Abstract
Robust mechanisms to control cell proliferation have evolved to maintain the integrity of organ architecture. Here, we investigated how two critical proliferative pathways, Myc and E2f, are integrated to control cell cycles in normal and Rb deficient cells using a murine intestinal model. We show that Myc and E2f1-3 have little impact on normal G1-S transitions. Instead, they synergistically control an S-G2 transcriptional program required for normal cell divisions and maintaining crypt-villus integrity. Surprisingly, Rb deficiency results in the Myc-dependent accumulation of E2f3 protein and chromatin repositioning of both Myc and E2f3, leading to the ‘super activation’ of a G1-S transcriptional program, ectopic S phase entry and rampant cell proliferation. These findings reveal that Rb deficient cells hijack and redeploy Myc and E2f3 from an S-G2 program essential for normal cell cycles to a G1-S program that re-engages ectopic cell cycles, exposing an unanticipated addiction of Rb-null cells on Myc.
Spatial and temporal control of cell proliferation is vital for organogenesis and maintenance of tissue integrity. Normal proliferative stimuli impinge on cell surface receptors and engage multiple intracellular signaling cascades that converge on cell cycle control. Growth factor receptor activation culminates in the accumulation of Myc and E2f transcription factors, two central components believed to link external proliferative signals to early phases of the cell cycle1, 2.
In response to mitogenic signals such as Wnt, Notch, TGFβ and activation of receptor tyrosine kinases3–7, Myc transcriptionally orchestrates a broad range of biological processes, including macromolecule biosynthesis, energy production, induction of cyclin-dependent kinases (Cdks)7–17, and physically interacts with the DNA pre-replication complex18 to collectively prepare cells for S phase entry. Several studies have also linked Myc to the regulation of E2f expression and activity9, 19, 20. The E2f family consists of eight related members that have transcription activation and repression functions21. Phosphorylation of Rb by Cdks leads to the accumulation of E2f1-3 late in G1 and activation of a G1-S transcriptional program that licenses the entry into S phase and commitment through cell division22. E2f6-8 mediated transcriptional repression late in S phase contributes to the oscillatory expression of target genes as cells march through the cell cycle23. The fact that MYC and E2F1-3, or components that regulate them such as RB, CDKs and p16INK4A, are invariably disrupted in human cancer highlights their central role in the control of cellular proliferation23, 24.
Surprisingly, knockout mouse models of Myc and E2fs reveal a paucity of major cell cycle defects25–29. This has been attributed to redundancy within these two families of transcription factors30, 31. Recent work showed that mouse retinal precursors can proliferate in the absence of N-Myc or E2f1-3, but not in the absence of both sets of factors27, suggesting further redundancy among the two transcription factor families. Whether Myc and E2fs collaborate in other cell types and tissues, and how they might do so, remains to be determined.
Here we evaluated the roles of Myc and E2f1-3 in the control of cell proliferation in the small intestine of mice. The functional unit of the small intestine consists of pouch-like invaginations called crypts and finger-like projections called villi. Pluripotent stem cells at the base of crypts continuously divide to generate transit amplifying progenitor cells, which undergo several rounds of proliferation before they exit the cell cycle, differentiate and repopulate villi. Here we show that the combined loss of Myc and E2f1-3 has little impact on G1-S transitions in the small intestine of mice. Rather, Myc and E2f1-3 engage an S-G2 transcriptional program required for the completion of S phase and progression through mitosis. When Rb is inactivated, however, we show that Myc and E2fs are redeployed and engage a distinct G1-S program that promotes ectopic cell cycles. These findings distinguish how Myc and E2f control the proliferation of normal versus Rb deficient cells, and expose a molecular mechanism for the unexpected dependency of Rb deficient cells on Myc.
RESULTS
Combined ablation of Myc and E2f1-3 results in disruption of crypt-villus integrity
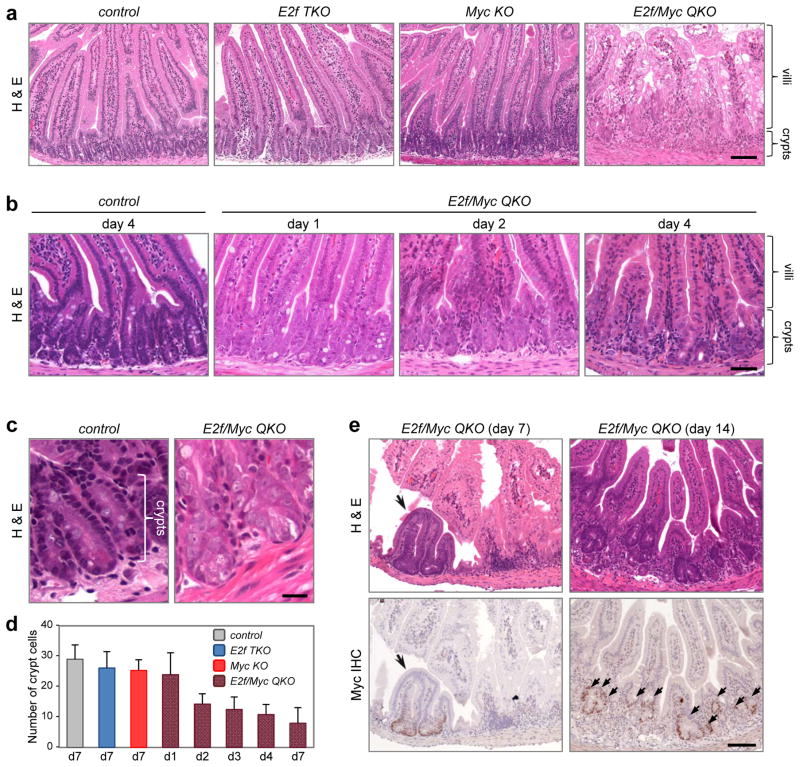
To explore whether Myc and E2f activities collaborate in the control of normal cell cycles in vivo, we examined the small intestine of mice containing an inducible intestinal-specific Cre transgene (Ah-cre) and conditional alleles of Myc and/or E2f332. Because of the potential functional redundancy between E2f3 and the other two E2f activators, we also introduced null alleles of E2f1 and E2f2 into experimental animals. Ah-cre expression in crypts was induced by intraperitoneal administration of β-naphthoflavone (β-NF) and tissue histopathology was examined 7 days later by haematoxylin-and-eosin staining. Ablation of either E2f1-3 (Ah-cre;E2f1−/−;E2f2−/−;E2f3loxP/loxP, E2f TKO) or Myc (Ah-cre;MycloxP/loxP, Myc KO) had little effect on intestinal architecture (Fig. 1a), consistent with recent studies showing active crypt cell proliferation in the absence of either E2f1-3 or Myc25, 28, 33. The simultaneous deletion of E2f1-3 and Myc (Ah-cre;E2f1−/−;E2f2−/−;E2f3loxP/loxP;MycloxP/loxP, E2f/Myc QKO), however, resulted in the complete collapse of crypt-villus structure. Examination at earlier time points following β-NF injection revealed that alterations in crypt architecture preceded the changes observed in associated villi (Fig. 1b). By two days post β-NF injection, E2f/Myc QKO crypt cells had enlarged nuclei with reduced basophilic staining, and appeared overall larger than controls (Fig. 1b, c). By four days post β-NF injection, the number of cells in E2f/Myc QKO crypts decreased to less than 50% of control animals, leading to marked crypt atrophy and deterioration of villus integrity (Fig. 1d). While mice became moribund within 1–2 weeks of β-NF treatment, they subsequently recovered, groomed and appeared healthy. Inspection of their small intestines showed that residual crypts escaping Cre-mediated deletion had repopulated the intestinal epithelium (Fig. 1e), as similarly observed in other studies using this system33, 34.
Figure 1. Disruption of the small intestine by combined loss of Myc and E2f1-3.
(a) Haematoxylin-and-eosin (H&E) stained tissue sections from control, E2f TKO, Myc KO and E2f/Myc QKO intestines collected 7 days after induction of Ah-cre expression. (b) Progressive degeneration of E2f/Myc QKO crypts from day 1 to day 4 after induction of Ah-cre expression. (c) H&E stained tissue sections from control and E2f/Myc QKO intestines collected 2 days after induction of Ah-cre expression. Data in a–c are representative images from n=3 mice per genetic group at each indicated time point. (d) Quantification of average number of crypt cells. Data presented as mean ± s.d., n=3 mice per genetic group at each indicated time point. (e) E2f/Myc QKO tissue sections stained by H&E and immunohistochemistry (IHC) of Myc to show the regeneration of intestinal epithelium by Myc-positive cells at 7 and 14 days after induction of Ah-cre expression (arrows). Data are representative images from n=3 mice at each time point. Scale bars represent 100 μm (a, e), 50 μm (b) and 25 μm (c).
Combined Myc and E2f1-3 deficiency leads to S-G2 cell cycle arrest
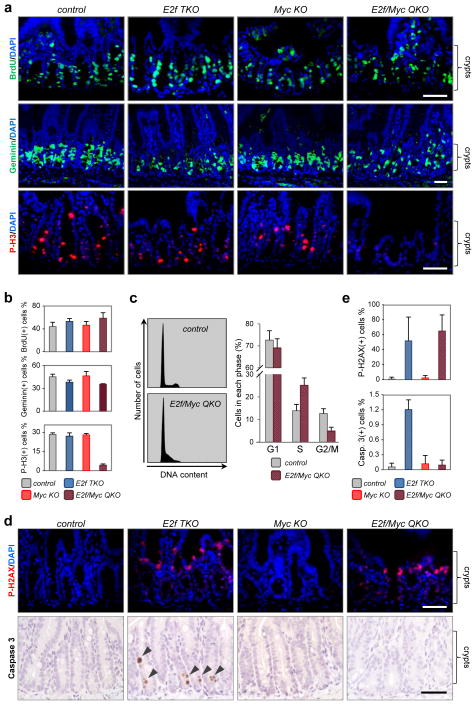
We reasoned that the acute degeneration of E2f/Myc QKO crypts could be due to decreased cell proliferation. Surprisingly, DNA synthesis was unaffected in progenitor cells at a time when Myc and E2f1-3 proteins were clearly depleted (Fig. 2a, b and Supplementary Fig. 1a–c). Expression of geminin, a protein involved in blocking the re-replication of the genome late in S phase and G235, was also normal in E2f/Myc QKO cells (Fig. 2a, b and Supplementary Fig. 1a, b). However, progression through cell division was severely impaired in E2f/Myc QKO cells as indicated by the absence of mitotic figures and Serine 10-phosphorylated histone 3 (P-H3) staining (Fig. 2a, b). Fluorescence-activated cell sorting analysis showed an accumulation of E2f/Myc QKO crypt cells in S phase and a reduction in G2-M compared to control littermates (Fig. 2c). Despite the late cell cycle arrest in E2f/Myc QKO samples, cell type-specific marker analysis revealed an appropriate number of paneth and goblet cells along the crypt-villus unit (Supplementary Fig. 1d), probably reflecting pre-existing non-deleted cells that persist beyond the experimental time frame analyzed here (i.e. paneth cells live for several weeks)36. Together, these findings suggest that E2f/Myc QKO progenitor cells were able to enter S phase but failed to fully progress through S-G2.
Figure 2. S-G2 cell cycle arrest in E2f/Myc QKO progenitor cells.
(a) Immunofluorescence (IF) staining of BrdU (green), geminin (green) and P-H3 (red) in crypts from intestines harvested 2 days after induction of Ah-cre expression. Nuclei were stained with DAPI (blue). Data are representative images from n=3 mice per genetic group. (b) Quantification of BrdU, geminin and P-H3 staining. Data presented as mean ± s.d., n=3 mice per genetic group. (c) Fluorescence-activated cell sorting analysis of cell cycle status in control and E2f/Myc QKO crypts from intestines harvested 2 days after induction of Ah-cre expression. Left, representative histograms from control (n=3 mice) and E2f/Myc QKO (n=5 mice) crypts. Right, quantification of histograms; data presented as mean ± s.d., n=3 (control) or n=5 (E2f/Myc QKO) mice. (d) IF staining of P-H2AX (red) and IHC staining of cleaved caspase-3 (brown) in crypts. Intestines were harvested 2 days (P-H2AX) or 4 days (cleaved caspase-3) after induction of Ah-cre expression. Data are representative images from n=3 mice per genetic group. (e) Quantification of P-H2AX and cleaved caspase-3 staining. Data presented as mean ± s.d., n=3 mice per genetic group. Scale bars in a and d represent 50 μm.
DNA integrity was compromised in E2f/Myc QKO progenitor cells as indicated by increased phosphorylated H2AX (P-H2AX) staining (Fig. 2d, e). This increase in DNA damage was a consequence from the specific ablation of E2fs since E2f TKO, but not Myc KO intestines, displayed higher levels of P-H2AX. To determine whether cell death, possibly due to incurred DNA damage, contributed to E2f/Myc QKO crypt degeneration, tissue sections were processed for immunohistochemistry (IHC) using cleaved caspase-3 specific antibodies. This analysis showed that E2f TKO crypts, but not Myc KO or E2f/Myc QKO crypts, contained apoptotic cells (Fig. 2d, e). We considered the possibility that loss of Myc might accelerate the elimination of E2f1-3 deficient apoptotic cells in E2f/Myc QKO crypts; however, this seems unlikely since a similar analysis at one and two days following β-NF injection also failed to detect apoptotic cells in these samples. Thus, the execution of programmed cell death in E2f1-3 deficient crypts is dependent on Myc. Interestingly Myc was recently shown to be required for DNA damage induced apoptosis of crypt cells37. From these findings we conclude that cell cycle arrest in S-G2 underlies crypt atrophy caused by combined E2f-Myc deficiency.
Synergistic regulation of an S-G2 transcriptional program by Myc and E2f1-3
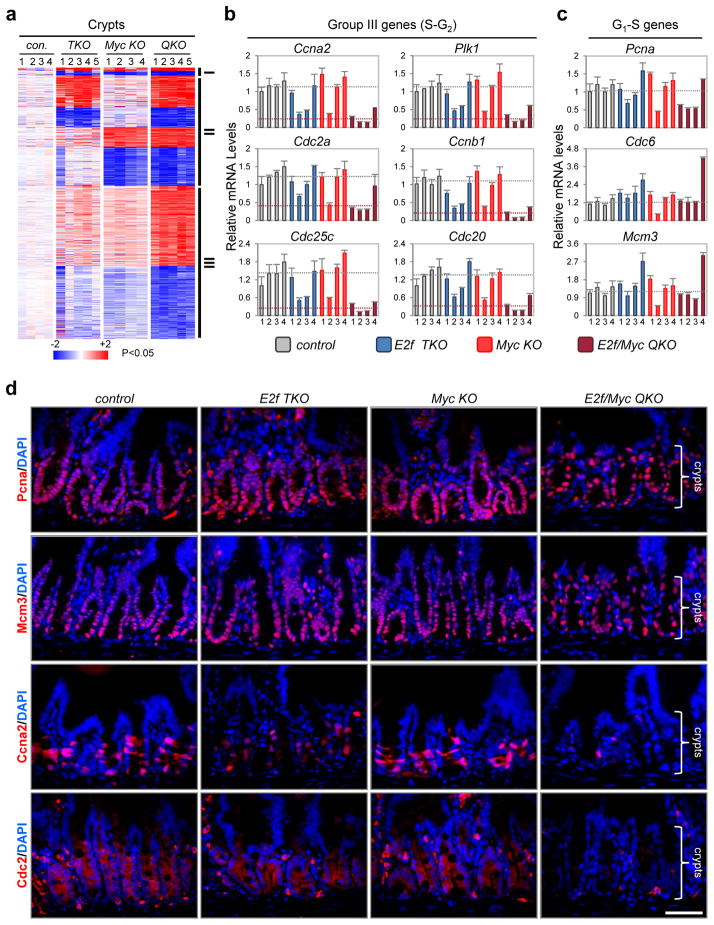
Given the established roles of Myc and E2f as transcription factors, we reasoned that changes in gene expression might underlie the observed S-G2 block in E2f/Myc QKO cells. We thus examined the transcriptome from β-NF treated control, E2f TKO, Myc KO and E2f/Myc QKO crypts using an Affymetrix platform. This comparison revealed three main categories of differentially expressed genes (Fig. 3a and Supplementary Table 1). One category (group I) includes genes dysregulated in all three mutant genetic groups, indicating a requirement for E2f1-3 and Myc in regulating the expression of these genes. A second category (group II) includes genes dysregulated in E2f/Myc QKO samples and either E2f TKO or Myc KO samples, suggesting that these target genes are uniquely regulated by E2f1-3 or Myc. The third and perhaps most interesting category (group III) includes genes that are unaffected or only marginally dysregulated in E2f TKO and Myc KO cells, but are profoundly dysregulated in E2f/Myc QKO crypts, suggesting that this group of genes are synergistically regulated, albeit not necessarily directly, by E2f1-3 and Myc. Gene ontology (GO) analysis of each group failed to detect any significant enrichment for classic G1-S regulated mRNAs. However, group III genes were dramatically enriched for mitotic related functions (Supplementary Table 2). Quantitative gene expression and immunofluorescence assays confirmed the acute and specific down-regulation of S-G2 related genes in E2f/Myc QKO crypts (Fig. 3b–d). We conclude that Myc and E2f synergistically regulate an S-G2 expression program required for normal cell cycle progression in vivo.
Figure 3. Synergistic regulation of an S-G2 transcriptional program by Myc and E2f1-3.
(a) Heatmap representation for clustering of differentially expressed genes between mutant genetic groups compared to control samples. Crypts were collected 2 days after induction of Ah-cre expression. n=4 for control and Myc KO mice, n=5 for E2f TKO and E2f/Myc QKO mice. P<0.05, Student’s t-test. (b) Quantitative polymerase chain reaction with reverse transcription (RT-qPCR) analysis for a subset of Group III (S-G2 related) genes. Normal expression levels are illustrated as grey dotted lines and dysregulated expression levels in E2f/Myc QKO crypts are illustrated as red dotted lines. (c) RT-qPCR analysis for a subset of G1-S related genes. Normal expression levels are illustrated as grey dotted lines. In b and c, expression levels from individual mice are plotted (4 per genetic group) and error bars represent mean ± s.d. from n=3 technical replicates. (d) IF staining of Pcna, Mcm3, Ccna2 and Cdc2. Note that degenerating E2f/Myc QKO crypts with less dense cells have comparable protein levels of Pcna and Mcm3, yet significantly less Ccna2 and Cdc2, compared to other genetic groups. Data are representative images from n=3 mice per genetic group. Scale bars in d represent 50 μm.
Myc and E2f3 bind to both G1-S and S-G2 target genes in crypts
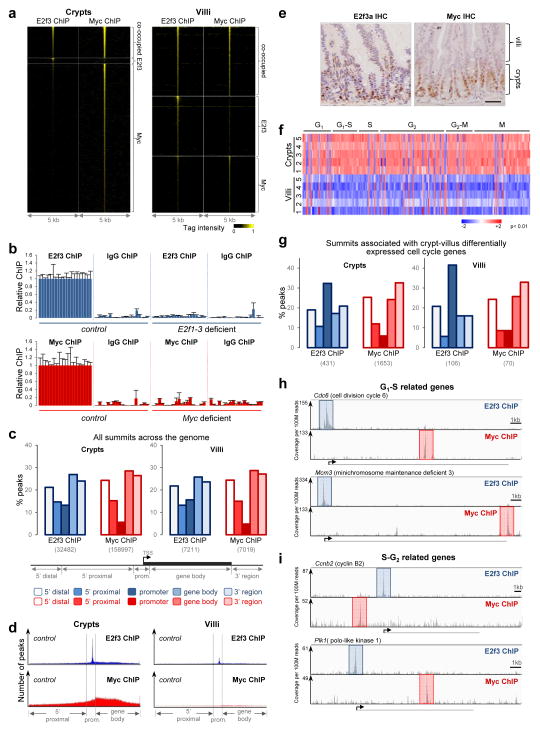
We then queried the genome-wide occupancy of E2f3 and Myc on chromatin of intestinal cells using chromatin immunoprecipitation combined with exonuclease digestion, followed by next generation sequencing (ChIP-exo-seq)38, 39. The ChIP-exo-seq approach provides enhanced sensitivity and specificity over traditional ChIP-seq methods, improving the detection of chromatin occupancy by non-abundant transcription factors, such as E2fs and Myc. E2f3 and Myc DNA binding peak summits were identified using two algorithms (MACS2 & GEM) with a false discovery rate (FDR) of less than 1%. A summary of the ChIP-exo-seq data is presented in Figure 4a and Supplementary Table 3. Twenty randomly selected E2f3- and twenty Myc-specific peak summits were validated by ChIP-PCR assays using control, E2f3-deficient and Myc-deficient tissues (Fig. 4b). Peak summits were then mapped to genes based on the minimal distance between summits and proximal transcription start sites (Fig. 4c). The patterns of E2f3 and Myc chromatin binding in crypts were distinct, with E2f3 occupancy biased towards promoter regions and Myc occupancy distributed more broadly across genes (Fig. 4d). E2f3 and Myc binding was reduced in villi, consistent with lower levels of these two proteins in this compartment (Fig. 4e and Supplementary Fig. 2a).
Figure 4. Chromatin binding of E2f3 and Myc in wild type tissues.
(a) Heatmap of tag intensity for all E2f3 and Myc binding locations in wild type intestines. Data collected from pooled crypts (n=32 mice) or villi (n=7 mice). (b) E2f3 ChIP-PCR validation: control (Rb KO, n=3 villi) and E2f1-3 deficient (Rb/E2f QKO, n=3 villi); Myc ChIP-PCR validation: control (wild type, n=4 crypts) and Myc deficient (Myc KO, n=3 crypts). Data presented as mean ± s.d. For detailed peak location and primer sequence information see Supplementary Table 11. (c) Genomic spatial distribution of all E2f3 and Myc peak summits in wild type intestines. The number of summits in each tissue compartment is shown in parentheses. 5′ distal: 5′ region more than −50 kilobases (kb) from transcription start sites (TSSs). 5′ proximal: 5′ region within −50kb to −5kb of TSSs. Promoter: −5kb to +2kb of TSSs. Gene body: from +2kb of TSSs to end of transcripts. 3′ region: 3′ region starting from end of transcripts. (d) Density plots of all E2f3 and Myc peak summits across genomic regions in wild type intestines. Gene bodies for individual genes were divided into 100 bins and summit locations were accordingly assigned a genomic position. Data in c and d were collected from pooled crypts (n=32 mice) or villi (n=7 mice). (e) IHC staining of E2f3a and Myc in wild type intestines. Data are representative images from n=3 mice. Scale bars, 50 μm. (f) Heatmap of differential expression of cell cycle related genes, as annotated in Cyclebase database, in wild type crypts (n=5 mice) and villi (n=5 mice). P<0.01, empirical Bayes method. (g) Genomic spatial distribution of E2f3 and Myc peak summits associated with differentially expressed cell cycle related genes in wild type intestines. The data represent a subset of c and classification of genomic regions is the same as in c. (h, i) ChIP-exo-seq track examples showing E2f3 and Myc binding to selected G1-S related genes (h) and S-G2 related genes (i) in wild type crypts. E2f3 and Myc peaks are highlighted in blue and red, respectively. Examples are derived from pooled crypts (n=32 mice). Scale bars in h and i represent 1kb.
We then profiled and compared gene expression in wild type crypts and villi. As expected, cell cycle related genes were preferentially expressed in crypts and differentiation related genes were preferentially expressed in villi (Fig. 4f, Supplementary Fig. 2b and Supplementary Table 4, 5). Integration of ChIP-exo-seq and gene expression data sets revealed a marked enrichment of E2f3 binding to promoter regions of differentially expressed cell cycle related genes (compare Fig. 4c and 4g)40. In crypts, E2f3 and Myc bound to both G1-S and S-G2 related genes (Fig. 4h, i, Supplementary Fig. 2c and Supplementary Table 5). Since only the expression of S-G2 related genes was disrupted in E2f/Myc QKO crypts, we suggest that the regulation of S-G2 targets requires direct binding by E2f or Myc, whereas the regulation of G1-S targets likely involves additional factors beyond E2f and Myc.
Myc drives ectopic proliferation of Rb deficient intestinal cells
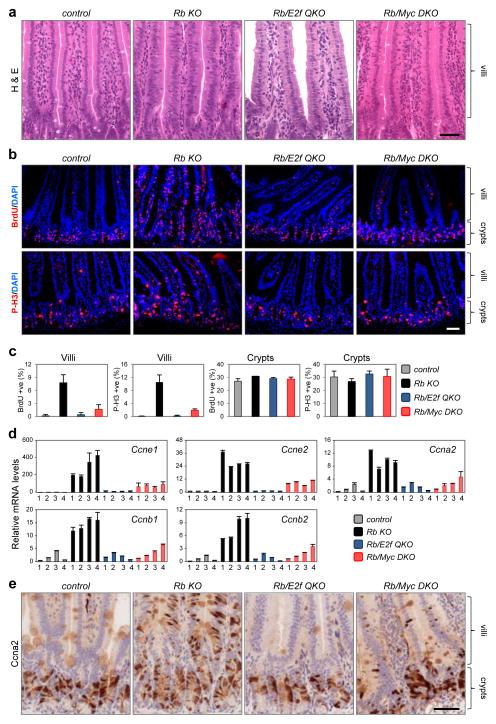
Previous work suggested that distinct mechanisms regulate the proliferation of normal and cancer cells41–43. Rb loss results in excessive proliferation of transit amplifying progenitor cells, even after they migrate, differentiate and begin to populate the length of the villus28. To explore the roles of E2f and Myc in an abnormal hyper-proliferative context, we initially queried expression profiles derived from β-NF treated RbloxP/loxP (control) and Ah-cre;RbloxP/loxP (Rb KO) villus-enriched fractions. In addition to the expected upregulation of E2f target genes in Rb deficient villi, there was a striking increase in the expression of many known Myc target genes, as defined by previous gene-expression, reporter and ChIP assays (Supplementary Table 6). We thus tested whether Myc may be playing a role in driving the ectopic proliferation caused by Rb deficiency. Remarkably, ablation of Myc suppressed the ectopic proliferation of Rb deficient villus cells to a similar extent as loss of E2f1-3 (Fig. 5a-c; Ah-cre;RbloxP/loxP;MycloxP/loxP (Rb/Myc DKO), Ah-cre;RbloxP/loxP;E2f1−/−;E2f2−/−;E2f3loxP/loxP (Rb/E2f QKO)), without impacting the proliferation of progenitor cells in crypts.
Figure 5. Rb deficient cells require Myc to drive ectopic cell cycles.
(a) H&E stained tissue sections from control, Rb KO, Rb/E2f QKO and Rb/Myc DKO intestines. Note the hyperplastic feature of Rb KO villi. (b) IF staining of BrdU and P-H3. Note the non-specific staining of blood cells in the lumen of villi. Data in a and b are representative images from n=3 mice (BrdU) or n=4 mice (H&E, P-H3) per genetic group. (c) Quantification of BrdU and P-H3 staining. Data presented as mean ± s.d., BrdU (n=3 mice), P-H3 (n=4 mice). (d) RT-qPCR analysis for indicated cyclins in control, Rb KO, Rb/E2f QKO and Rb/Myc DKO villi. Expression levels from individual animals are plotted (4 per genetic group) and error bars represent mean ± s.d. from n=3 technical replicates. (e) IHC staining of Ccna2. Data are representative images from n=3 mice per genetic group. Scale bars in a, b and e represent 50 μm.
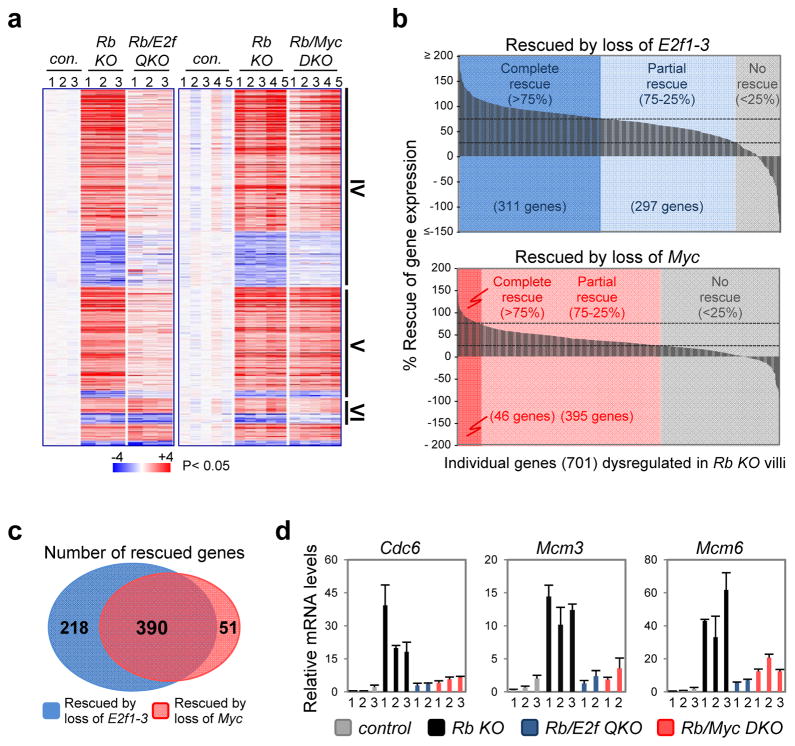
Evaluation of a panel of cell cycle markers showed that loss of Myc or E2f1-3 in Rb deficient villi prevented the aberrant accumulation of critical cell cycle regulators (Fig. 5d, e). We also profiled global gene expression in control, RbKO, Rb/E2f QKO and Rb/Myc DKO villi. The heatmaps and waterfall plots shown in Figure 6a and 6b illustrate the magnitude by which loss of Myc or E2f1-3 in Rb deficient villi restored expression programs to normal (>75% rescue) or near normal levels (25–75% rescue). Interestingly, the expression of the majority of dysregulated target genes was corrected by loss of either E2f1-3 or Myc (390 from a total 701 genes; Fig. 6c, d, Supplementary Fig. 3 and Supplementary Table 7). This 390 gene-set included classic G1-S and S-G2 regulated mRNAs (Supplementary Table 8). Together, these findings distinguish the roles of Myc and E2f in controlling transcription and cell proliferation in wild type crypts and Rb mutant villi.
Figure 6. Myc and E2f1-3 regulate an overlapping G1-S transcriptional program in Rb-null cells.
(a) Heatmap representation of dysregulated expression in Rb KO villi (701 genes) (left panel is from previously published data28 and is included here for comparison). Group IV: genes with expression levels rescued by loss of either E2f1-3 or Myc. Group V: genes with expression levels rescued by loss of E2f1-3. Group VI: genes with expression levels rescued by loss of Myc. P<0.05, Student’s t-test. (b) Waterfall plots illustrating the extent to which gene expression dysregulation in RbKO villi (701 genes) is ameliorated in Rb/E2f QKO and Rb/Myc DKO villi. (c) Venn diagram showing the overlap between genes with expression levels rescued by loss of E2f1-3 or Myc (complete and partial rescue). Data in a–c were collected from n=3 mice per genetic group for E2f-rescue experiments and n=5 mice per genetic group for Myc-rescue experiment. (d) RT-qPCR analysis for a subset of G1-S related genes. Expression levels for individual mice are plotted (2 or 3 per genetic group as indicated) and error bars represent mean ± s.d. from n= 3 technical replicates.
Redeployment of Myc and E2f3 in Rb deficient cells
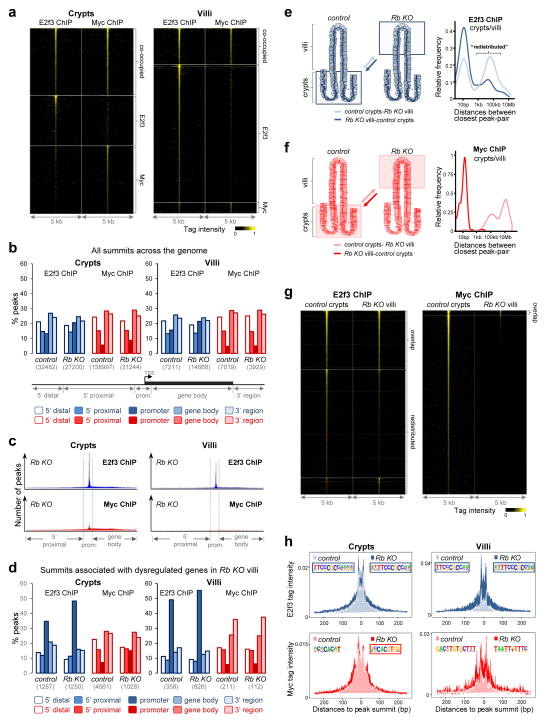
We then profiled Myc and E2f3 chromatin occupancy in Rb deficient crypts/villi. A summary of the ChIP-exo-seq data is presented in Figure 7a and Supplementary Table 3. In crypts, the number of E2f3 peak summits was unaffected by loss of Rb, whereas the number of Myc summits was significantly decreased (Fig. 7b, left panel). In villi, the number of E2f3 peak summits was increased by loss of Rb but the number of Myc summits was decreased (Fig. 7b, right panel). The spatial distribution of peak summits across gene regions is illustrated in Figure 7c.
Figure 7. Rb loss redefines the chromatin binding landscape of E2f3 and Myc.
(a) Heatmap representation of tag intensity for all E2f3 and Myc binding locations in Rb KO intestines. Data were collected from pooled crypts (n=27 mice) or villi (n=7 mice). (b) Genomic spatial distribution of all E2f3 and Myc peak summits in control and Rb KO tissues (data for control tissues from Fig. 4c is included here for comparison). The number of summits in each tissue compartment is shown in parentheses. Data were collected from pooled crypts (n=32 mice for control, n=27 mice for Rb KO) or villi (n=7 mice for control, n=7 mice for Rb KO). (c) Density plots of all E2f3 and Myc peaks across genomic regions in Rb KO crypts (pooled from n=27 mice) and villi (pooled from n=7 mice). Gene bodies for individual genes were divided into 100 bins and summit locations were accordingly assigned a genomic position. (d) Genomic spatial distribution of E2f3 and Myc peak summits associated with dysregulated genes in Rb KO villi. The number of summits in each compartment is shown in parentheses. The data represent a subset of b and classification of genomic regions is the same as in b. (e, f) Peak summit-distance plots for E2f3 summits (e) and Myc summits (f) in control crypts and Rb KO villi that are associated with the 701 dysregulated genes in Rb KO villi. (g) Heatmap representation of tag intensity for all E2f3 and Myc binding locations in control crypts and Rb KO villi. (h) Tag intensity plots (tags per bp per peak per 100M reads) around the peak summits associated with genes dysregulated in Rb KO villi. The canonical DNA binding motifs for E2f3 (TTCCCGCC) and Myc (CACGTG) are highlighted in blue and red boxes, respectively. Data in d–h were collected from pooled crypts (n=32 mice for control, n=27 mice for Rb KO) or villi (n=7 mice for control, n=7 mice for Rb KO).
Three major parameters were analyzed and compared between wild type and Rb deficient crypts and villi: DNA binding location, DNA binding strength and DNA binding sequence motifs.
DNA binding location
We focused further analysis on the binding of E2f3 and Myc to target genes dysregulated by Rb deficiency (Supplementary Table 9). E2f3 binding, but not Myc binding, was highly enriched in promoter regions (Fig. 7d). We then compared E2f3 binding in the two proliferative compartments, wild type crypts and Rb deficient villi, by measuring the distance between a given summit in wild type crypts (as a reference; Fig. 7e, light blue line) and its corresponding summit in Rb KO villi; the same comparison was performed with the Rb KO villi as the reference compartment (Fig. 7e, dark blue line). A distance of 0–100bp between corresponding summits in crypts and villi indicates overlapping peaks and suggests that binding of E2f3 was not altered between compartments; a distance greater than 100bp between corresponding summits suggests a movement or disappearance of E2f3 occupancy from its original position. The ‘peak summit-distance plots’ shown in Figure 7e revealed that approximately half of the E2f3 summits in control crypts had corresponding summits in Rb deficient villi at distal locations greater than 100bp (light blue line). Consistent with this analysis, heatmaps of sequence tags showed that a portion of E2f3 summits in control crypts and Rb deficient villi were identical, whereas a large portion of summits present in control crypts were reduced/absent in the corresponding positions in Rb deficient villi (Fig. 7g). Moreover, a significant number of E2f3 summits in Rb deficient villi were reduced/absent in control crypts. Summit-distance plots and heatmaps comparing E2f3 peak summits across other possible compartment and genotype combinations are presented in Supplementary Figure 4a–d. Examples of E2f3 binding to specific genes are shown in Supplementary Figure 5a–c. Together, these findings show that Rb deficiency in villi results in the recruitment of E2f3 to some of the same chromatin positions it normally occupies in control crypts, but also to new chromatin positions. A similar analysis failed to show a redistribution of Myc binding from wild type crypts to Rb deficient villi (Fig. 7f, g). However, comparison of control and Rb deficient crypts revealed the appearance of ‘new’ Myc-specific summits in Rb deficient crypts (Supplementary Fig. 4e–h; also see below). Thus, E2f3 is redistributed in Rb deficient villi and Myc is redistributed in Rb deficient crypts.
DNA binding strength
The intensity plots shown in Figure 7h and Supplementary Figure 6a illustrate an increase in the E2f3- and Myc-sequence tag intensity in Rb KO versus control tissues, particularly in villi, indicating that E2f3 and Myc binding strength is increased upon Rb loss.
DNA binding sequence motifs
Sequence analysis by de novo motif algorithms shows that E2f3 predominantly utilizes E2f canonical DNA binding elements irrespective of Rb deletion status or tissue compartment being analyzed (Fig. 7h top panels and Supplementary Fig. 6–7). In contrast, Myc utilizes a variety of non-canonical motifs in villi and control crypts but utilizes canonical E-box elements in Rb KO crypts (Fig. 7h bottom panels and Supplementary Fig. 6–7).
From the combined analysis described above, which includes changes in the number of binding events as well as changes in DNA binding location/distribution, DNA binding strength, and DNA binding sequence motifs, we conclude that loss of Rb results in a major spatial redeployment of E2f3 binding in villi and a refocusing of Myc binding in crypts.
Myc is required for the accumulation of E2f3 in Rb deficient villi
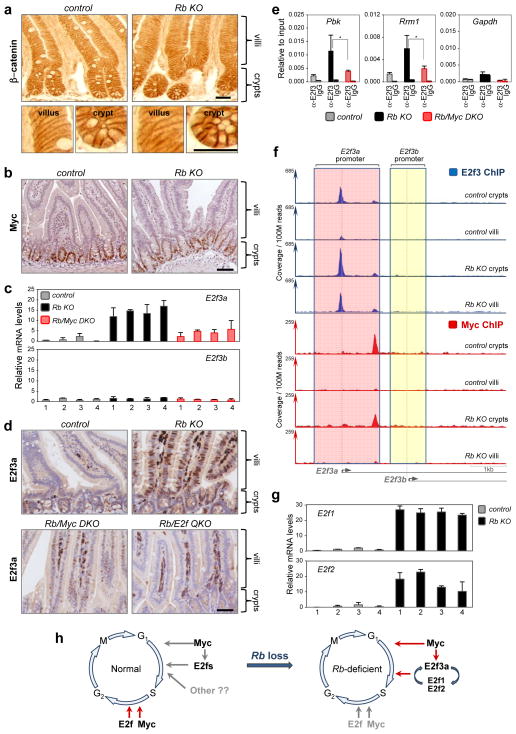
We then explored the mechanistic relationship between Myc and E2fs in regulating gene expression. Previous studies established Myc as a critical downstream effector of Apc/β-catenin signaling34. Several nodes of crosstalk between canonical Apc/β-catenin/Myc and Cdk/Rb/E2f pathways have also been described44–49. Thus, we investigated the possibility that Apc/β-catenin/Myc signaling might be increased in Rb deficient intestinal cells and contribute to their ectopic proliferation. Immunostaining of intestinal sections showed identical levels and localization of β-catenin protein in control and Rb KO small intestines (Fig. 8a). Moreover, comparison of global mRNA profiles from control and Rb KO villi (Fig. 6a) showed no change in the expression of a large cadre of known β-catenin target genes, including Myc itself (Supplementary Fig. 8a and Supplementary Table 10). Indeed, IHC of tissue sections showed that Myc protein levels were unchanged by loss of Rb (Fig. 8b). These results suggest that loss of Rb does not cause a general increase in Apc/β-catenin/Myc signaling in villi.
Figure 8. Myc regulates E2f3a expression in Rb deficient villi.
(a) IHC staining of β-catenin. (b) IHC staining of Myc. Data in a and b are representative images from n=3 mice for each genetic group. (c) RT-qPCR analysis for E2f3a and E2f3b in control, Rb KO and Rb/Myc DKO villi. Expression levels for individual mice are plotted (4 per genetic group) and error bars represent mean ± s.d. from n=3 technical replicates. (d) IHC staining of E2f3a in control, Rb KO, Rb/Myc DKO and Rb/E2f QKO samples. Note the non-specific staining of blood cells in the lumen of the villi. Data are representative images from n=3 mice per genetic group. (e) ChIP-PCR analysis showing E2f3 loading to target genes (Pbk and Rrm1) in control, Rb KO and Rb/Myc DKO villi. n=4 mice per genetic group. The 5′ region ~1kb away from TSS of Gapdh was used as the negative control. *P<0.05, one-tailed Student’s t-test. (f) ChIP-exo-seq tracks showing E2f3 and Myc occupancy on the E2f3 locus. Distinct promoter regions for E2f3a and E2f3b are shaded in red and yellow, respectively. Data were collected from pooled crypts (n=32 mice for control, n=27 mice for Rb KO) or villi (n=7 mice for control, n=7 mice for Rb KO). (g) RT-qPCR analysis for E2f1 and E2f2 in control and Rb KO villi. Expression levels for individual mice are plotted (4 for each genetic group) and error bars represent mean ± s.d. from n=3 technical replicates. (h) Diagrams summarizing the regulation of cell cycles by Myc and E2f1-3 in wild type and Rb deficient cells. Scale bars represent 25 μm (a) and 50 μm (b, d). Scale bars in f represent 1kb.
We then considered the possibility that Myc may regulate E2f expression as previous cell culture studies have shown9, 19, 20. The E2f3 locus encodes two isoforms, E2f3a and E2f3b, driven by distinct promoters50. RT-qPCR and IHC assays showed that E2f3a (but not E2f3b) expression was markedly increased in Rb KO villi compared to wild type controls, and that this increase was suppressed by loss of Myc (Fig. 8c, d). Furthermore, E2f3-ChIP assays showed increased recruitment of E2f3 to target promoters in Rb KO villi compared to wild type controls, and that this increase was attenuated by loss of Myc (Fig. 8e).
We then evaluated Myc binding to the E2f3 locus in both crypts and villi. ChIP-exo-seq data from both control and Rb deficient crypts revealed an abundance of Myc protein residing close to the TSS of the E2f3a gene (Fig. 8f)50. Myc binding to the E2f3 locus was absent in villi. Binding of E2f3 to a region just upstream of the E2f3a TSS (Fig. 8f), which contains E2f binding elements that positively regulate its expression20, was increased in Rb KO villi compared to controls. Furthermore, the expression of E2f1 and E2f2, whose promoters are occupied by E2f3 and are also E2f auto-regulated23, was increased in Rb deficient villi (Fig. 8g and Supplementary Fig. 8b). Together, these findings suggest that Myc binding to the E2f3a promoter in Rb deficient crypts contributes to the ectopic accumulation of ‘free’ E2f3a protein in villi. We propose that Myc-mediated E2f3a accumulation engages a positive feed-back loop culminating in the execution of an E2f gene expression program that includes G1-S genes and drives unregulated cell proliferation.
DISCUSSION
Cell proliferation is orchestrated by developmental and environmental mitogenic cues that are transmitted through signaling pathways controlling key events in the cell cycle. The convergence of multiple pathways provides cells with the necessary robustness (redundancy) to withstand perturbations in any single pathway that might otherwise compromise cell cycle progression. We show here that Myc and E2f converge on the control of S-G2 in normal proliferating cells of the small intestine, but switch to the control of G1-S when Rb is inactivated (Fig. 8h). These experiments uncover a molecular mechanism to explain how normal Myc and E2f pathways may be exploited and redirected by inactivation of the Rb tumor suppressor to foster unregulated cell proliferation, and highlight the unprecedented addiction of these Rb deficient cells on Myc.
We show an unexpected collaboration between Myc and E2f in the synergistic regulation of an S-G2 transcriptional program required for the cell cycle progression of progenitor cells. The immediate consequence of ablating Myc and E2f1-3 from progenitor cells is an arrest in S and/or G2. Whether intestinal stem cells have a similar requirement for Myc and E2f1-3 requires further rigorous evaluation. The exact mechanisms that underlie the cooperative roles of Myc and E2f in regulating S-G2 transcription remain to be determined but likely involve the co-recruitment of Myc and E2f3 to target promoters and interactions with chromatin remodeling factors as shown previously in vitro51, 52.
The observation that G1-S programs are refractory to the combined loss of Myc and E2f1-3 is surprising. Recently, the combined ablation of N-Myc with E2f1-3 in mouse embryos was shown to impede DNA replication in retinal progenitor cells27. The retina is a non-regenerating tissue where cell proliferation is restricted to a narrow developmental window late in embryogenesis. The caveat in studying non-regenerating tissues is that in the absence of a continuous replenishing pool of progenitor cells, an arrest at any cell cycle stage would exhibit reduced expression of G1-S target genes and score negative by assays that measure DNA replication (i.e. BrdU), precluding an understanding of the underlying nature of the arrest. Thus, it remains possible that Myc and E2f may also converge to regulate S-G2 in the mouse retina as we show here in the small intestine. While G1-S transcription is insensitive to the combined loss of Myc and E2f1-3, we show that Myc and E2f do indeed bind to G1-S related targets. Thus, it is possible and perhaps likely that Myc and E2f may contribute to G1-S transcriptional control, but only in collaboration with additional transcriptional inputs yet-to-be identified53. Despite extensive functional redundancy, the Myc and E2f pathways also appear to have unique roles. For example, we show here that E2f1-3 helps maintain genomic integrity, whereas Myc supports the necessary biosynthetic capacity to fuel robust cell proliferation33.
In contrast to normal cells, ectopic cell cycle progression of Rb deficient intestinal cells is acutely dependent on Myc and E2f1-3. The analysis described here provides molecular insights into how loss of Rb subverts the existing Myc transcription machinery to support unregulated proliferation. While physiological levels of Myc in normal crypts utilize a variety of non-canonical DNA binding sequences54, loss of Rb in crypts leads to the recruitment of Myc to target genes through the classical E-box sequence motif, as recently described for over-expressed or amplified Myc55–58. The mechanisms for how this shift to the use of E-box DNA binding elements in crypts is established and how this might impact transcription in villi remain to be determined. It is possible that loss of Rb results in a global reconfiguration of chromatin59–62, which exposes cryptic DNA elements that enhance Myc binding and activity. Myc may contribute to the transcriptional output in Rb deficient villi in two ways that are not mutually exclusive. On one hand, Myc binding to regulatory sequences of E2f3a in crypts may facilitate its expression, which in the absence of Rb-mediated repression, would lead to the accumulation of ‘free’ E2f3a protein. This event would then engage a positive feed-forward loop to activate expression of all three E2f activators and increase total E2f transcriptional activity. In this view, Myc in crypts indirectly participates to amplify the output of the E2f program. On the other hand, the redirecting of Myc binding to E-box elements in Rb deficient crypts may impose lasting chromatin structural changes that facilitate increased binding of E2f and possibly other transcription factors to their target sites, even in subsequent cell cycles. This may provide an additional molecular mechanism for how Rb’s function in crypts is essential to suppress ectopic proliferation in villi.
Loss of Rb also subverts the existing E2f transcription machinery to support unregulated proliferation. ChIP-exo-seq analysis shows that Rb deficiency results in the accumulation and recruitment of E2f3 in villi to some of the same chromatin positions it normally occupies in control crypts, but also to new chromatin locations not normally used in proliferating crypts. Binding of E2f3 to chromatin, like that of other transcription factors, is a dynamic process that is continuously in flux due to on-off binding kinetics. In addition, E2f3 protein acutely decreases in late S-G2 and must be remade and reloaded on chromatin at the next G1-S. The broad ectopic redeployment of E2f3 to ‘old’ and ‘new’ chromatin locations in Rb deficient villi culminates in the precocious expression of a transcriptional program that drives unregulated proliferation. In summary, we show that Myc conspires with inactivation of Rb to regulate and redeploy E2f3 across the entire genome, providing a molecular mechanism that explains how Rb deficient cells rely on or become addicted to Myc.
METHODS
Mice
Mouse usage has been approved by Institutional Animal Care and Use Committee at the Ohio State University. Mouse strains used in this study (Ah-cre, RbloxP/loxP, E2f1−/−, E2f2−/−, E2f3loxP/loxP and MycloxP/loxP) have been described before28, 63 and maintained in a mixed background (C57BL/6 × 129 × FVB/N). Primers used for genotyping the mice are listed in Supplementary Table 11. Both male and female mice were used in the studies. Within each mutant genetic group, the animals were randomly utilized. Litter-mates with appropriate genotypes were utilized as control group. The number of mice used for analysis is described in the legends and no statistical method was used to predetermine sample size. The Investigators were blinded to group allocation during experiments and outcome assessment. No criteria were used to exclude samples or animals from the analysis.
β-naphthoflavone, BrdU and EdU injection
To induce Ah-cre expression, 80mg kg−1 body weight of β-naphthoflavone (Sigma-Aldrich; N3633) dissolved in corn oil (Sigma-Aldrich; C8267) was administrated into 2-month old mice with 5 injections within 30 hours. For DNA synthesis detection, 100mg kg−1 body weight of BrdU (Sigma-Aldrich; B5002) or 5mg kg−1 body weight of EdU (Life Technologies; C10337) dissolved in sterile phosphate-buffered saline (PBS) was intraperitoneally injected 2 hours or 1 hour before the mice were sacrificed, respectively. EdU staining was performed following manufacturer’s protocol (Life Technologies; C10337).
Tissue preparation and histology
The tissue used for RNA and histology was collected from proximal 10cm intestine. For chromatin immunoprecipitation (ChIP), the intestine was divided into three parts of equal length and the first 1/3 proximal to stomach was used. Intestinal tissues were divided into ~1cm pieces and fixed with 10% pH-buffered formalin (Fisher Scientific; 23-245-685) for 48h at room temperature, embedded in paraffin and cut into 4μm sections for histological staining. H&E images were representative of n=3 or 4 animals per genetic group with minimal 3 fields per animal.
Isolation of villi and crypts
Isolation of villi and crypts were processed similarly as previous described64. Immediately after euthanizing the mice, the small intestine was dissected with all mesentery and adipose tissue removed, cut longitudinally and gently washed in PBS. The cleaned tissue was incubated in 25mL PBS with 0.5mM EDTA and 1mM dithiothreitol (DTT) for 30min at room temperature. Villus fractions were shaken off from the intestine sequentially in multiple tubes containing 10mL ice-cold PBS with 1mM DTT. The fractions were incubated on ice for 5min to allow spontaneous precipitation of intact villi by gravity. The supernatant containing broken pieces of villi was removed. After most villi were collected, the remaining intestinal tissue was incubated in 25mL PBS with 0.9mM EDTA and 1mM DTT for another 30min at room temperature. To remove residual amount of villi and collect crypts, the tissue was further shaken sequentially in multiple tubes containing 10mL ice-cold PBS with 1mM DTT until complete dissociation of epithelial cells from the mesenchyme/muscle tissues. Crypt-enriched fractions were filtered with 70μm cell strainer (Fisher Scientific; 22363548) to minimize the potential contamination from villus fractions and increase the yields of crypts (more available crypt factions after the removal of mixed villi). Each fraction was examined microscopically to ensure the purity of enriched villi/crypts for further experiments. After brief centrifuge, the cell pellet was washed in ice-cold PBS and frozen with liquid nitrogen or processed further for ChIP assays.
RNA and quantitative PCR
Total RNA from purified villi/crypts was isolated using TRIzol reagent (Invitrogen; 15596-018) and further cleaned up with RNeasy Mini Kit (Qiagen). First strand cDNA was synthesized using 100ng total RNA with Superscipt III Reverse Transcriptase Kit (Invitrogen; 18080-044) following manufacturer’s protocol. Quantitative PCR (qPCR) was performed using SYBR Green master mix (Bio-Rad; 170-8884) with Bio-Rad iQ5 Multicolor Real-Time PCR Detection System or Applied Biosystems StepOnePlus Real-Time PCR System. Samples were analyzed with triplicates and the relative expression levels of genes were calculated with 2−ΔΔCt method. Gapdh mRNA level was used for normalization. Primers used for qPCR are listed in Supplementary Table 11.
Affymetrix microarray analysis
Affymetrix microarray analysis was performed at the Ohio State University Shared Resources (http://cancer.osu.edu/research/cancerresearch/sharedresources/microarray) with Affymetrix Mouse 430 2.0 Array. Four or five independent samples of each genetic group were utilized for analysis. The p-value for expression level changes was calculated using empirical Bayes method or Student’s t-test (t.test(val1,val2)$p.value under R). The Gene Ontology analysis was performed by DAVID Bioinformatics Resources (Database for Annotation, Visualization and Integrated Discovery) at National Institute of Allergy and Infectious Diseases (NIAID), NIH. The degree of rescue (D) for mRNA level of a given gene in the waterfall plot in Figure 6b is defined as:
A: mRNA level fold change between Rb KO and control;
B: mRNA level fold change between Rb/E2f QKO (or Rb/Myc DKO) and control.
Immuno-staining
After deparaffinization, antigen retrieval was performed with boiling citrate buffer (DAKO; S169984) for 30min except that proteinase K was used for BrdU and lysozyme. Specifically, 2N HCl was further applied to tissue sections for exposing and detecting BrdU and then neutralized with 0.1M sodium tetraborate (pH 8.0). For immunohistochemistry staining, the endogenous peroxidase was inactivated by incubation with 3% H2O2 for 15min at room temperature. Normal horse serum was applied as the blocking reagent except that Mouse on Mouse (M.O.M.) Blocking Reagent (Vector Labs; MKB-2213) was applied for primary antibodies raised in mice. Antibodies and dilutions used in this study were as follows: BrdU (Dako; M0744, 1:50), geminin (Santa Cruz; sc-13015, 1:100), Serine 10-phosphorylated histone 3 (Millipore; 06-570, 1:250), cleaved caspase-3 (Cell Signaling; 9661, 1:100), phosphorylated H2AX (Cell Signaling; 9718, 1:100), Ccna2 (Santa Cruz; sc-596, 1:250), Cdc2 (Santa Cruz; sc-54, 1:100), Mcm3 (Santa Cruz; sc-9850, 1:500), Pcna (Santa Cruz; sc-56, 1:500), lysozyme (DAKO; A0099, 1:100), Myc (Santa Cruz; sc-764, 1:500), E2f3 (Millipore; 05-551, 1:100) and β-catenin (AbD Serotec; OBT1683, 1:250). Signals were visualized with biotinylated goat secondary anti-rabbit IgG antibody (Vector Labs; BA-1000, 1:250) and DAB substrate kit (Vector Labs; SK-4100), donkey anti-mouse Alexa Fluor 488 or Alexa Fluor 594, donkey anti-goat Alexa Fluor 594, donkey anti-rabbit Alexa Fluor 594 (Invitrogen, 1:250). TSA™ Plus Fluorescein kit (Perkin Elmer; NEL741001KT) was utilized for visualizing geminin signals. The nuclei were counterstained with haematoxylin or 4′,6-diamidino-2-phenylindole (DAPI). The images were collected using Nikon Eclipse 50i (immunohistochemistry) or Zeiss Axioskop 40 (immunofluorescence) microscopes with a Zeiss AxioCam HRc camera and AxioVision software. IHC and IF images were representative of n=3 or 4 animals per genetic group with minimal 3 fields per animal, except n=2 animals per genetic group were utilized for Supplementary Figure 2a.
ChIP-PCR
Freshly isolated crypts/villi were incubated in PBS with 1% formaldehyde at 37°C for 15min. The DNA-protein crosslink reaction was terminated by incubation with 0.125M glycine for 5min. The sample was washed in ice-cold PBS, resuspended in cytosolic lysis buffer (5mM PIPES (pH 8.0), 85mM KCl, 0.5% NP-40 and proteinase inhibitors) and nuclear lysis buffer (50mM Tris (pH 8.0), 10mM EDTA, 1% SDS and proteinase inhibitors). The chromatin was sonicated with majority of DNA fragments between 100-300bp. The fragmented chromatin was diluted 1:10 with IP dilution buffer (16.7mM Tris (pH 8.0), 167mM NaCl, 1.2mM EDTA, 1.1% Triton X-100 and proteinase inhibitors) and pre-cleared by incubating with Protein G Plus/Protein A beads (Calbiochem; IP05) at 4°C for 1 hour on rotator prior to adding antibodies. Antibodies used were E2f3 (Santa Cruz; sc-878 X), Myc (Santa Cruz; sc-764 X) and normal rabbit IgG (Santa Cruz; sc-2027). The DNA-protein-antibody-bead complex was washed twice with each following buffer in the presence of proteinase inhibitors: low-salt buffer (20mM Tris (pH 8.0), 2mM EDTA, 1% Triton X-100, 0.1% SDS and 150mM NaCl), high-salt buffer (20mM Tris (pH 8.0), 2mM EDTA, 1% Triton X-100, 0.1% SDS and 500mM NaCl), LiCl buffer (10mM Tris (pH 8.0), 1mM EDTA, 250mM LiCl, 1% NP-40 and 1% deoxycholic acid) and Tris-EDTA buffer (pH 8.0). Immunoprecipitated chromatin was retrieved from Protein G Plus/Protein A beads in 1% SDS plus 0.1M NaHCO3 and reverse-crosslinked with 0.5M NaCl and 0.1mg/mL RNase A overnight at 65°C (including non-immunoprecipitated inputs). The product was further treated with 0.1mg/mL Proteinase K and the DNA was purified with Qiagen kits (QIAquick Purification Kit (Qiagen; 28104) for DNA detected by quantitative PCR. Primers used for ChIP quantitative PCR are listed in Supplementary Table 11. One-tailed Student’s t-test was performed to determine whether E2f3 binding to target genes was decreased in Rb/Myc DKO villi compared to Rb KO villi in Figure 8e.
ChIP-exo-seq library construction and Illumina sequencing
Crypts from 32 control mice and 27 Rb KO mice and villi from 7 control mice and 7 Rb KO mice were utilized for ChIP-exo-seq. Crypts collected from every 4–5 mice of the same genotype were combined and processed as one sample. The crosslink, cytosolic lysis, nuclear lysis, sonication, dilution with IP dilution buffer, pre-clean and immunoprecipitation steps were performed as described above except Pierce Protein A/G Magnetic Beads (Thermo Scientific; 88802) were used. Before immunoprecipitation, the pooled chromatin derived from each genetic/tissue compartment was divided into two halves for E2f3- and Myc-ChIP, respectively. The library construction steps including all on-bead enzymatic reactions (end polishing, P7 exo-adapter ligation, nick repair, λ-exonuclease digestion, RecJf exonuclease digestion), elution and reverse cross-linking, primer extension and P5 exo-adaptor ligation were performed as described39. The resulting DNA was enriched by 13 cycles of PCR using NEBNext High-Fidelity PCR Master Mix (NEB; M0541S) before measuring the library concentration and DNA size distribution. Cluster generation on Illumina cBot and single-end high throughput sequencing on Illumina HiSeq 2500 platform were performed at the Ohio State University Shared Resources.
ChIP-exo-seq data analysis
The 50bp sequencing reads were de-multiplexed based on the barcodes in index adaptors utilized in the library construction, followed by mapping the reads to reference mouse genome (GRCm37/mm9) with Bowtie2 aligner (version 2.2.1)65. Only the reads uniquely mapped to the genome were utilized for further analysis. E2f3 and Myc DNA binding peaks were identified using Genome wide Event finding and Motif discovery algorithm GEM (version 2.4.1)66 with a false discovery rate (FDR) less than 1% and Model-based Analysis of ChIP-Seq algorithm MACS2 (version 2.0.10)67 with a FDR less than 5%. The intersection of GEM and MACS2 results (when the distance between corresponding summit coordinates predicted by these two algorithms was less than 1kb) were used for all downstream analysis. The DNA binding peaks were associated to genes based on the minimal distance between peak summits and proximal transcription start sites. Motif discovery and matching de novo to known motifs were performed with HOMER algorithm (version 4.6)68 using DNA sequences around peak summits (±50bp). Integrative Genomics Viewer algorithm IGV69 was used to visualize the binding events and reads coverage. The BED files with detailed information for identified peaks have been deposited in GEO database with accession number GSE56009. The computer code for ChIP-exo-seq analysis can be accessed upon request.
Flow cytometry
Isolated crypts were homogenized using a Dounce homogenizer in ice-cold suspension buffer (25mM Tris (pH 7.5), 50mM KCl, 2mM MgCl2, 1mM EDTA and 1mM phenylmethylsulfonyl fluoride (PMSF)). After brief centrifuge, the pellet was washed twice with the suspension buffer and once with PBS. Single cell suspension was then prepared in PBS containing 0.1% Triton X-100, 25μg/mL propidium iodide (Roche; 11348639001) and 20μg/mL RNase A (Invitrogen; 12091-021) and incubated 1 hour at room temperature at dark. The suspension was then filtered through 35μm cell strainer caps (BD Falcon; 352235). The DNA content measured by fluorescence intensity of propidium iodide was analyzed using BD LSR II Flow Cytometer at the Ohio State University Shared Resources. The data was analyzed with FlowJo cytometric analytical software.
Statistics
Student’s t-test and empirical Bayes method were performed. The data have normal distributions that meet the assumptions of the tests. Whether the variance is similar between the groups that are being statistically compared was not estimated. All data are presented as mean ± s.d.
Data deposition
Affymetrix gene expression and ChIP-exo-seq data has been deposited in GEO database with accession number GSE56009.
Supplementary Material
Acknowledgments
We thank M. L. Rawahneh, N. Lovett and J. Moffitt for assistance with histology. We also thank assistance from C. Martin for reviewing histological slides, S. Bae for advice on sequencing library construction, Dr. V. Jin, Z. Ye and J. McElroy for suggestions in the analysis of sequencing data. This work was supported by the Microarray, Nucleic Acid and Analytical Cytometry Core Shared Resources at Ohio State University. We are thankful to V. Gopalan and A. Simcox for critical suggestions. This work was funded by NIH grants to G.L. (R01CA121275 and R01HD047470) and NIH grant to J.M.P. (R01CA098956). H.L., T.P. and B.H. were recipients of Graduate, Postdoctoral and Undergraduate Pelotonia Fellowships, respectively.
Footnotes
AUTHOR CONTRIBUTIONS
H.L. and G.L. designed the experiments. H.L., P.D., B.H., S.R., N.F., A.B., R.K., C.K. and M.T.S.R. performed the experiments, collected and analyzed data. X.T., A.S., T.P., K.H., R.M. and P.C. performed bioinformatic and statistical analysis for the gene expression and ChIP-exo-seq data. Z.C. and Q.W. advised for the ChIP-exo-seq experiments. J.M.P. and G.L. supervised the study. H.L. and G.L. wrote the manuscript with inputs from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–28. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 7.Seoane J, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–8. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 8.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–9. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 9.Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–6. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard C, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–33. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermeking H, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97:2229–34. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 13.Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–10. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 14.Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–8. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 15.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 19.Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–35. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams MR, Sears R, Nuckolls F, Leone G, Nevins JR. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000;20:3633–9. doi: 10.1128/mcb.20.10.3633-3639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 22.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–26. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 23.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettess MD, et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–78. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, et al. Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 2006;66:5608–12. doi: 10.1158/0008-5472.CAN-05-4242. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, et al. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462:925–9. doi: 10.1038/nature08544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong JL, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–4. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wenzel PL, et al. Cell proliferation in the absence of E2F1-3. Dev Biol. 2011;351:35–45. doi: 10.1016/j.ydbio.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malynn BA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–9. [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SY, et al. Mouse development with a single E2F activator. Nature. 2008;454:1137–41. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ireland H, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–46. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Muncan V, et al. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26:8418–26. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–9. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 35.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–53. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 36.Ireland H, Houghton C, Howard L, Winton DJ. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev Dyn. 2005;233:1332–6. doi: 10.1002/dvdy.20446. [DOI] [PubMed] [Google Scholar]
- 37.Phesse TJ, et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21:956–66. doi: 10.1038/cdd.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serandour AA, Brown GD, Cohen JD, Carroll JS. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauthier NP, et al. Cyclebase.org--a comprehensive multi-organism online database of cell-cycle experiments. Nucleic Acids Res. 2008;36:D854–9. doi: 10.1093/nar/gkm729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Yu Q, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 44.Rustgi AK, Dyson N, Bernards R. Amino-terminal domains of c-myc and N-myc proteins mediate binding to the retinoblastoma gene product. Nature. 1991;352:541–4. doi: 10.1038/352541a0. [DOI] [PubMed] [Google Scholar]
- 45.Goodrich DW, Lee WH. Abrogation by c-myc of G1 phase arrest induced by RB protein but not by p53. Nature. 1992;360:177–9. doi: 10.1038/360177a0. [DOI] [PubMed] [Google Scholar]
- 46.Heinen CD, et al. The APC tumor suppressor controls entry into S-phase through its ability to regulate the cyclin D/RB pathway. Gastroenterology. 2002;123:751–63. doi: 10.1053/gast.2002.35382. [DOI] [PubMed] [Google Scholar]
- 47.Morris EJ, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–6. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson G, et al. Cell cycle control of wnt receptor activation. Dev Cell. 2009;17:788–99. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Cole AM, et al. Cyclin D2-cyclin-dependent kinase 4/6 is required for efficient proliferation and tumorigenesis following Apc loss. Cancer Res. 2010;70:8149–58. doi: 10.1158/0008-5472.CAN-10-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leone G, et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–32. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 52.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–6. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 53.Yang ZF, Mott S, Rosmarin AG. The Ets transcription factor GABP is required for cell-cycle progression. Nat Cell Biol. 2007;9:339–46. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 54.Uribesalgo I, et al. E-box-independent regulation of transcription and differentiation by MYC. Nat Cell Biol. 2011;13:1443–9. doi: 10.1038/ncb2355. [DOI] [PubMed] [Google Scholar]
- 55.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabo A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–7. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–5. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 60.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 61.Siddiqui H, Fox SR, Gunawardena RW, Knudsen ES. Loss of RB compromises specific heterochromatin modifications and modulates HP1alpha dynamics. J Cell Physiol. 2007;211:131–7. doi: 10.1002/jcp.20913. [DOI] [PubMed] [Google Scholar]
- 62.Petrella LN, et al. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development. 2011;138:1069–79. doi: 10.1242/dev.059501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trumpp A, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–73. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 64.Saenz-Robles MT, et al. Intestinal hyperplasia induced by simian virus 40 large tumor antigen requires E2F2. J Virol. 2007;81:13191–9. doi: 10.1128/JVI.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.