP63 mutation has been repaired (see above) and the original patient iPSC will be differentiated into keratinocytes. Besides transcriptome analysis and promoter binding studies, proteomic studies will be required to understand how TP63-AEC and TP63-wt proteins function in these cells. In addition, protein stability, the ratio of TP63-AEC and TP63-wt proteins, as well as the ability of TP63-AEC to contribute to multimeric protein complexes will be assessed. All of these data combined will yield a more complete picture of the disease process and will enable us to characterize the pathological mechanisms underlying AEC. Importantly, similar approaches can be used for modeling and characterizing of other TP63-related ED, such as EEC. Further, a comparative transcriptome analysis of different ED caused by TP63 mutations will significantly advance our understanding of TP63 function both in normal development and in disease.
OUTLOOK—STEM CELL BASED THERAPIES
There is currently no cure or effective treatment for skin erosions that occur in AEC patients. iPSC technology holds the promise to generate genetically corrected autologous keratinocytes with a low likelihood of rejection. However, further characterization of the immune responses to these transplanted keratinocytes should be a focus of future research [Pick et al., 2009; Apostolou and Hochedlinger, 2011; Araki et al., 2013]. Further, iPSC derived for therapy must be free of viral vectors, which means that more efficient and safe reprogramming methods still need to be developed. Another concern is the potential of accumulating mutations in iPSC, either because patient cells that already carry mutations are converted into iPSC or because prolonged cell culture has the potential to generate mutations [Lowry and Quan, 2010; Mayshar et al., 2010; Gore et al., 2011].
However, it is likely that these remaining technical challenges will be overcome. iPSC technology offers a unique opportunity to both elucidate the molecular and cellular mechanisms underlying AEC and to eventually develop novel methods to treat the skin manifestations of this disorder.
Acknowledgments
We would like to thank Dr. Mayumi Fujita, University of Colorado School of Medicine, for providing the histological specimen of normal human skin. We would also like to thank the University of Colorado School of Medicine Histology Core (www.medschool.ucdenver.edu/histology) and iPSC Core (www.medschool.ucdenver.edu/iPS) for technical support in the generation of cells and tissues used in this study. We would like to thank the National Foundation for Ectodermal Dysplasias (NFED) for providing patient photographs for this article. Research reported in this publication was supported by a grant from the NFED and by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), part of the National Institutes of Health, under Award Numbers R00 AR054696 (M.I.K.), R01 AR061506 (M.I.K.), RO1 AR053892 (P.J.K.), and P30 AR057212.
Abbreviations
- ED
ectodermal dysplasia
- AEC
ankyloblepharon-ectodermal defects-cleft lip/palate syndrome
- TP63-AEC
TP63 containing a mutation identified in AEC patients
- iPSC
induced pluripotent stem cell
- TALEN
transcription activator-like effector nucleases
- FACS
fluorescence activated cell sorting
Footnotes
Conflict of interest: none.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Aasen T, Izpisua Belmonte JC. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc. 2010;5:371–382. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Aberdam E, Barak E, Rouleau M, de LS, Berrih-Aknin S, Suter DM, Krause KH, Amit M, Itskovitz-Eldor J, Aberdam D. A pure population of ectodermal cells derived from human embryonic stem cells. Stem Cells. 2008;26:440–444. doi: 10.1634/stemcells.2007-0588. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Stem cells: iPS cells under attack. Nature. 2011;474:165–166. doi: 10.1038/474165a. [DOI] [PubMed] [Google Scholar]
- Araki R, Uda M, Hoki Y, Sunayama M, Nakamura M, Ando S, Sugiura M, Ideno H, Shimada A, Nifuji A, Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Beaudry VG, Pathak N, Koster MI, Attardi LD. Differential PERP regulation by TP63 mutants provides insight into AEC pathogenesis. Am J Med Genet A. 2009;149A:1952–1957. doi: 10.1002/ajmg.a.32760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition of p63 by the E3 ligase ITCH: Effect of an ectodermal dysplasia mutant. Cell Cycle. 2010;9:3754–3763. [PubMed] [Google Scholar]
- Bertola DR, Kim CA, Albano LM, Scheffer H, Meijer R, van Bokhoven H. Molecular evidence that AEC syndrome and Rapp-Hodgkin syndrome are variable expression of a single genetic disorder. Clin Genet. 2004;66:79–80. doi: 10.1111/j.0009-9163.2004.00278.x. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Chen J, Roop DR. Differentiation of mouse induced pluripotent stem cells into a multipotent keratinocyte lineage. J Invest Dermatol. 2011;131:857–864. doi: 10.1038/jid.2010.364. [DOI] [PubMed] [Google Scholar]
- Bree AF. Clinical lessons learned from the International Research Symposium on Ankyloblepharon-Ectodermal Defects-Cleft Lip/Palate (AEC) syndrome. Am J Med Genet A. 2009;149A:1894–1899. doi: 10.1002/ajmg.a.32788. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Browne G, Cipollone R, Lena AM, Serra V, Zhou H, van Bokhoven H, Doetsch V, Merico D, Mantovani R, Terrinoni A, Knight RA, Candi E, Melino G. Differential altered stability and transcriptional activity of DeltaNp63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124:2200–2207. doi: 10.1242/jcs.079327. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Chung J, Grant RI, Kaplan DR, Irwin MS. Special AT-rich binding protein-2 (SATB2) differentially affects disease-causing p63 mutant proteins. J Biol Chem. 2011;286:40671–40680. doi: 10.1074/jbc.M111.271189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements SE, Techanukul T, Holden ST, Mellerio JE, Dorkins H, Escande F, McGrath JA. Rapp Hodgkin and Hay Wells ectodermal dysplasia syndromes represent a variable spectrum of the same genetic disorder. Br J Dermatol. 2010;163:624–629. doi: 10.1111/j.1365-2133.2010.09859.x. [DOI] [PubMed] [Google Scholar]
- Clements SE, Techanukul T, Lai-Cheong JE, Mee JB, South AP, Pourreyron C, Burrows NP, Mellerio JE, McGrath JA. Mutations in AEC syndrome skin reveal a role for p63 in basement membrane adhesion, skin barrier integrity and hair follicle biology. Br J Dermatol. 2012;167:134–144. doi: 10.1111/j.1365-2133.2012.10888.x. [DOI] [PubMed] [Google Scholar]
- Cole P, Hatef DA, Kaufman Y, Magruder A, Bree A, Friedman E, Sindwani R, Hollier LH. Facial clefting and oroauditory pathway manifestations in ankyloblepharon ectodermal defects-cleft lip/palate (AEC) syndrome. Am J Med Genet A. 2009;149A:1910–1915. doi: 10.1002/ajmg.a.32836. [DOI] [PubMed] [Google Scholar]
- Di Costanzo A, Festa L, Duverger O, Vivo M, Guerrini L, La MG, Morasso MI, Calabro V. Homeodomain protein Dlx3 induces phosphorylation-dependent p63 degradation. Cell Cycle. 2009;8:1185–1195. doi: 10.4161/cc.8.8.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianzani I, Garelli E, Gustavsson P, Carando A, Gustafsson B, Dahl N, Anneren G. Rapp-Hodgkin and AEC syndromes due to a new frameshift mutation in the TP63 gene. J Med Genet. 2003;40:e133. doi: 10.1136/jmg.40.12.e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishop MK, Bree AF, Hicks MJ. Pathologic changes of skin and hair in ankyloblepharon-ectodermal defects-cleft lip/palate (AEC) syndrome. Am J Med Genet A. 2009;149A:1935–1941. doi: 10.1002/ajmg.a.32826. [DOI] [PubMed] [Google Scholar]
- Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, Dotsch V, Brunner HG, van Bokhoven H. Gain-of-function mutation in ADULT syndrome reveals the presence of a second trans-activation domain in p63. Hum Mol Genet. 2002;11:799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- Duverger O, Morasso MI. Epidermal patterning and induction of different hair types during mouse embryonic development. Birth Defects Res Part C. 2009;87:263–272. doi: 10.1002/bdrc.20158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Mollo MR, Thomason HA, Antonini D, Zhou H, Ambrosio R, De Rosa L, Salvatore D, Getsios S, van Bokhoven H, Dixon J, Missero C. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum Mol Genet. 2013;22:531–543. doi: 10.1093/hmg/dds464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, Zhou H, Ambrosio R, Rice DP, Acampora D, van Bokhoven H, Del Vecchio L, Koster MI, Tadini G, Spencer-Dene B, Dixon M, Dixon J, Missero C. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol Med. 2012;4:192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin B, Touitou E. Transdermal skin delivery: Predictions for humans from in vivo, ex vivo and animal models. Adv Drug Delivery Rev. 2007;59:1152–1161. doi: 10.1016/j.addr.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee JH, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LS, Zhang K. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargus G, Cooper O, Deleidi M, Levy A, Lee K, Marlow E, Yow A, Soldner F, Hockemeyer D, Hallett PJ, Osborn T, Jaenisch R, Isacson O. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc Natl Acad Sci U S A. 2010;107:15921–15926. doi: 10.1073/pnas.1010209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Metry DW, Barrish J, Levy M. Uncombable hair (cheveux incoiffables, pili trianguli et canaliculi) syndrome: Brief review and role of scanning electron microscopy in diagnosis. Ultrastruct Pathol. 2001;25:99–103. [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. AbnormalmorphogenesisbutintactIKKactivationinmice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. PNAS. 2011;108:8797–8802. doi: 10.1073/pnas.1100332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julapalli MR, Scher RK, Sybert VP, Siegfried EC, Bree AF. Dermatologic findings of ankyloblepharon-ectodermal defects-cleft lip/palate (AEC) syndrome. Am J Med Genet A. 2009;149A:1900–1906. doi: 10.1002/ajmg.a.32797. [DOI] [PubMed] [Google Scholar]
- Kantaputra PN, Hamada T, Kumchai T, McGrath JA. Heterozygous mutation in the SAM domain of p63 underlies Rapp-Hodgkin ectodermal dysplasia. J Dent Res. 2003;82:433–437. doi: 10.1177/154405910308200606. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Marinari B, Payne AS, Kantaputra PN, Costanzo A, Roop DR. DeltaNp63 knockdown mice: A mouse model for AEC syndrome. Am J Med Genet A. 2009;149A:1942–1947. doi: 10.1002/ajmg.a.32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. p63 and epithelial appendage development. Differentiation. 2004;72:364–370. doi: 10.1111/j.1432-0436.2004.07208002.x. [DOI] [PubMed] [Google Scholar]
- Koster MI. p63 in skin development and ectodermal dysplasias. J Invest Dermatol. 2010;130:2352–2358. doi: 10.1038/jid.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Ann Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Sorting Out the p63 signaling network. J Invest Dermatol. 2008;128:1617–1619. doi: 10.1038/jid.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MI, Wendy-Yeo WY, Ramasamy R, Nordin N, Rosli R, Veerakumarasivam A, Abdullah S. Advancements in reprogramming strategies for the generation of induced pluripotent stem cells. J Assist Reprod Genet. 2011;28:291–301. doi: 10.1007/s10815-011-9552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 2000;60:4016–4020. [PubMed] [Google Scholar]
- Lopardo T, Lo IN, Marinari B, Giustizieri ML, Cyr DG, Merlo G, Crosti F, Costanzo A, Guerrini L. Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS ONE. 2008;3:e2715. doi: 10.1371/journal.pone.0002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Quan WL. Roadblocks en route to the clinical application of induced pluripotent stem cells. J Cell Sci. 2010;123:643–651. doi: 10.1242/jcs.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinari B, Ballaro C, Koster MI, Giustizieri ML, Moretti F, Crosti F, Papoutsaki M, Karin M, Alema S, Chimenti S, Roop DR, Costanzo A. IKK[alpha] Is a p63 transcriptional target involved in the pathogenesis of ectodermal dysplasias. J Invest Dermatol. 2008;129:60–69. doi: 10.1038/jid.2008.202. [DOI] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Ji L, de Pablo JJ, Palecek SP. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 2008;26:372–380. doi: 10.1634/stemcells.2007-0501. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. Genetic basis of skin appendage development. Sem Cell Dev Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo IN, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, Ballaro C, Alema S, Chimenti S, Guerrini L, Costanzo A. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotech. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo M. Ectodermal dysplasias: An overview and update of clinical and molecular-functional mechanisms. Am J Med Genet A. 2009;149A:2003–2013. doi: 10.1002/ajmg.a.32804. [DOI] [PubMed] [Google Scholar]
- Qin J, Guo X, Cui GH, Zhou YC, Zhou DR, Tang AF, Yu ZD, Gui YT, Cai ZM. Cluster characterization of mouse embryonic stem cell-derived pluripotent embryoid bodies in four distinct developmental stages. Biologicals. 2009;37:235–244. doi: 10.1016/j.biologicals.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Rinne T, Brunner HG, van Bokhoven H. p63-associated disorders. Cell Cycle. 2007;6:262–268. doi: 10.4161/cc.6.3.3796. [DOI] [PubMed] [Google Scholar]
- Rinne T, Bolat E, Meijer R, Scheffer H, van Bokhoven H. Spectrum of p63 mutations in a selected patient cohort affected with ankyloblepharon-ectodermal defects-cleft lip/palate syndrome (AEC) Am J Med Genet A. 2009;149A:1948–1951. doi: 10.1002/ajmg.a.32793. [DOI] [PubMed] [Google Scholar]
- Rinne T, Clements SE, Lamme E, Duijf PHG, Bolat E, Meijer R, Scheffer H, Rosser E, Tan TY, McGrath JA, Schalkwijk J, Brunner HG, Zhou H, van Bokhoven H. A novel translation re-initiation mechanism for the p63 gene revealed by amino-terminal truncating mutations in Rapp-Hodgkin/Hay-Wells-like syndromes. Hum Mol Genet. 2008;17:1968–1977. doi: 10.1093/hmg/ddn094. [DOI] [PubMed] [Google Scholar]
- Salinas CF, Montes GM. Rapp-Hodgkin syndrome: Observations on ten cases and characteristic hair changes (pili canaliculi) Birth Defects Orig Artic Ser. 1988;24:149–168. [PubMed] [Google Scholar]
- Schambach A, Cantz T, Baum C, Cathomen T. Generation and genetic modification of induced pluripotent stem cells. Expert Opin Biol Ther. 2010;10:1089–1103. doi: 10.1517/14712598.2010.496775. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Paus R. Molecular principles of hair follicle induction and morphogenesis. Bioessays. 2005;27:247–261. doi: 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Plane JM, Williams AJ, Deng W. Switching cell fate: The remarkable rise of induced pluripotent stem cells and lineage reprogramming technologies. Trends Biotechnol. 2010;28:214–223. doi: 10.1016/j.tibtech.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried E, Bree A, Fete M, Sybert VP. Skin erosions and wound healing in ankyloblepharon-ectodermal defect-cleft lip and/or palate. Arch Dermatol. 2005;141:1591–1594. doi: 10.1001/archderm.141.12.1591. [DOI] [PubMed] [Google Scholar]
- Simpson C, Kojima Si, Getsios S. RNA Interference in Keratinocytes and an Organotypic Model of Human Epidermis. Methods Mol Biol. 2010;585:127–146. doi: 10.1007/978-1-60761-380-0_10. [DOI] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKKalpha. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- Testoni B, Mantovani R. Mechanisms of transcriptional repression of cell-cycle G2/M promoters by p63. Nucl Acids Res. 2006;34:928–938. doi: 10.1093/nar/gkj477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, Titeux M, Osborn MJ, Lund TC, Hovnanian A, Wagner JE, Blazar BR. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2011;131:848–856. doi: 10.1038/jid.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, Merkx GF, Tenconi R, Fryns JP, Verloes A, Newbury-Ecob RA, Raas-Rotschild A, Majewski F, Beemer FA, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates JR, Neri G, Brunner HG. p63 genemutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001;69:481–492. doi: 10.1086/323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhooft SL, Stephan MJ, Sybert VP. Severe skin erosions and scalp infections in AEC syndrome. Pediatr Dermatol. 1993;10:334–340. doi: 10.1111/j.1525-1470.1993.tb00394.x. [DOI] [PubMed] [Google Scholar]
- Veraitch O, Kobayashi T, Imaizumi Y, Akamatsu W, Sasaki T, Yamanaka S, Amagai M, Okano H, Ohyama M. Human induced pluripotent stem cell-derived ectodermal precursor cells contribute to hair follicle morphogenesis in vivo. J Invest Dermatol. 2013;133:1479–1488. doi: 10.1038/jid.2013.7. [DOI] [PubMed] [Google Scholar]
- Visinoni AF, Lisboa-Costa T, Pagnan NAB, Chautard-Freire-Maia EA. Ectodermal dysplasias: Clinical and molecular review. Am J Med Genet A. 2009;149A:1980–2002. doi: 10.1002/ajmg.a.32864. [DOI] [PubMed] [Google Scholar]
- Wong VW, Sorkin M, Glotzbach JP, Longaker MT, Gurtner GC. Surgical approaches to create murine models of human wound healing. J Biomed Biotechnol. 2011;2011:969618. doi: 10.1155/2011/969618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41:51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming toa pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Webster DE, Lopez-Pajares V, Vander Stoep HB, Qu K, Yan KJ, Berk DR, Sen GL, Khavari PA. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am J Hum Genet. 2012;91:435–443. doi: 10.1016/j.ajhg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]