Abstract
Chemokines are small, secreted proteins that have been shown to be important regulators of leukocyte trafficking and inflammation. All the known effects of chemokines are transduced by action at a family of G protein coupled receptors. Two of these receptors, CCR5 and CXCR4, are also known to be the major cellular receptors for HIV-1. Consideration of the evolution of the chemokine family has demonstrated that the chemokine Stromal cell Derived Factor-1 or SDF1 (CXCL12) and its receptor CXCR4 are the most ancient members of the family and existed in animals prior to the development of a sophisticated immune system. Thus, it appears that the original function of chemokine signaling was in the regulation of stem cell trafficking and development. CXCR4 signaling is important in the development of many tissues including the nervous system. Here we discuss the manner in which CXCR4 signaling can regulate the development of different structures in the central and peripheral nervous systems and the different strategies employed to achieve these effects.
Keywords: Chemokine, Chemokine receptor, Development, Nervous system
Introduction
During the development of the nervous system neural stem cells born in the environment of the ventricular neuroepithelium must migrate to their final destinations where they develop into specific subsets of neurons and glia. This complex trafficking process has been shown to be under the control of several important messenger molecules that promote or inhibit migration. Importantly, when the adult brain is damaged, attempts at brain repair appear to recapitulate many of the features of this developmental program. One signaling system that has been shown to be of great importance for the regulation of both embryological and adult neurogenesis is that directed by chemokines, and the chemokine SDF1/CXCL12 in particular. Indeed, SDF1 signaling through its receptor CXCR4 is important in the development of nearly every tissue. Because this is the case it is also now clear that the same strategies are used time and again and that similarities are observed in the way that CXCR4 signaling controls neural development and that of the hematopoietic system and other tissues. Here we discuss the role of chemokines in neural development and how CXCR4 signaling is employed in the nervous system as well as other tissues.
Chemokine families and subfamilies
Chemokines are a highly conserved class of secreted signaling molecules, initially discovered for their central role in leukocyte trafficking. However, it is now known that the actions of chemokines span a wide range of purposes including implantation of the embryo, cancer metastasis and the regulation of pain. The essential chemokine signaling complex includes a secreted chemokine protein that binds to a transmembrane G protein coupled receptor (GPCR). Both these peptide and receptor families exhibit a number of conserved structural features.
The term chemokine is derived from their initial discovery as chemotactic cytokines. Each molecule has a molecular mass between 8–10 kDa. A hallmark of this family of proteins is four conserved cysteine residues that form disulfide bonds. The first and third cysteine, as well as the second and fourth, form the disulfide bonds creating the basis of the molecule's tertiary structure (Clark-Lewis et al. 1995; Fernandez and Lolis 2002). Chemokines are subdivided into 4 subfamilies based on the amino acid sequence around the first two cysteine residues (Fernandez and Lolis 2002; Huising et al. 2003; Bonecchi et al. 2009). CXC chemokines or α-chemokines contain one amino acid between the first two cysteine residues. CC chemokines or β-chemokines have no intervening amino acid. Two other subfamilies are the CX3C or γ-chemokines that contains one family member and the C or δ-chemokine that only has 2 of the 4 conserved cysteine residues. Together, there are nearly 50 distinct chemokines (Fernandez and Lolis 2002; Huising et al. 2003). Each chemokine has a trivial name reflecting the circumstances of its original discovery as well as an official name reflecting its structure. For example the chemokine CXCL12 is also known as SDF1 (Stromal cell Derived Factor 1), CCL2 is also known as MCP1 (Monocyte Chemotactic Protein 1) and so on.
Chemokine receptors are members of the family of G protein coupled receptors (GPCRs). The receptors are also classified into four subfamilies, based on the subfamily of ligands within which they interact (Table 1). Thus, CXCR1-7, CCR1-11, CX3CR1 and CR1 represent analogous receptor subfamilies (Thelen 2001; Fernandez and Lolis 2002; Huising et al. 2003). Within each subfamily of chemokines and receptors, there is significant cross reactivity as each ligand may usually bind to a number of receptors. Chemokines rarely bind receptors outside of their analogous receptor subfamily (Mackay 2001; Thelen 2001; Fernandez and Lolis 2002).
Table 1.
Classification of chemokines and chemokine receptors
| Family | Receptors | Ligands |
|---|---|---|
| CXCR | CXCR1 | IL-8, GCP-2 |
| CXCR2 | IL-8, GCP-2, GRO α, β, γ, NAP-2, ENA78 | |
| CXCR3 | IP-10, MIG, I-TAC | |
| CXCR4 | SDF-1α, β, γ | |
| CXCR5 | BLC/BCA | |
| CXCR6 | CXCL16 | |
| CXCR7 | I-TAC, SDF-1 | |
| CCR | CCR1 | MIP-1α, δ, RANTES, MCP-2, MCP-3, MCP-4, HCC-1, MPIF-1 |
| CCR2 | MCP-1, MCP-2, MCP-3, MCP-4 | |
| CCR3 | Eotaxin, Eotaxin-2, Eotaxin-3, RANTES, MIP-3, MIP-1δ, MCP-2, MCP-3, MCP-4 | |
| CCR4 | RANTES, MDC, TARC | |
| CCR5 | RANTES, MCP-2, MIP-1α, β | |
| CCR6 | LARC | |
| CCR7 | SLC, ELC | |
| CCR8 | I-309, TARC, MIP-1β | |
| CCR9 | TECK | |
| CCR10 | ESkine | |
| CCR11 | MCP-2, MCP-4 | |
| CX3CR | CX3CR1 | Fractalkine |
| XCR | XCR1 | Lymphotactine α, β |
Chemokine signaling
Upon activation of a chemokine receptor, GTP replaces GDP in the α subunit of the G protein. The G protein heterotrimer then dissociates from the ligand/receptor complex and signals through a number of downstream pathways. Both α as well as β/γ subunit complexes influence downstream signals. Many signaling pathways can be activated, including MAPK, FAK and PKB through α, and PKC and PyK-2 through β/γ subunits. A combination of these diverse pathways mediates a range of chemokine dependent cellular functions.
Chemokine receptors are also thought to dimerize or to form other types of higher order arrays which may additionally regulate the binding selectivity of ligands and the signaling options resulting from receptor activation (Springael et al. 2005; Bonecchi et al. 2010; Kramp et al. 2011). Chemokine receptors have been observed to form homo- and heterodimers, as well as higher order oligomers (Lagane et al. 2008). The signaling mechanism of a specific receptor complex may be different depending on what kind of multimer it forms (Thelen 2001; Springael et al. 2006; Sohy et al. 2009). Finally, recent evidence shows that chemokine receptors may also interact and form complexes with other G protein receptors such as opioid receptors (Thelen 2001; Pello et al. 2008, Toth et al. 2003; Burbassi et al. 2010; Heinisch et al. 2011). However, although receptor multimers of this type have been observed in cell culture studies their significance for chemokine action in vivo is not clear.
As with most GPCRs, binding of a chemokine to its receptor results in desensitization and downregulation of receptor signaling. In many cases the ligand/receptor complex undergoes endocytosis (Mackay 2001; Thelen 2001; Neel et al. 2005; Borroni et al. 2010). In some cases recycling of the receptor to the plasma membrane has been reported. Endocytosis and recycling can therefore regulate the availability of chemokine receptors at the cell surface (Neel et al. 2005; Borroni et al. 2010). The endocytosis mechanism can also result in degradation of the ligand. As a result, these mechanisms can also regulate the level of chemokines in the extracellular environment. Clearly, endocytosis and recycling are potentially important mechanisms for regulating chemokine signaling (Jones et al. 2006; Borroni et al. 2010). This possibility has recently become a subject of considerable interest owing to the discovery of the CXCR7 receptor, which is the only other receptor apart from CXCR4 that binds SDF1. CXCR7 is a member of a particular subgroup of chemokine “receptors”, which also include DARC, D6, and CCXCKR, whose properties are somewhat unusual for GPCRs because, even though they bind chemokines, they don't actually activate G proteins. An examination of their sequences reveals that these receptors don't contain the amino acid motif that has been typically associated with the activation of G proteins by chemokine receptors (Borroni et al. 2008). Hence, it has not been clear whether activation of these receptors can lead to cell signaling. However the repertoire of known signaling pathways associated with GPCRs is very large and so non-G protein-related functions such as the activation of β-arrestin mediated signaling can certainly be envisaged for these receptors (Rajagopal et al. 2010a, b). Another idea is that these molecules primarily function as “decoy” receptors. That is to say they can bind chemokines and remove them from the external environment through receptor-mediated endocytosis. Once internalized by a decoy receptor, a particular chemokine may be degraded or even perhaps rereleased intact from another part of the cell, a process known as transcytosis (Borroni et al. 2008). Previously, receptors like DARC and D6 have been shown to bind and internalize numerous chemokines-but not SDF1. SDF1 has been shown to be internalized via CXCR7 within the adult CNS endothelium (Cruz-Orengo et al. 2011).
In addition to endogenous chemokine signals, both viral and bacterial proteins have been shown to signal through chemokine receptors. For example, Cytomegalovirus (CMV) contains a number of genes that are homologous to mammalian chemokines and chemokine receptors (Penfold et al. 1999; Saederup et al. 1999). It is generally thought that infected cells may express these proteins as a mechanism for CMV to evade detection by the immune system. This is thought to play a role in the establishment of long-term persistence of CMV in mammalian hosts (Penfold et al. 1999; Saederup et al. 1999; Vink et al. 2001; Saederup and Mocarski 2002; Alcendor et al. 2009). In addition to CMV, the Epstein-Barr virus (EBV) contains genes that encode chemokine receptor like proteins (Nijmeijer et al. 2010). These EBV proteins can actually heterodimerize with native chemokine receptors to impair their function (Vischer et al. 2008; Nijmeijer et al. 2010).
The most notable and widely examined example of human pathology involves the Human Immunodeficiency Virus 1 (HIV1). HIV1 viral particles utilize chemokine receptors to enter a target cell (Doranz et al. 1996; Wells et al. 1996). Viral entry is an essential step in HIV1 infection. The HIV1 coat protein gp120 binds CD4 in combination with either CCR5 or CXCR4, stimulating endocytosis of the complex and subsequently HIV1 is able to infect the cell (Lapham et al. 1996; Dittmar et al. 1997; Simmons et al. 1998). Blockade of CXCR4 or CCR5 has proven to be an efficacious method of preventing HIV1 entry, and thus prevents infection of target cells (Cocchi et al. 1996; Clapham et al. 1999). Hence chemokine receptor signaling appears to have widespread functions in normal physiology and development as well as mediating the effects of numerous pathogens and the immune response to these organisms.
Rethinking the purpose of chemokines and their receptors: SDF1 and CXCR4
The numerous possible chemokine receptor interactions and downstream signal-transduction cascades, underscore the potential variety of functions for this signaling axis. As further evidence has emerged on the diversity of chemokine signaling, the essential nature of chemokine function has been rethought. In fact, the notion that chemokines are primarily chemotactic cytokines involved in inflammation is clearly not entirely correct. Certainly the diversity of chemokine signals evolved as the mammalian immune system expanded in complexity. The evolution of microbial genes also suggests that chemokine genes have coevolved through host-pathogen interactions. Yet, the origin of chemokine signaling is likely to have roots outside the immune system (Huising et al. 2003).
The most ancient of the α-chemokines is SDF1, which likely represents the original chemokine. Of all the α-chemokines, it shares the closest homology to β-chemokines. The receptor for SDF1 is CXCR4, the HIV1 co-receptor, which also appears to be the original chemokine receptor (Huising et al. 2003). Both SDF1 and CXCR4 are very widely expressed in the developing embryo (Fig. 1). Initial examination of mice null for either SDF1 or CXCR4 revealed profound developmental impairments in a number of organ systems including the gastro-intestinal vasculature and bone marrow (Nagasawa et al. 1996; Tachibana et al. 1998; Zou et al. 1998). In the past decade, CXCR4 signaling has been demonstrated to play a role in implantation of the embryo (Dominguez et al. 2003; Valles and Domínguez 2006), germ cell migration (Ara et al. 2003; Knaut et al. 2003), regulating hematopoietic homeostasis (Suratt et al. 2004; Lapidot et al. 2005; Dar et al. 2006; Sugiyama et al. 2006), regulating the development of neuropathic pain syndromes (Oh et al. 2001; Bhangoo et al. 2007a, b; Bhangoo et al. 2009; Wilson et al. 2011) and the development of many cancers including gliobastoma (Sehgal et al. 1998a, b), breast (Müller et al. 2001; Sloan and Anderson 2002), medulloblastoma (Rubin et al. 2003; Yang et al. 2007) and colon cancers (Mitra et al. 1999; Zeelenberg et al. 2003). These findings, especially in light of their high level of conservation, suggest that SDF1/CXCR4 signaling actually predated the evolution of the immune system (Huising et al. 2003).
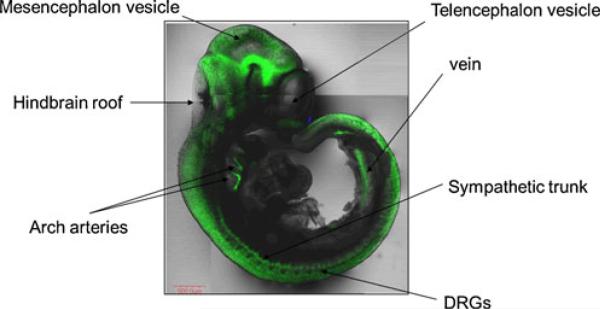
Fig. 1.
The figure illustrates the distribution of EGFP expression in an E 11 embryo from a CXCR4-EGFP BAC transgenic reporter mouse, showing the wide distribution of CXCR4 expression in the developing nervous system and in other structures (unpublished observations)
Discovery of chemokines in the nervous system
Over the past decade, chemokines have been demonstrated to mediate a number of functions in the nervous system from the regulation of development and neural activity to neuropathology. These findings underscore a fundamental aspect of the chemokine biology, namely that chemokines perform functions well beyond the limits of the immune system. Chemokines were not originally considered to have functions in the brain, although these were initially revealed through studies on the molecular mechanism of HIV1 dementia (Miller and Meucci 1999; Miller 2008).
As mentioned above, the viral coat protein gp120 mediates viral entry into immune cells. In vitro studies of the effect of HIV1 isolates on cultured neurons indicated that even in the absence of systemic infection, HIV1 could induce apoptosis of neurons (Brenneman et al. 1988), implying that neurons must express the HIV1 receptor. The viral coat protein gp120 from two separate HIV1 isolates was shown to exert its toxic effect on neurons by binding CD4, in conjunction with other factors (Brenneman et al. 1988). Moreover, subsequent investigations have also suggested that some versions of gp120 can produce neurotoxic effects in the absence of CD4 binding. These findings implicated HIV1 coat proteins (Hill et al. 1993), particularly gp120 (Toggas et al. 1994), as neurotoxic components in the genesis of AIDS dementia.
Initial work concerning the possible neuropathogenic effects of gp120 indicated the involvement of intracellular calcium (Dreyer et al. 1990; Kaiser et al. 1990). Subsequently NMDA receptors (Lipton et al. 1991; Savio and Levi 1993), and inflammatory cytokines (Yeung et al. 1995; Westmoreland et al. 1996), were shown to regulate gp120-induced neurotoxicity. In 1996, it was demonstrated that TGF-β, a protective cytokine, could decrease the toxic effect of gp120 (Meucci and Miller 1996). This effect was shown to be mediated by regulating intracellular Ca signals (Meucci and Miller 1996).
Independently of work on HIV1 neuropathogenesis, a putative chemokine receptor was cloned from the Locus Coeruleus. This group published evidence that the receptor was expressed in both neurons and glia of the adult rat brain (Wong et al. 1996). The receptor was eventually shown to be identical to CXCR4. Further work with fetal human tissue demonstrated the presence of several chemokine receptors expressed by neurons (Halks-Miller et al. 1997; Hesselgesser et al. 1997; Horuk et al. 1997; Hesselgesser et al. 1998).
In keeping with these observations, it was demonstrated that a range of chemokines could induce calcium influx in cultured hippocampal neurons (Meucci et al. 1998). This work also showed that chemokines were able to modify the toxic effects of gp120. This was the first unambiguous demonstration that chemokines could signal in neurons (Meucci et al. 1998). Rapidly, data emerged that indicated a role for chemokines in the nervous system in the case of HIV1 infection (Lavi et al. 1998).
Role of chemokines in neural development
Data from SDF1 and CXCR4 null mice reinforced the idea that chemokines and their receptors have important functions in the brain, and in brain development in particular. The first CNS defects were shown in the cerebellum of CXCR4 deficient mice (Zou et al. 1998). The same phenotype was observed for both SDF1 and CXCR4 null mice, suggesting that SDF1 acted through CXCR4 to regulate cerebellar development (Ma et al. 1998). Further brain abnormalities were observed in the dentate gyrus (Lu et al. 2002), cerebral cortex (Stumm et al. 2003) and dorsal root ganglia (DRG) (Belmadani et al. 2005) all of which were shown to require CXCR4 expression for proper development. In some areas of the brain, such as the cortex, multiple cellular subtypes utilize SDF1/CXCR4 signaling for proper migration (Stumm et al. 2003; Borrell and Marín 2006; Paredes et al. 2006; Tiveron et al. 2006). Moreover, SDF1/CXCR4 signals regulate other CNS developmental processes such as axon guidance (Chalasani et al. 2003; Kreibich et al. 2004; Lieberam et al. 2005; Chalasani et al. 2007). The idea that SDF1/CXCR4 signals subserve essential developmental functions in the CNS supports the idea that this signal originated to control the CNS, and not the immune system (Huising et al. 2003).
SDF1/CXCR4 signaling in the development of the nervous system
It is now recognized that SDF1/CXCR4 signaling regulates the development of nervous tissue in a number of different ways (Lazarini et al. 2003; Tran and Miller 2003a, b), particularly due to its effects on cell migration and axon guidance.
As noted above, mice null for either SDF1 or CXCR4 show pronounced defects in brain development. Some of the earliest data on specific roles for SDF1 and CXCR4 showed a role for this signal in regulating cerebellar granule cell migration (Ma et al. 1998). Expression analysis showed that SDF1 is present in the meninges of the outer layer of the cerebellum perinatally (Ma et al. 1998; Klein et al. 2001). During the first weeks of postnatal life, CXCR4 is expressed by cells immediately below the meninges in the external granular layer (EGL), which is a germinal zone (Reiss et al. 2002). SDF1/CXCR4 signaling anchors these cells, the progenitors for cerebellar granule neurons, in the EGL where they are exposed to critical mitogens such as sonic hedgehog (Klein et al. 2001; Reiss et al. 2002). Indeed, SDF1 itself may be a mitogen (Bajetto et al. 2001a, b; Klein et al. 2001; Barbero et al. 2002; Khan et al. 2003).
Eventually, the maturing granule neurons migrate radially, through the pyramidal cell layer, to populate the inner granule cell layer (IGL). In mice null for either SDF1 or CXCR4, the radial migration occurs prematurely, preventing these cells from receiving the proper developmental cues in the EGL (Klein et al. 2001). This mechanism demonstrates how SDF1 can time the migration of newly born neurons, keeping them in a germinal zone until they receive appropriate cues from their final destination.
Meningeal SDF1 also regulates the migration of Cajal-Retzius (CR) cells in the cortex (Borrell and Marín 2006; Paredes et al. 2006). CRs comprise a population of transient neurons that regulate CNS development. SDF1/CXCR4 signaling retains CRs near the meninges as they migrate across the surface of the cortex (Borrell and Marín 2006). Eventually these cells populate the superficial cortical layers just below the pia mater, and produce the molecule reelin (Borrell and Marín 2006; Paredes et al. 2006). Reelin gradients are fundamental to the laminar structure of the cortex (Caviness and Sidman 1973; Caviness and Rakic 1978; Ogawa et al. 1995; Rakic and Caviness 1995). Without a proper reelin gradient, the 6 layers of the cortex become disorganized, and proper cortical circuitry is not established (Ogawa et al. 1995). Thus, the role of SDF1 in regulating CR cell migration is essential for proper cortical development (Borrell and Marín 2006). Importantly, CRs utilize meningeal SDF1 in a similar manner to the cerebellar granule cells. Migration of the CR does not require SDF1, but retention near the meninges is SDF1-dependent (Borrell and Marín 2006). Disruption of SDF1/CXCR4 signaling leads to aberrant positioning in deeper layers of cortex (Borrell and Marín 2006; Paredes et al. 2006). This results in disorganized layers of cortex, and CNS circuitry is fundamentally corrupted. Thus, the migration of CRs away from the meninges is regulated by an SDF1-timing mechanism similar to that of cerebellar granule neurons.
Even though chemokines are known to act as chemoattractants in the immune system, one essential function of SDF1/CXCR4 signaling is in bone marrow homeostasis (Sugiyama et al. 2006). Like the cerebellar meninges, stromal cells in the bone marrow use SDF1/CXCR4 signals to retain hematopoietic progenitors within the bone marrow in close proximity to the hematopoietic germinal matrix (Sugiyama et al. 2006). As these cells mature, they are released into circulation. Pharmacologic blockade of CXCR4 results in emigration of many bone marrow cells into circulation and progenitor cells preferentially emigrate at a much higher rate than normal (Broxmeyer et al. 2005). Thus, the concept of SDF1 as an “anchor” that regulates the timing of cells within a germinal zone is recapitulated in the immune system as well as in neural structures such as the cerebellum.
Moreover, the concept of meningeal-derived SDF1 in retaining CXCR4-expressing cells in the subpial region is recapitulated in multiple brain regions. Aside from the cerebellum and cortex, pontine pre-cerebellar neurons migrate using a subpial, meningeal-derived SDF1-guidance mechanism, and disruption of this signal results in ectopic formation of pontine nuclei, deep within the brainstem (Vilz et al. 2005; Zhu et al. 2009).
SDF1/CXCR4 signals have quite a different mode of action in the developing hippocampal dentate gyrus (DG) where SDF1 acts primarily as a chemoattractant rather than an anchor. Granule cell precursors migrate from the wall of the lateral ventricle in response to meningeal-derived SDF1 signals (Bagri et al. 2002; Lu et al. 2002; Li et al. 2009) that is from their germinal zone to the developing DG. In the DG, they form a temporary subpial zone (SPZ) where further development occurs (Li et al. 2009). The transient SPZ eventually gives rise to the sub-granular zone (SGZ), through a reelin-dependent process. In this case, an SDF1 gradient secreted by the meninges allows the formation of a temporary germinal zone, which eventually gives rise to a permanent DG structure in the hippocampus (Bagri et al. 2002; Li and Pleasure 2005; Li et al. 2009). Thus, the precise temporal sequence of SDF1/CXCR4 expression is important for the final formation of the SGZ and interruption of CXCR4 signaling results in lack of formation of the normal DG (Fig. 2).
Fig. 2.
CXCR4 signaling in the development of the mouse dentate gyrus (DG). Left hand panel: Expression of SDF1 and CXCR4 in developing neocortex and hippocampus. A–G: Coronal sections through mouse forebrains on E15.5, E17.5, and the day of birth (P0) processed for in situ hybridization. Panel G shows a higher magnification of the DG (dg) region. SDF1 is expressed in the intermediate zone (iz) of the neocortex (ncx) (A), in the meninges (mng) overlying the hippocampus (hp) (A, C, E, and G), and in a zone close to the hippocampal fissure also occupied by Cajal–Retzius cells (A, C, E, and G; see text). White arrowhead in G points to the opening of the hippocampal fissure. White asterisk in G marks some of the SDF1-expressing cells lining the hippocampal fissure. CXCR4 also is expressed in the iz at E15.5 (B). At all ages, strongest CXCR4 expression appears in the developing DG and in a migratory stream (ms) of cells moving toward the DG (D and F). Right hand panel: Defects in the secondary proliferative cell population that forms the DG. A-D: Coronal sections through the E18.5 hippocampus of wild-type (A and C) or CXCR4 mutant mice (B and D), processed to show expression of Prox1 (blue) or Prox1 together with BrdUrd-labeled dividing cells (orange). In the wild-type mouse, Prox1 is expressed in the forming DG (A and C). By contrast, in the mutant, Prox1 is expressed in the vestigial DG but also along the migratory stream (ms) (arrows in D) of dividing cells running along the ventral surface of the hippocampus into the DG. BrdUrd-labeled cells of the ms can be seen between the two arrows in C. (E) A higher magnification of the migrating stream of cells shown in D. Numerous blue, Prox1-expressing cells appear among the brown, BrdUrd-labeled cells, but the populations appear largely distinct. Arrows in E point to two single-labeled cells. (F and G) High magnification views of BrdUrd-labeled dividing cells (dark blue) coursing through the ms in a wild-type (F) and a CXCR4 mutant mouse (G). About 30% fewer BrdUrd-labeled cells appear in the ms of the mutant (G) than in the wild type (Lu et al. 2002)
Further underscoring the important role for SDF1/ CXCR4 in hippocampal DG development is the fact that the DG is a site of active neurogenesis in the adult (Miller et al. 2008). SDF1/CXCR4 is active in at least two subsets of cells in the adult hippocampus (Bhattacharyya et al. 2008; Marchionni et al. 2010). CXCR4 receptors are expressed by DG neural progenitor cells and on immature DG granule cells both situated in the SGZ (Bhattacharyya et al. 2008; Kolodziej et al. 2008) (Fig. 3). In the adult, SDF1 is expressed by neurons in the DG (Tran et al. 2007). Recent work has shown that SDF1 exerts effects in the adult DG by modulating GABA-ergic inputs to CXCR4+ progenitor cells (Bhattacharyya et al. 2008). This line of evidence indicates a potential role for SDF1 and CXCR4 in adult neurogenesis. In the adult DG, it appears SDF1/CXCR4 has a separate regulatory role unrelated to migration. Thus, a gradient of SDF1 in the developing DG is supplanted by synaptic release of SDF1 in the adult DG, underscoring the temporal significance of each developmental cue.
Fig. 3.
Heterogeneity of EGFP-expressing cells in the dentate gyrus (DG) of CXCR4-EGFP BAC transgenic mice during postnatal development. EGFP is expressed in the DG as well as in a population of cells with the position and morphology of Cajal-Retzius cells. At 1 (A) and 2 (B) weeks, CXCR4 is expressed throughout the entire DG, more numerous cells being observed in the inner layer of the granular layer as well as in the SGZ. During development, the concentration of EGFP-expressing cells decreased in the outermost parts of the granule cell layer. At 3–5 weeks (D–I), CXCR4-EGFP-expressing cells are more localized to the SGZ and internal aspects of the granule cell layer. F: The expression pattern of CXCR4-EGFP cells in Cajal-Retzius cells. At 6 weeks (J–M), CXCR4 is expressed mainly in immature granule cells. In addition, it is also expressed in some neural progenitors (type 2 cells, insert in M) and radial astrocyte-like cells (type 1 cells) localized in the SGZ and extending long processes into the granular cell layer, as shown at higher magnification in M. At 3 months (N, O), the expression of CXCR4 is more restricted. Scale bars: 200 μm in A, B; 100 μm in C, D; 50 μm in E, F, G, H, I, J, K, L, N, O; 20 μm in M (Tran et al. 2007)
A different example of gradient formation in SDF1/CXCR4 signaling in neural development comes from the neocortex. Cortical GABA-ergic interneurons that migrate from the basal forebrain use the SDF1/CXCR4 signal to position themselves appropriately (Stumm et al. 2003; Liapi et al. 2008; Tanaka et al. 2010). In contrast to the cerebellum, cortical interneurons migrate along a gradient of increasing SDF1 towards their final position. Deletion of either SDF1 or CXCR4 prevents proper migration, in this case leading to inappropriate ultimate positioning of the interneurons. As a result, a range of different GABA-ergic interneurons remains in deeper cortical layers, failing to migrate to their proper location in the superficial layers (Stumm et al. 2003; Tanaka et al. 2010). This is very similar to how most chemokines work in the immune system, guiding cells to their final destination along a diffusible gradient.
The regulation of SDF1/CXCR4 in cortical interneuron migration is complicated by the presence of CXCR7 in the cortex during the same embryonic window as SDF1 and CXCR4 (Sánchez-Alcañiz et al. 2011; Wang et al. 2011). Recent work indicates that CXCR7 works to modulate the SDF1/CXCR4 signaling gradient in several ways. One report indicates that CXCR7 may act to modify the SDF1 gradient in the cortex (Wang et al. 2011). This is based on the finding that CXCR7 can act as a scavenger or “decoy” receptor, binding SDF1 in regions to which CXCR4 expressing cells do not normally migrate (Boldajipour et al. 2008; Wang et al. 2011). Another proposed mechanism for CXCR7 is to modulate CXCR4 responsiveness for the interneurons in which both are expressed (Sánchez-Alcañiz et al. 2011). CXCR7 binds and removes extracellular SDF1 by endocytosis and so precisely regulates the extracellular levels of chemokine available for activating CXCR4 (Sánchez-Alcañiz et al. 2011). Furthermore, as CXCR7 is widely expressed in the developing embryo it is likely that it may modify the action of SDF1 in other cases as well (Schönemeier et al. 2008; Zhao et al. 2008; Olesnicky Killian et al. 2009; Göttle et al. 2010).
As mentioned above, the cortical meninges also regulate the development of cortical CR cells by anchoring them in the subpial zone (Borrell and Marín 2006; Paredes et al. 2006). Thus, SDF1 expression from a single source regulates CXCR4 expressing cells in two different ways (Liapi et al. 2008). While cortical interneurons utilize the gradient formed by meningeal SDF1, CR cells only require SDF1 as an anchor to remain in the subpial zone. This divergent function of a single signal illustrates the importance of multiple SDF1/CXCR4 signaling mechanisms even within the same brain sub-region.
Similar to the cortex, the brainstem also utilizes SDF1 in multiple ways but for the same cellular population. Precerebellar pontine nuclei are formed after proper migration of neurons along a subpial route, which is dependent on SDF1 (Vilz et al. 2005; Zhu et al. 2009). In CXCR4 null mice, many migrating cells moved deeply into the hind-brain, away from the meninges. A second migratory defect, however, was present as some cells failed to migrate anteriorly, from the dorsal germinal region. One group has provided evidence for an SDF1 gradient that is responsible for this anterior migration (Zhu et al. 2009). Gradients of diffusible SDF1 are a powerful and conserved mechanism for guiding chemotaxis. SDF1 that is present in the extra-cellular matrix can influence any CXCR4 expressing cell, providing a broad application for this mechanism. In vitro data often indicate this as the primary role for SDF1 in chemotaxis. Gradients are limited, however, by the diffusible distance of SDF1 and the existence of other sources of SDF1, perturbing gradient formation. Thus, it is clear that gradients comprise one of multiple signaling processes. Moreover, spatiotemporal regulation of diffusible gradients is essential such that they do not interfere with other SDF1/CXCR4 signals.
Guideposts: lining the chosen path
Another important mechanism for SDF1/CXCR4 signaling involves guiding migration along a specific pathway. This can be accomplished through multiple mechanisms. One such example is migration of neural crest cells (NCC) to either the DRG (Belmadani et al. 2005) or the sympathetic ganglia (SG) (Kasemeier-Kulesa et al. 2010). NCC express CXCR4 receptors and deletion of CXCR4 results in ectopic positioning of DRG (Belmadani et al. 2005) (Figs. 4 and 5) and SG do not form properly (Kasemeier-Kulesa et al. 2010). The DRG and SG share an initial migratory path characterized by SDF1 expression. Upon reaching the DRG, certain NCC continue their migration to the SG, while others remain to populate the DRG (Kasemeier-Kulesa et al. 2010). Those that continue to the SG require SDF1/CXCR4 signaling, and migrate along another path lined by SDF1 expression. These results indicate that SDF1 is permissive for the SG progenitors that must continue past the DRG (Kasemeier-Kulesa et al. 2010). Proper utilization of SDF1/CXCR4 involves the formation of SDF1 expressing pathways deep into the periphery, well outside the CNS.
Fig. 4.
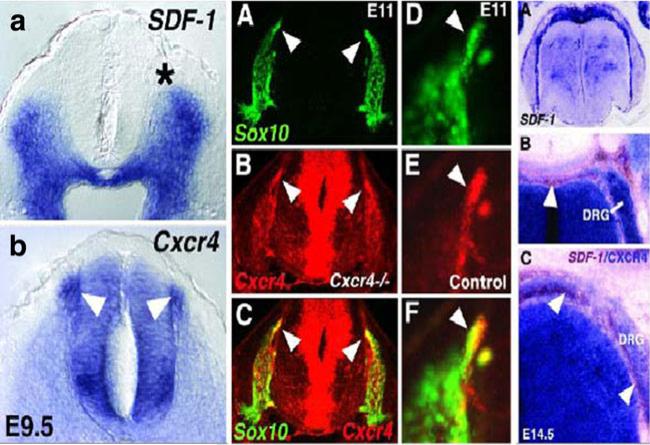
The expression patterns of SDF1, CXCR4, and Sox10 within the spinal cords of mouse embryos. P1, A, B, Coronal sections through the wild-type mouse mid-spinal cord at E9.5, processed with in situ hybridization to show mRNA expression of SDF1 and CXCR4. SDF1 expression extends close to the site at which neural crest cells emerge from the neural tube (A, asterisk). The latter cells are marked by prominent expression of CXCR4 (B, arrowheads). Marked expression also appears in a distinct region of the ventral spinal cord, which may approximately correspond to the position of developing motor neurons. P2, A–F, Migrating DRG neural crest cells are CXCR4 positive. Sections through the spinal cord of a CXCR4 mutant (A–C) and a littermate control (D–F) at E11. Sections were processed for double-fluorescence in situ hybridization (Sox10, green, encodes a transcription factor expressed in neural crest cells; CXCR4, red). A–C, In a CXCR4 homozygous mutant, single fluorescence for Sox10 marks neural crest cells in and migrating to the DRGs on either side of the spinal cord (A). Arrowheads point to a patch of migrating cells also labeled for CXCR4 fluorescence (B, C). C is an overlay of A and B. D–F, Higher magnification of a Sox10- (D, F; arrowhead) and CXCR4-(E, F) expressing cell group migrating toward the DRG (F is an overlay of D, E). P3, A–C, Coronal sections through wild-type E14.5 spinal cord processed with one- or two-color in situ hybridization for SDF1 (blue in A) or both SDF1 (brown in B, C) and CXCR4 (blue in B, C). SDF1 expression is strong over the dorsal neural tube and runs ventrally in two streams that appear to surround the DRGs (B, C; arrowheads). Note that at this later age, E14.5, neural crest migration to the DRGs is near completion (Belmadani et al. 2005)
Fig. 5.
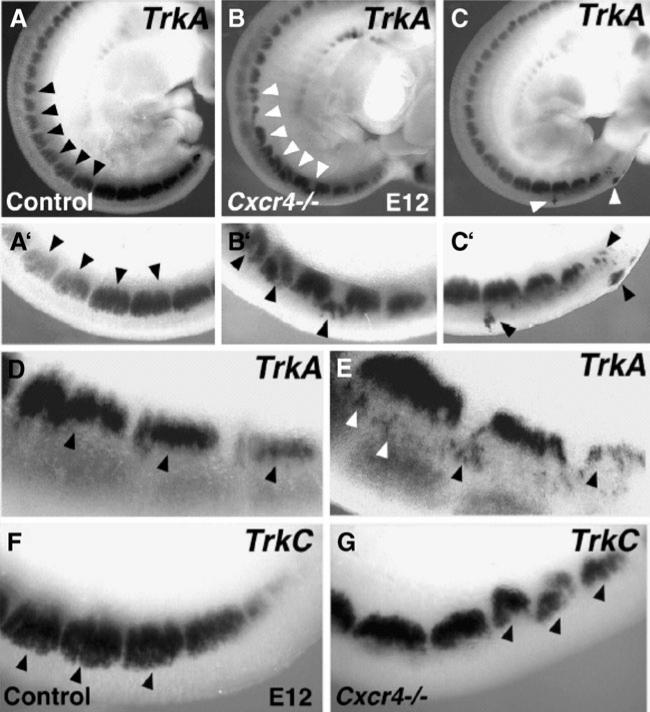
Abnormal DRG development in the absence of CXCR4 receptors. Whole-mount E12 embryos processed to show DRG expression of TrkA (A–C, D, E) and TrkC (F, G). A’–C’ are high-magnification views of the bottom third of A–C. In contrast with the regularly shaped DRGs forming in control embryos (A), DRGs in CXCR4 null mutants are misshapen, disorganized, or small (B, C). Arrowheads indicate normal DRGs (A), compared with split, misshapen DRGs and cell islands in B and C. Ectopic cells dorsal to the DRGs are indicated in E but are absent from the more regular DRGs in a control mouse (D). Similarly, DRGs marked by expression of TrkC are regularly shaped in a control mouse (F) but split or malformed in a CXCR4 null mouse (G) (Belmadani et al. 2005)
Another example using SDF1-lined pathways is the migration of interneurons in the Subventricular Zone/Intermediate Zone (SVZ/IZ) path. These interneurons migrate in response to expression of SDF1 by motor neuron progenitors of the cortex (Tiveron et al. 2006). This population of interneurons is distinct from other cortical interneurons, in that they migrate tangentially along a transient pattern of SDF1 expression that is specific to the SVZ/IZ. Without requiring a gradient formed by the meninges, the interneurons stay within the stereotyped SVZ/IZ to reach the motor neurons with which they make cortical motor circuits (Tiveron et al. 2006). This is a unique example of motor neurons guiding the migration of interneurons, which is notable because the interneurons stay in close proximity to their targets throughout their migration.
A slightly different method of creating a migratory path involves a non-diffusible gradient (Schwarting et al. 2007). Essentially, CXCR4 expressing cells migrate along a gradient that is formed not by diffusion, but by the actual level of SDF1 expression by target cells. An example of this is in the migration of Gonatropin-releasing Hormone (GnRH) neurons to the forebrain, a migration that is impaired in CXCR4 null mice (Schwarting et al. 2006). The GnRH cells migrate as differentiated neurons, from the vomeronasal organ (VNO) to the forebrain (FB). This migratory pathway contains mesenchyme that expresses SDF1. GnRH neurons transiently express CXCR4 as they migrate towards the FB, through the SDF1 expressing mesenchyme (Schwarting et al. 2007). Critically, the level of expression in mesenchymal cells increases until the border of the mesenchyme with the FB. Thus, CXCR4 expressing cells utilize a gradient of cellular expression to find their path to the FB (Schwarting et al. 2006, 2007). The mechanisms described here differ from the previous mechanisms in specific ways. This guide-post mechanism does not anchor CXCR4 expressing cells in a germinal or subpial zone. Moreover, the guidepost system does not rely on diffusible gradients of SDF1. Thus, this mechanism is a distinct mechanism that can work in parallel with the other mechanisms described above.
CXCR4 signaling and axonal guidance
In contrast to the regulation of progenitor cell migration, SDF1/CXCR4 can also act as a guidance cue. The following illustrations serve to underscore the importance of regulating the spatio-temporal distribution of SDF1 and CXCR4. Ectopic or dysregulated expression of either the ligand or the receptor can lead to developmental defects that include not only cellular migration, but axon guidance and circuit formation as well (Odemis et al. 2005; Toba et al. 2008; McIntyre et al. 2010).
Axons almost always involve long complicated projection paths. Failure of proper axon guidance may result in rendering a circuit useless. Moreover, axons relay signals along their length to their cell body, influencing neural activity at a distance. The classic example of SDF1 guiding axons is from the embryonic spinal cord, where growth cones of ventral motor neurons migrate towards a mesenchymal source of SDF1 (Lieberam et al. 2005). These motor neurons exit the spinal cord ventrally, and grow to their peripheral targets, usually muscles. The initial ventral trajectory is defined by SDF1, as absence of the signal causes the axons to grow dorsally and exit through a different portion of the spinal cord (Lieberam et al. 2005). Moreover, CXCR4 expressing motor neurons will evade sensory ganglia, while axons that lack CXCR4 expression will invade the DRG (Lieberam et al. 2005).
While motor neurons use SDF1/CXCR4 to determine their initial trajectory, other neurons utilize SDF1/CXCR4 as a modulatory signal for axon pathfinding. SDF1 is considered modulatory because in these cases it does not exert a guidance effect, either attractive or repulsive, in isolation. The effects of known guidance cues, in this case repellants such as Slit-2 or Semaphorins, are altered in the presence of SDF1/CXCR4 signals. The mechanism for this modulatory effect utilizes G protein mediated cyclic AMP elevation in the growth cone of target axons (Chalasani et al. 2003, 2007).
Another example of SDF1 guiding axon growth occurs after SDF1 guides cells to their final location. As mentioned above, both DRG and SG neurons require SDF1 to reach their positions within the ganglia. After reaching their position within the ganglia, neurons from both the Dorsal Root Ganglion and Sympathetic Ganglion grow axons into the spinal cord (Chalasani et al. 2003). In mice null for CXCR4, axons from the Dorsal Root and Sympathetic ganglia grow into the spinal cord aberrantly and are hyper-fasciculated (Chalasani et al. 2003). Thus, SDF1/CXCR4 is not required for every aspect of axon guidance for the DRG and SG, but does regulate some aspect of spinal entry.
The effect of SDF1/CXCR4 on guiding axons reinforces the concept that SDF1/CXCR4 signals regulate multiple aspects of the developing brain. Clearly, improper distribution of SDF1 can lead to aberrant patterns, not just for migrating cells, but for developing axons as well. Thus, while gradients and other expression patterns of SDF1 are critical for cellular migration, these signals can also influence axons that grow into the developing tissue. As such, the distribution of SDF1 and CXCR4 must be tightly regulated for proper brain patterning.
SDF1/CXCR4 in non-mammalian systems: insight for further work in mammals
The various examples of brain development described above serve to illustrate the importance of specificity in SDF1/CXCR4 expression. All of the examples describe components of mammalian vertebrate brain development. Both the sites of SDF1 release and the timing of CXCR4 expression result in tightly regulated expression patterns, which the complex mammalian brain requires. The basic patterning motifs of the mammalian brain, however, derive from simpler vertebrate systems. SDF1 regulation of axon guidance (Miyasaka et al. 2007), SDF1/CXCR4 cellular chemotaxis (Sapède et al. 2005) and interaction between CXCR4 and CXCR7 (Cubedo et al. 2009) have been reported in the nervous system of lower vertebrates. Thus, SDF1/CXCR4 activity in brain development predates mammals and is present in earlier vertebrates (Huising et al. 2003; Alonso et al. 2009). What can we learn from non-mammalian systems that have not already been examined in the mammalian brain? Many of the findings discussed above have analogous findings in non-mammalian systems. Entire nervous structures in the zebrafish, from GnRH neurons (Palevitch et al. 2010) to peripheral ganglia (Sapède et al. 2005; Dambly-Chaudiere et al. 2007), have analogous developmental requirements for SDF1/CXCR4 in mammalian vertebrates. While most of the examples given above have parallels in zebrafish, one instance remains unique to zebrafish, and may hold some insight for further mammalian studies on the roles of SDF1/CXCR4.
A recent study by Diotel et al. (2010) focuses on a population of cells that are present throughout the lifetime of the zebrafish, but are only present transiently in the developing mammalian brain. These cells, called radial glia, are shown to express CXCR4 throughout the adult zebrafish neuraxis (Diotel et al. 2010). Moreover, SDF1 expression is detected at low levels in some of the radial glia as well. This study demonstrates the existence of CXCR4 in the cells that are slow dividing but retain some capability for assisting in the regenerative capacity of the adult zebrafish brain (Diotel et al. 2010). The exact role of SDF1/CXCR4 for zebrafish radial glia, however, is unclear.
Importantly, two studies have published the existence of CXCR4 expression in radial glia, but neither has identified a function for SDF1/CXCR4 in this cellular population. Berger et al. (2007) demonstrate that the post-natal hippocampus has CXCR4 cells that label with glial fibrillary acidic protein (GFAP), a marker for both mature astrocytes and radial glia. Morphologically, their CXCR4-expressing GFAP + cells resemble radial glia (Berger et al. 2007). Thus, they hypothesize that SDF1 may play a role in the reorganization of the postnatal dentate gyrus by effects mediated on CXCR4-expressing radial glia. Stumm and Höllt (2007) demonstrate the presence of CXCR4 in telencephalic radial glia. Importantly they note that SDF1 is “not detectable” in the ganglionic eminences, and hypothesize that the meninges may be a source of SDF1 for radial glia (Stumm and Höllt 2007). As a central player in brain development, radial glia are essential for a number of highly coordinate processes in brain development, including the proliferation and migration of many cell types. Yet, as non-migratory, non-axonal cells, radial glia would represent fundamentally different aspect of chemokine activity. Given the myriad effects of SDF1/CXCR4 in the development of the mammalian brain, it is somewhat surprising that radial glia have not been examined in greater detail.
Conclusion
The SDF1/CXCR4 signal is active throughout development of the neuraxis. The diversity of this signaling modality is evident, and suggests it has been integrated as a fundamental aspect of neural development. Many aspects of this signaling modality indicate the potential of chemokines as regulators of neural development. The dogmatic notion that limits the role of chemokines to chemotaxis is no longer adequate. Further work with receptor dimerization, mechanisms of cellular activity, regulation under pathological conditions and neuro-immune interactions promise to unveil fundamental aspects of chemokine activity in brain development.
Contributor Information
Divakar S. Mithal, Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Ghazal Banisadr, Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Richard J. Miller, Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Chicago, IL, USA Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Medical School, 303 E Chicago Ave, Chicago, IL 60611, USA.
References
- Alcendor DJ, Zong J, Dolan A, Gatherer D, Davison AJ, Hayward GS. Patterns of divergence in the vCXCL and vGPCR gene clusters in primate cytomegalovirus genomes. Virology. 2009;395:21–32. doi: 10.1016/j.virol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso E, Gomez-Santos L, Madrid JF, Saez F. The expression of a novel cxcr4 gene in xenopus embryo. Histol Histopathol. 2009;24:1097–1103. doi: 10.14670/HH-24.1097. [DOI] [PubMed] [Google Scholar]
- Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, Matsui Y, Nagasawa T. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF1). Proc Natl Acad Sci U S A. 2003;100:5319–5323. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagri A, Gurney T, He X, Zou YR, Littman DR, Tessier-Lavigne M, Pleasure SJ. The chemokine SDF regulates migration of dentate granule cells. Development. 2002;129:4249–4260. doi: 10.1242/dev.129.18.4249. [DOI] [PubMed] [Google Scholar]
- Bajetto A, Barbero S, Bonavia R, Piccioli P, Pirani P, Florio T, Schettini G. Stromal cell-derived factor 1 alpha induces astrocyte proliferation through the activation of extracellular signal-regulated kinases 1/2 pathway. J Neurochem. 2001a;77:1226–1236. doi: 10.1046/j.1471-4159.2001.00350.x. [DOI] [PubMed] [Google Scholar]
- Bajetto A, Bonavia R, Barbero S, Florio T, Schettini G. Chemokines and their receptors in the central nervous system. Front Neuroendocrinol. 2001b;22:147–84. doi: 10.1006/frne.2001.0214. [DOI] [PubMed] [Google Scholar]
- Barbero S, Bajetto A, Bonavia R, Porcile C, Piccioli P, Pirani P, Ravetti JL, Zona G, Spaziante R, Florio T, Schettini G. Expression of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1 in human brain tumors and their involvement in glial proliferation in vitro. Ann N Y Acad Sci. 2002;973:60–69. doi: 10.1111/j.1749-6632.2002.tb04607.x. [DOI] [PubMed] [Google Scholar]
- Belmadani A, Tran PB, Ren D, Assimacopoulos S, Grove EA, Miller RJ. The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors. J Neurosci. 2005;25:3995–4003. doi: 10.1523/JNEUROSCI.4631-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger O, Li G, Han SM, Paredes M, Pleasure SJ. Expression of SDF1 and cxcr4 during reorganization of the postnatal dentate gyrus. Dev Neurosci. 2007;29:48–58. doi: 10.1159/000096210. [DOI] [PubMed] [Google Scholar]
- Bhangoo S, Ren D, Miller RJ, Henry KJ, Lineswala J, Hamdouchi C, Li B, Monahan PE, Chan DM, Ripsch MS, White FA. Delayed functional expression of neuronal chemokine receptors following focal nerve demyelination in the rat: a mechanism for the development of chronic sensitization of peripheral nociceptors. Mol Pain. 2007a;3:38. doi: 10.1186/1744-8069-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo SK, Ren D, Miller RJ, Chan DM, Ripsch MS, Weiss C, McGinnis C, White FA. Cxcr4 chemokine receptor signaling mediates pain hypersensitivity in association with antiretroviral toxic neuropathy. Brain Behav Immun. 2007b;21:581–591. doi: 10.1016/j.bbi.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo SK, Ripsch MS, Buchanan DJ, Miller RJ, White FA. Increased chemokine signaling in a model of HIV1-associated peripheral neuropathy. Mol Pain. 2009;5:48. doi: 10.1186/1744-8069-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya BJ, Banisadr G, Jung H, Ren D, Cronshaw DG, Zou Y, Miller RJ. The chemokine stromal cell-derived factor-1 regulates GABAergic inputs to neural progenitors in the postnatal dentate gyrus. J Neurosci. 2008;28:6720–6730. doi: 10.1523/JNEUROSCI.1677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Bonecchi R, Galliera E, Borroni EM, Corsi MM, Locati M, Mantovani A. Chemokines and chemokine receptors: an overview. Front Biosci. 2009;14:540–551. doi: 10.2741/3261. [DOI] [PubMed] [Google Scholar]
- Bonecchi R, Savino B, Borroni EM, Mantovani A, Locati M. Chemokine decoy receptors: structure-function and biological properties. Curr Top Microbiol Immunol. 2010;341:15–36. doi: 10.1007/82_2010_19. [DOI] [PubMed] [Google Scholar]
- Borrell V, Marín O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- Borroni EM, Bonecchi R, Buracchi C, Savino B, Mantovani A, Locati M. Chemokine decoy receptors: new players in reproductive immunology. Immunol Invest. 2008;37:483–497. doi: 10.1080/08820130802191318. [DOI] [PubMed] [Google Scholar]
- Borroni EM, Mantovani A, Locati M, Bonecchi R. Chemokine receptors intracellular trafficking. Pharmacol Ther. 2010;127:1–8. doi: 10.1016/j.pharmthera.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Brenneman DE, Westbrook GL, Fitzgerald SP, Ennist DL, Elkins KL, Ruff MR, Pert CB. Neuronal cell killing by the envelope protein of HIV and its prevention by vasoactive intestinal peptide. Nature. 1988;335:639–642. doi: 10.1038/335639a0. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, Liles WC, Li X, Graham-Evans B, Campbell TB, Calandra G, Bridger G, Dale DC, Srour EF. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbassi S, Sengupta R, Meucci O. Alterations of CXCR4 function in μ-opioid receptor-deficient glia. Eur J Neurosci. 2010;32:1278–1288. doi: 10.1111/j.1460-9568.2010.07402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness VS, Jr, Sidman RL. Time of origin or corresponding cell classes in the cerebral cortex of normal and reeler mutant mice: an autoradiographic analysis. J Comp Neurol. 1973;148:141–151. doi: 10.1002/cne.901480202. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Rakic P. Mechanisms of cortical development: a view from mutations in mice. Annu Rev Neurosci. 1978;1:297–326. doi: 10.1146/annurev.ne.01.030178.001501. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Sabelko KA, Sunshine MJ, Littman DR, Raper JA. A chemokine, SDF-1, reduces the effectiveness of multiple axonal repellents and is required for normal axon pathfinding. J Neurosci. 2003;23:1360–1371. doi: 10.1523/JNEUROSCI.23-04-01360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani SH, Sabol A, Xu H, Gyda MA, Rasband K, Granato M, Chien CB, Raper JA. Stromal cell-derived factor-1 antagonizes slit/robo signaling in vivo. J Neurosci. 2007;27:973–980. doi: 10.1523/JNEUROSCI.4132-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham PR, Reeves JD, Simmons G, Dejucq N, Hibbitts S, McKnight A. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol Membr Biol. 1999;16:49–55. doi: 10.1080/096876899294751. [DOI] [PubMed] [Google Scholar]
- Clark-Lewis I, Kim KS, Rajarathnam K, Gong JH, Dewald B, Moser B, Baggiolini M, Sykes BD. Structure-activity relationships of chemokines. J Leukoc Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The v3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- Cruz-Orengo L, Holman DW, Dorsey D, Zhou L, Zhang P, Wright M, McCandless EE, Patel JR, Luker GD, Littman DR, Russell JH, Klein RS. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J Exp Med. 2011;208:327–339. doi: 10.1084/jem.20102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubedo N, Cerdan E, Sapede D, Rossel M. CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons. Mol Cell Neurosci. 2009;40:474–484. doi: 10.1016/j.mcn.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;29:7–23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A, Kollet O, Lapidot T. Mutual, reciprocal SDF1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in nod/scid chimeric mice. Exp Hematol. 2006;34:967–975. doi: 10.1016/j.exphem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Diotel N, Vaillant C, Gueguen MM, Mironov S, Anglade I, Servili A, Pellegrini E, Kah O. CXCR4 and CXCL12 expression in radial glial cells of the brain of adult zebrafish. J Comp Neurol. 2010;518:4855–4876. doi: 10.1002/cne.22492. [DOI] [PubMed] [Google Scholar]
- Dittmar MT, McKnight A, Simmons G, Clapham PR, Weiss RA, Simmonds P. Hiv-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- Dominguez F, Galan A, Martin JJL, Remohi J, Pellicer A, Simón C. Hormonal and embryonic regulation of chemokine receptors cxcr1, cxcr4, ccr5 and ccr2b in the human endometrium and the human blastocyst. Mol Hum Reprod. 2003;9:189–198. doi: 10.1093/molehr/gag024. [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors ckr-5, ckr-3, and ckr-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Dreyer EB, Kaiser PK, Offermann JT, Lipton SA. HIV-1 coat protein neurotoxicity prevented by calcium channel antagonists. Science. 1990;248:364–367. doi: 10.1126/science.2326646. [DOI] [PubMed] [Google Scholar]
- Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- Göttle P, Kremer D, Jander S, Odemis V, Engele J, Hartung HP, Küry P. Activation of CXCR7 receptor promotes oligodendroglial cell maturation. Ann Neurol. 2010;68:915–924. doi: 10.1002/ana.22214. [DOI] [PubMed] [Google Scholar]
- Halks-Miller M, Hesselgesser J, Miko IJ, Horuk R. Chemokine receptors in developing human brain. Methods Enzymol. 1997;288:27–38. doi: 10.1016/s0076-6879(97)88005-6. [DOI] [PubMed] [Google Scholar]
- Heinisch S, Palma J, Kirby LG. Interactions between chemokine and mu-opioid receptors: anatomical findings and electrophysiological studies in the rat periaqueductal grey. Brain Behav Immun. 2011;25:360–372. doi: 10.1016/j.bbi.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper SC, Hoxie J, Kolson DL, Taub D, Horuk R. Cd4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- Hesselgesser J, Taub D, Baskar P, Greenberg M, Hoxie J, Kolson DL, Horuk R. Neuronal apoptosis induced by HIV-1 gp120 and the chemokine SDF1 alpha is mediated by the chemokine receptor CXCR4. Curr Biol. 1998;8:595–598. doi: 10.1016/s0960-9822(98)70230-1. [DOI] [PubMed] [Google Scholar]
- Hill JM, Mervis RF, Avidor R, Moody TW, Brenneman DE. HIV envelope protein-induced neuronal damage and retardation of behavioral development in rat neonates. Brain Res. 1993;603:222–233. doi: 10.1016/0006-8993(93)91241-j. [DOI] [PubMed] [Google Scholar]
- Horuk R, Martin AW, Wang Z, Schweitzer L, Gerassimides A, Guo H, Lu Z, Hesselgesser J, Perez HD, Kim J, Parker J, Hadley TJ, Peiper SC. Expression of chemokine receptors by subsets of neurons in the central nervous system. J Immunol. 1997;158:2882–2890. [PubMed] [Google Scholar]
- Huising MO, Stet RJM, Kruiswijk CP, Savelkoul HFJ, Lidy Verburg-van Kemenade BM. Molecular evolution of CXC chemokines: extant CXC chemokines originate from the CNS. Trends Immunol. 2003;24:307–313. doi: 10.1016/s1471-4906(03)00120-0. [DOI] [PubMed] [Google Scholar]
- Jones MC, Caswell PT, Norman JC. Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol. 2006;18:549–557. doi: 10.1016/j.ceb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Kaiser PK, Offermann JT, Lipton SA. Neuronal injury due to HIV-1 envelope protein is blocked by anti-gp120 antibodies but not by anti-cd4 antibodies. Neurology. 1990;40:1757–1761. doi: 10.1212/wnl.40.11.1757. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, McLennan R, Romine MH, Kulesa PM, Lefcort F. CXCR4 controls ventral migration of sympathetic precursor cells. J Neurosci. 2010;30:13078–13088. doi: 10.1523/JNEUROSCI.0892-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MZ, Brandimarti R, Musser BJ, Resue DM, Fatatis A, Meucci O. The chemokine receptor CXCR4 regulates cell-cycle proteins in neurons. J Neurovirol. 2003;9:300–314. doi: 10.1080/13550280390201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, Luster AD. SDF1 alpha induces chemo-taxis and enhances sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- Knaut H, Werz C, Geisler R, Tübingen 2000 Screen Consortium. Nüsslein-Volhard C. A zebrafish homologue of the chemokine receptor CXCR4 is a germ-cell guidance receptor. Nature. 2003;421:279–282. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- Kolodziej A, Schulz S, Guyon A, Wu DF, Pfeiffer M, Odemis V, Höllt V, Stumm R. Tonic activation of CXC chemokine receptor 4 in immature granule cells supports neurogenesis in the adult dentate gyrus. J Neurosci. 2008;28:4488–4500. doi: 10.1523/JNEUROSCI.4721-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramp BK, Sarabi A, Koenen RR, Weber C. Heterophilic chemokine receptor interactions in chemokine signaling and biology. Exp Cell Res. 2011;317:655–663. doi: 10.1016/j.yexcr.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Kreibich TA, Chalasani SH, Raper JA. The neurotransmitter glutamate reduces axonal responsiveness to multiple repellents through the activation of metabotropic glutamate receptor 1. J Neurosci. 2004;24:7085–7095. doi: 10.1523/JNEUROSCI.0349-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagane B, Chow KYC, Balabanian K, Levoye A, Harriague J, Planchenault T, Baleux F, Gunera-Saad N, Arenzana-Seisdedos F, Bachelerie F. CXCR4 dimerization and beta-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in whim syndrome. Blood. 2008;112:34–44. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]
- Lapham CK, Ouyang J, Chandrasekhar B, Nguyen NY, Dimitrov DS, Golding H. Evidence for cell-surface association between fusin and the cd4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- Lavi E, Kolson DL, Ulrich AM, Fu L, González-Scarano F. Chemokine receptors in the human brain and their relationship to HIV infection. J Neurovirol. 1998;4:301–311. doi: 10.3109/13550289809114531. [DOI] [PubMed] [Google Scholar]
- Lazarini F, Tham TN, Casanova P, Arenzana-Seisdedos F, Dubois-Dalcq M. Role of the alpha-chemokine stromal cell-derived factor (SDF1) in the developing and mature central nervous system. Glia. 2003;42:139–148. doi: 10.1002/glia.10139. [DOI] [PubMed] [Google Scholar]
- Li G, Pleasure SJ. Morphogenesis of the dentate gyrus: what we are learning from mouse mutants. Dev Neurosci. 2005;27:93–99. doi: 10.1159/000085980. [DOI] [PubMed] [Google Scholar]
- Li G, Kataoka H, Coughlin SR, Pleasure SJ. Identification of a transient subpial neurogenic zone in the developing dentate gyrus and its regulation by CXCL12 and reelin signaling. Development. 2009;136:327–335. doi: 10.1242/dev.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapi A, Pritchett J, Jones O, Fujii N, Parnavelas JG, Nadarajah B. Stromal-derived factor 1 signaling regulates radial and tangential migration in the developing cerebral cortex. Dev Neurosci. 2008;30:117–131. doi: 10.1159/000109857. [DOI] [PubMed] [Google Scholar]
- Lieberam I, Agalliu D, Nagasawa T, Ericson J, Jessell TM. A CXCL12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron. 2005;47:667–679. doi: 10.1016/j.neuron.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Sucher NJ, Kaiser PK, Dreyer EB. Synergistic effects of HIV coat protein and NMDA receptor-mediated neurotoxicity. Neuron. 1991;7:111–118. doi: 10.1016/0896-6273(91)90079-f. [DOI] [PubMed] [Google Scholar]
- Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci U S A. 2002;99:7090–7095. doi: 10.1073/pnas.092013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired b-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4-and SDF1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay CR. Chemokines: immunology's high impact factors. Nat Immunol. 2001;2:95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- Marchionni I, Takács VT, Nunzi MG, Mugnaini E, Miller RJ, Maccaferri G. Distinctive properties of CXC chemokine receptor 4-expressing Cajal-Retzius cells versus GABAergic interneurons of the postnatal hippocampus. J Physiol. 2010;588:2859–2878. doi: 10.1113/jphysiol.2010.190868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Titlow WB, McClintock TS. Axon growth and guidance genes identify nascent, immature, and mature olfactory sensory neurons. J Neurosci Res. 2010;88:3243–3256. doi: 10.1002/jnr.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meucci O, Miller RJ. gp120-induced neurotoxicity in hippocampal pyramidal neuron cultures: protective action of tgf-beta1. J Neurosci. 1996;16:4080–4088. doi: 10.1523/JNEUROSCI.16-13-04080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meucci O, Fatatis A, Simen AA, Bushell TJ, Gray PW, Miller RJ. Chemokines regulate hippocampal neuronal signaling and gp120 neurotoxicity. Proc Natl Acad Sci U S A. 1998;95:14500–14505. doi: 10.1073/pnas.95.24.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RJ. Chemokine signaling in the central and peripheral nervous systems and its role in development and neuropathology. In: Meucci O, editor. Chemokine receptors and neuroAIDS; Beyond co-receptor function and links to other neuropathologies. Springer-Verlag; 2008. pp. 191–220. [Google Scholar]
- Miller RJ, Meucci O. AIDS and the brain: is there a chemokine connection? Trends Neurosci. 1999;22:471–479. doi: 10.1016/s0166-2236(99)01408-3. [DOI] [PubMed] [Google Scholar]
- Miller RJ, Banisadr G, Bhattacharyya BJ. CXCR4 signaling in the regulation of stem cell migration and development. J Neuroimmunol. 2008;198:31–38. doi: 10.1016/j.jneuroim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Shibuta K, Mathai J, Shimoda K, Banner BF, Mori M, Barnard GF. CXCR4 mRNA expression in colon, esophageal and gastric cancers and hepatitis C infected liver. Int J Oncol. 1999;14:917–925. doi: 10.3892/ijo.14.5.917. [DOI] [PubMed] [Google Scholar]
- Miyasaka N, Knaut H, Yoshihara Y. CXCL12/CXCR4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134:2459–2468. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verástegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of b-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine pbsf/SDF1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Neel NF, Schutyser E, Sai J, Fan GH, Richmond A. Chemokine receptor internalization and intracellular trafficking. Cytokine Growth Factor Rev. 2005;16:637–658. doi: 10.1016/j.cytogfr.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijmeijer S, Leurs R, Smit MJ, Vischer HF. The Epstein-bar virus-encoded g protein-coupled receptor bilf1 hetero-oligomerizes with human CXCR4, scavenges Gi proteins, and constitutively impairs CXCR4 functioning. J Biol Chem. 2010;285:29632–29641. doi: 10.1074/jbc.M110.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odemis V, Lamp E, Pezeshki G, Moepps B, Schilling K, Gierschik P, Littman DR, Engele J. Mice deficient in the chemokine receptor CXCR4 exhibit impaired limb innervation and myogenesis. Mol Cell Neurosci. 2005;30:494–505. doi: 10.1016/j.mcn.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Oh SB, Tran PB, Gillard SE, Hurley RW, Hammond DL, Miller RJ. Chemokines and glycoprotein120 produce pain hypersensitivity by directly exciting primary nociceptive neurons. J Neurosci. 2001;21:5027–5035. doi: 10.1523/JNEUROSCI.21-14-05027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky Killian EC, Birkholz DA, Artinger KB. A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev Biol. 2009;333:161–172. doi: 10.1016/j.ydbio.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevitch O, Abraham E, Borodovsky N, Levkowitz G, Zohar Y, Gothilf Y. CXCL12a-CXCR4b signaling is important for proper development of the forebrain GnRH system in zebrafish. Gen Comp Endocrinol. 2010;165:262–268. doi: 10.1016/j.ygcen.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Paredes MF, Li G, Berger O, Baraban SC, Pleasure SJ. Stromal-derived factor-1 (CXCL12) regulates laminar position of Cajal-Retzius cells in normal and dysplastic brains. J Neurosci. 2006;26:9404–9012. doi: 10.1523/JNEUROSCI.2575-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pello OM, Martínez-Muñoz L, Parrillas V, Serrano A, Rodríguez-Frade JM, Toro MJ, Lucas P, Monterrubio M, Martínez AC, Mellado M. Ligand stabilization of CXCR4/delta-opioid receptor heterodimers reveals a mechanism for immune response regulation. Eur J Immunol. 2008;38:537–549. doi: 10.1002/eji.200737630. [DOI] [PubMed] [Google Scholar]
- Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. Cytomegalovirus encodes a potent alpha chemokine. Proc Natl Acad Sci U S A. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, Gerard C, Lefkowitz RJ. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci U S A. 2010a;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010b;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Caviness VS., Jr Cortical development: view from neurological mutants two decades later. Neuron. 1995;14:1101–1104. doi: 10.1016/0896-6273(95)90258-9. [DOI] [PubMed] [Google Scholar]
- Reiss K, Mentlein R, Sievers J, Hartmann D. Stromal cell-derived factor 1 is secreted by meningeal cells and acts as chemotactic factor on neuronal stem cells of the cerebellar external granular layer. Neuroscience. 2002;115:295–305. doi: 10.1016/s0306-4522(02)00307-x. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, Kieran MW, Luster AD, Segal RA. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Lin YC, Dairaghi DJ, Schall TJ, Mocarski ES. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc Natl Acad Sci U S A. 1999;96:10881–10886. doi: 10.1073/pnas.96.19.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Mocarski ES., Jr Fatal attraction: cytomegalovirus-encoded chemokine homologs. Curr Top Microbiol Immunol. 2002;269:235–256. doi: 10.1007/978-3-642-59421-2_14. [DOI] [PubMed] [Google Scholar]
- Sánchez-Alcañiz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, López-Bendito G, Stumm R, Marín O. CXCR7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sapède D, Rossel M, Dambly-Chaudière C, Ghysen A. Role of SDF1 chemokine in the development of lateral line efferent and facial motor neurons. Proc Natl Acad Sci U S A. 2005;102:1714–1718. doi: 10.1073/pnas.0406382102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio T, Levi G. Neurotoxicity of HIV coat protein gp120, NMDA receptors, and protein kinase c: a study with rat cerebellar granule cell cultures. J Neurosci Res. 1993;34:265–272. doi: 10.1002/jnr.490340303. [DOI] [PubMed] [Google Scholar]
- Schönemeier B, Kolodziej A, Schulz S, Jacobs S, Hoellt V, Stumm R. Regional and cellular localization of the CXCL12/SDF-1 chemokine receptor CXCR7 in the developing and adult rat brain. J Comp Neurol. 2008;510:207–220. doi: 10.1002/cne.21780. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Henion TR, Nugent JD, Caplan B, Tobet S. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J Neurosci. 2006;26:6834–6840. doi: 10.1523/JNEUROSCI.1728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting GA, Wierman ME, Tobet SA. Gonadotropin-releasing hormone neuronal migration. Semin Reprod Med. 2007;25:305–312. doi: 10.1055/s-2007-984736. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Keener C, Boynton AL, Warrick J, Murphy GP. CXCR-4, a chemokine receptor, is overexpressed in and required for proliferation of glioblastoma tumor cells. J Surg Oncol. 1998a;69:99–104. doi: 10.1002/(sici)1096-9098(199810)69:2<99::aid-jso10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Ricks S, Boynton AL, Warrick J, Murphy GP. Molecular characterization of cxcr-4: a potential brain tumor-associated gene. J Surg Oncol. 1998b;69:239–248. doi: 10.1002/(sici)1096-9098(199812)69:4<239::aid-jso9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, McKnight A, Dejucq N, Hibbitts S, Power CA, Aarons E, Schols D, De Clercq E, Proudfoot AE, Clapham PR. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan EKI, Anderson RL. Genes involved in breast cancer metastasis to bone. Cell Mol Life Sci. 2002;59:1491–1502. doi: 10.1007/s00018-002-8524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohy D, Yano H, de Nadai P, Urizar E, Guillabert A, Javitch JA, Parmentier M, Springael JY. Hetero-oligomerization of CCR2, CCR5, and CXCR4 and the protean effects of “selective” antagonists. J Biol Chem. 2009;284:31270–31279. doi: 10.1074/jbc.M109.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, Urizar E, Parmentier M. Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev. 2005;16:611–623. doi: 10.1016/j.cytogfr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Springael JY, Le Minh PN, Urizar E, Costagliola S, Vassart G, Parmentier M. Allosteric modulation of binding properties between units of chemokine receptor homo- and hetero-oligomers. Mol Pharmacol. 2006;69:1652–1661. doi: 10.1124/mol.105.019414. [DOI] [PubMed] [Google Scholar]
- Stumm R, Höllt V. CXC chemokine receptor 4 regulates neuronal migration and axonal pathfinding in the developing nervous system: implications for neuronal regeneration in the adult brain. J Mol Endocrinol. 2007;38:377–382. doi: 10.1677/JME-06-0032. [DOI] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Höllt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Suratt BT, Petty JM, Young SK, Malcolm KC, Lieber JG, Nick JA, Gonzalo JA, Henson PM, Worthen GS. Role of the CXCR4/SDF1 chemokine axis in circulating neutrophil homeostasis. Blood. 2004;104:565–571. doi: 10.1182/blood-2003-10-3638. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- Tanaka DH, Mikami S, Nagasawa T, Miyazaki JI, Nakajima K, Murakami F. CXCR4 is required for proper regional and laminar distribution of cortical somatostatin-, calretinin-, and neuropeptide y-expressing GABAergic interneurons. Cereb Cortex. 2010;20:2810–2817. doi: 10.1093/cercor/bhq027. [DOI] [PubMed] [Google Scholar]
- Thelen M. Dancing to the tune of chemokines. Nat Immunol. 2001;2:129–34. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, König N, Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba Y, Tiong JD, Ma Q, Wray S. CXCR4/SDF-1 system modulates development of GnRH-1 neurons and the olfactory system. Dev Neurobiol. 2008;68:487–503. doi: 10.1002/dneu.20594. [DOI] [PubMed] [Google Scholar]
- Toggas SM, Masliah E, Rockenstein EM, Rall GF, Abraham CR, Mucke L. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature. 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- Toth PB, Ren DJ, Miller RJ. Regulation of CXCR4 receptor dimerization by the chemokine SDF-1α and the HIV-1 coat protein gp120; a fluorescence resonance energy transfer study. J Pharm Exp Ther. 2003;310:8–17. doi: 10.1124/jpet.103.064956. [DOI] [PubMed] [Google Scholar]
- Tran PB, Miller RJ. Chemokine receptors in the brain: a developing story. J Comp Neurol. 2003a;457:1–6. doi: 10.1002/cne.10546. [DOI] [PubMed] [Google Scholar]
- Tran PB, Miller RJ. Chemokine receptors: signposts to brain development and disease. Nat Rev Neurosci. 2003b;4:444–455. doi: 10.1038/nrn1116. [DOI] [PubMed] [Google Scholar]
- Tran PB, Banisadr G, Ren D, Chenn A, Miller RJ. Chemokine receptor expression by neural progenitor cells in neurogenic regions of mouse brain. J Comp Neurol. 2007;500:1007–1033. doi: 10.1002/cne.21229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles CS, Domínguez F. Embryo-endometrial interaction. Chang Gung Med J. 2006;29:9–14. [PubMed] [Google Scholar]
- Vilz TO, Moepps B, Engele J, Molly S, Littman DR, Schilling K. The SDF1/CXCR4 pathway and the development of the cerebellar system. Eur J Neurosci. 2005;22:1831–1839. doi: 10.1111/j.1460-9568.2005.04378.x. [DOI] [PubMed] [Google Scholar]
- Vink C, Smit MJ, Leurs R, Bruggeman CA. The role of cytomegalovirus-encoded homologs of g protein-coupled receptors and chemokines in manipulation of and evasion from the immune system. J Clin Virol. 2001;23:43–55. doi: 10.1016/s1386-6532(01)00184-6. [DOI] [PubMed] [Google Scholar]
- Vischer HF, Nijmeijer S, Smit MJ, Leurs R. Viral hijacking of human receptors through heterodimerization. Biochem Biophys Res Commun. 2008;377:93–97. doi: 10.1016/j.bbrc.2008.09.082. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JLR. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells TN, Proudfoot AE, Power CA, Marsh M. Chemokine receptors-the new frontier for aids research. Chem Biol. 1996;3:603–609. doi: 10.1016/s1074-5521(96)90126-x. [DOI] [PubMed] [Google Scholar]
- Westmoreland SV, Kolson D, González-Scarano F. Toxicity of TNF alpha and platelet activating factor for human nt2n neurons: a tissue culture model for human immunodeficiency virus dementia. J Neurovirol. 1996;2:118–126. doi: 10.3109/13550289609146545. [DOI] [PubMed] [Google Scholar]
- Wilson NM, Jung H, Ripsch MS, Miller RJ, White FA. CXCR4 signaling mediates morphine-induced tactile hyperalgesia. Brain Behav Immun. 2011;25:565–673. doi: 10.1016/j.bbi.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Xin WW, Duman RS. Rat lcr1: cloning and cellular distribution of a putative chemokine receptor in brain. Mol Psychiatry. 1996;1:133–140. [PubMed] [Google Scholar]
- Yang L, Jackson E, Woerner BM, Perry A, Piwnica-Worms D, Rubin JB. Blocking CXCR4-mediated cyclic AMP suppression inhibits brain tumor growth in vivo. Cancer Res. 2007;67:651–658. doi: 10.1158/0008-5472.CAN-06-2762. [DOI] [PubMed] [Google Scholar]
- Yeung MC, Pulliam L, Lau AS. The HIV envelope protein gp120 is toxic to human brain-cell cultures through the induction of interleukin-6 and tumor necrosis factor-alpha. AIDS. 1995;9:137–143. [PubMed] [Google Scholar]
- Zeelenberg IS, Ruuls-Van Stalle L, Roos E. The chemokine receptor cxcr4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63:3833–3839. [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Matsumoto T, Mikami S, Nagasawa T, Murakami F. SDF1/CXCR4 signaling regulates two distinct processes of pre-cerebellar neuronal migration and its depletion leads to abnormal pontine nuclei formation. Development. 2009;136:1919–1928. doi: 10.1242/dev.032276. [DOI] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]