Abstract
Nucleocytoplasmic trafficking of proteins/RNAs is essential to normal cellular function. Indeed, accumulating evidence suggests that cancer cells escape anti-neoplastic mechanisms and benefit from pro-survival signals via the dysregulation of this system. The nuclear exporter chromosome region maintenance 1 (CRM1) protein is the only protein in the karyopherin-β protein family that contributes to the trafficking of numerous proteins and RNAs from the nucleus. It is considered to be an oncogenic, anti-apoptotic protein in transformed cells, since it reportedly functions as a gatekeeper for cell survival, including affecting p53 function, and ribosomal biogenesis. Furthermore, abnormally high expression of CRM1 is correlated with poor patient prognosis in various malignancies. Therapeutic targeting of CRM1 has emerged as a novel cancer treatment strategy, starting with a clinical trial with leptomycin B, the original specific inhibitor of CRM1, followed by development of several next-generation small molecules. KPT-330, a novel member of the CRM1-selective inhibitors of nuclear export (SINE) class of compounds, is currently undergoing clinical evaluation for the therapy of various malignancies. Results from these trials suggest that SINE compounds may be particularly useful against hematological malignancies, which often become refractory to standard chemotherapeutic agents.
Keywords: CRM1, nuclear export, SINE, hematological malignancies, KPT-330, ribosomal biogenesis
Introduction
Nucleocytoplasmic trafficking of proteins and RNAs is mediated by the nuclear exporter chromosome region maintenance 1 (CRM1, also known as exportin 1 or XPO1) protein, which has over 230 identified cargos(Xu, Grishin, & Chook, 2012). The protein plays a critical role in nuclear export of growth and survival factors in eukaryotic cells, including transformed cells. Certain natural products (e.g., leptomycin B, ratjadone, anguinomycin, and goniothalamin (Bonazzi, Guttinger, Zemp, Kutay, & Gademann, 2007; Daelemans et al., 2002; Hamamoto, Uozumi, & Beppu, 1985; Hayakawa, Sohda, Shin-Ya, Hidaka, & Seto, 1995; Kau, Way, & Silver, 2004; Komiyama et al., 1985; Kudo et al., 1999; Kudo et al., 1998; Meissner, Krause, & Vinkemeier, 2004; Mutka et al., 2009; Turner, Dawson, & Sullivan, 2012; Wach, Guttinger, Kutay, & Gademann, 2010)) are inhibitors of CRM1. This discovery led to the development of CRM1-specific small molecules (e.g., N-azolylacrylates, KOS-2464, and CBS9106) as potentially useful, anti-cancer agents (Mutka, et al., 2009; Sakakibara et al., 2011; Turner, et al., 2012). These include the potent, second-generation CRM1-selective inhibitors of nuclear export (designated as SINE) class of agents that has added further promise of this targeted strategy for the elimination of malignant cells (Etchin, Sanda, et al., 2013). Indeed, in vitro experimental scenarios have shown that the blockade of CRM1 transport by these inhibitors can induce cancer cell death, which is believed to occur by the forced nuclear retention of tumor-suppressors, transcriptional factors that are inactive in these cells due to aberrant CRM1 transport into the cytoplasm. Furthermore, treatment of various solid tumors and hematological malignancies with SINE compounds has been shown to block transformed cell proliferation and induce apoptosis in these cells in vivo (Mutka, et al., 2009; Sakakibara, et al., 2011; Turner, et al., 2012). SINE compounds apparently have limited toxicity in normal human cells, which enhances the overall therapeutic index of these agents (Etchin, Sun, et al., 2013). In particular, KPT-330, with its well-established pharmacokinetic and pharmacodynamics properties, including high oral bioavailability, is a promising SINE that has recently entered into clinical trials. In this review, we present the cellular biology associated with the nuclear export of proteins/RNAs by CRM1, and outline the preclinical and potential clinical impact of the regulation of this protein function as a candidate therapeutic target in human malignancies.
Nuclear Export and the Functions of CRM1
The nuclear envelope provides a compartmentalized intracellular environment for DNA replication, the synthesis of RNA, and production of ribosomes, and, as such, it can regulate cellular biological processes including apoptosis and proliferation. Nucleocytoplasmic trafficking of RNAs, ribosomes, regulators of transcription, and cell cycle modulators is tightly regulated by the nuclear pore complex, and by the presence of transport receptor molecules including the karyopherin-β family proteins (Turner, et al., 2012). Each karyopherin-β protein recognizes a unique group of cargo proteins or RNAs, and conveys their nucleocytoplasmic import or export. The presence of either a nuclear localization signal/nuclear export signal (NES) amino acid sequence facilitates cargo molecule recognition by the transporter. CRM1 is among seven exportins, and the only one that mediates the transport of over 230 proteins including tumor suppressors (e.g., p53, p73, and FOXO1), growth regulator/pro-inflammatory (e.g., IkB, Rb, p21, p27, BRCA1, and APC), and anti-apoptotic proteins (e.g., NPM and AP-1) (Table 1, the aforementioned proteins are part of a comprehensive list appearing on the web page: http://prodata.swmed.edu/LRNes/Academics/IndexFiles/names.php) (Kau, et al., 2004; Turner, et al., 2012; Xu, et al., 2012). CRM1 is also required for the transport of several mRNAs, proteins, and rRNAs that are essential for ribosomal biogenesis (Bai, Moore, & Laiho, 2013; Golomb et al., 2012; Tabe et al., 2013; Thomas & Kutay, 2003).
Table 1.
CRM1 cargo proteins.
| β-Arrestin-2 | CPEB3 | hRio2 | MLH1 | PAK4 | Sox10 |
| γ134.5 Protein (HSV-1) | CPEB4 | hRpf1/Nedd4 | MoKA | PAK5 | SOX9 |
| λPKC | Crk | Hsc70/Hsc54 | MondoA | Pap1 | Spc27 |
| 4E-T | Cuf1 | HSCARG | mPER1 | PARP-10 | STAT1 |
| Actinin-4 | Cyclin B1 | HsfA2/HSF30 | mPER2 | Pat1b | STAT3 |
| ADAR1 | Cyclin B1 | Hsp105 | Mst1 | Paxillin | Stau2 |
| AhR | Cyclin D1 | Hsp70-Ssb1p | Mta | PBX1 | STRADα |
| AID | DAB1 | Hst2 | MTF-1 | PCNA | survivin |
| ALX | DARPP-32 | hTERT | N protein | PDK-1 | Tax |
| AMPKα2 | Dengue Virus NS5 | Huntingtin | N-WASP | Pericentrin | Tbx5 |
| An3 | DGKζ | Hxk2 | NADE | Php4 | TcADKn |
| ANCO-1 | Dpr1 | IκBα | NANOG | PKIα | TCF11 |
| APC Protein | Dsk1 | IκBε | Nap1p | PLC-δ1 | TDP-43 |
| APOBEC1 | Dysbindin | Id1 | NC2β | PP2A B56α | TFIIIA |
| Ataxin-7 | E1B-55K | Id2 | Neurogenin 3 | PP2Acα | Tgs1 LF |
| ATF-2 | E2F-4 | IPMK | NF-ATc1 | Protein 9b | TIS11 |
| Aven | E2F1 | IRF-3 | Nibrin | Protein UL84 | Topoisomerase 2-alpha |
| Bach1 | E4-34kD | IRF-5 | Nmd3 | Rabies virus P protein | Topoisomerase II-beta |
| Beclin 1 | Early E1A 32 kDa protein | Jab1/CSN5 | Nmd3p | RanBP1 | TRIP-Br2 |
| BICP27 | EDS1 | Keap1 | NOSTRINβ | RBCK1 | Trip6 |
| BMAL1 | Eps15 | KLF5 | NPM | RelA | Tropomodulin-1 |
| Bok | ESE-1 | KLF6 | NPM mutants | Rev | UL4 |
| BPV-E1 | Exd | LANA2 | Nrf2 | Rex Protein | UL47 (HSV-1) |
| BRCA1 | FAK | LCD1 | Nrf2 | rhTRIM5alpha | UL94 |
| BRCA2 | FANCA | LEI/L-DNase II | NS2-P (MVM) | RIP3 | VDUP1 |
| BRO-a | Fbxo7 | Liar | nsP2 (VEE) | RITA | VEEV Capsid protein |
| C | FMRP | LPP | NT-PGC-1α | RoXan | VIK-1 |
| CaMKIα | Foxo3 | Ltv1 | NURR1 | Rsp5 | VP19C |
| CCTα | Foxa2 | LZTS2 | NXF3 | RSV M protein | VP3 |
| Cdc14A | Fyn | Mad1p | OREBP | SBP2 | Vpr (HIV-1) |
| Cdc14p | GRTH | MAPKAP kinase 2 (MK2) | ORF45 of KSHV | SD | WDR42A |
| Cdc25 | HBx | MAPKK1/MEK1 | ORF9 | SENP2 | Wee1 |
| Cdc7 | HDAC1 | Mcm3 | OsNMD3 | SH2-Bβ | X11L2 |
| Chibby (Cby) | HDAC4 | mDia2 | p100 | Sima | XAB1/Gpn1 |
| CHP1 | HDAC5 | Menin | p120ctn | SIRT2 | Xp54 |
| ChREBP | HDM2 | Mia1p/Alp7p | p21Cip1 | Smad1 | Yap1p |
| cIAP1 | HIV-REV | MIER1-3A | p28GANK | Smad4 | ZAP |
| COMMD1 | hMSH4 | MK5 | p37 protein of ASFV | Smurf1 | Zinc finger protein RFP |
| COP1 | hMSH5 | MKP-3 | p38 (p40) | Snail | ZO-2 |
| CPEB1 | HPV11 E1 | MKP-7 | P53 | SNUPN | Zyxin |
| HPV16 E7 | p73 |
The CRM1 protein is encoded by the XPO1 gene and was originally identified by a genetic screen of S. pombe that revealed involvement of the protein in control of chromosomal structure (Adachi & Yanagida, 1989). CRM1 was later characterized and designated as a ubiquitous nuclear export receptor protein of the karyopherin-β family, which exports the cargo proteins harboring a specific NES into the cytoplasm (Fornerod, Ohno, Yoshida, & Mattaj, 1997; Fukuda et al., 1997; Ossareh-Nazari, Bachelerie, & Dargemont, 1997). CRM1 is upregulated in a variety of solid tumor types (e.g., osteosarcomas, gliomas, and pancreatic, ovarian, cervical, and renal carcinomas) (Huang et al., 2009; Inoue et al., 2013; Noske et al., 2008; Shen et al., 2009; van der Watt et al., 2009; Yao et al., 2009), as well as in hematological malignancies (e.g., acute myeloid/lymphoid leukemia (AML/ALL), chronic myeloid/lymphoid leukemia (CML/CLL), mantle cell lymphomas (MCL), and multiple myeloma [MM]) (Etchin, Sanda, et al., 2013; Etchin, Sun, et al., 2013; Kojima et al., 2013; Lapalombella et al., 2012; Ranganathan et al., 2012; Sakakibara, et al., 2011; Schmidt et al., 2013; Tai et al., 2014; Walker et al., 2013; Yoshimura et al., 2014; Zhang et al., 2013). In fact, the overexpression of CRM1 is positively correlated with poor prognosis in these malignancies (Huang, et al., 2009; Kojima, et al., 2013; Noske, et al., 2008; Shen, et al., 2009; Tai, et al., 2014; Yao, et al., 2009; Yoshimura, et al., 2014). Therefore, it has been suggested that alterations in nucleocytoplasmic trafficking, and hence the aberrant cytoplasmic localization of tumor suppressor proteins, cell cycle regulators, and/or pro-apoptotic proteins, as well as the deregulation of ribosomal biogenesis, is associated with oncogenesis and resistance to chemotherapy.
For CRM1-substrate binding to occur, the cargo molecule must have a leucine-rich NES (Fukuda, et al., 1997; Wen, Meinkoth, Tsien, & Taylor, 1995). The NES for CRM1 contains hydrophobic amino acids, including isoleucine, leucine, methionine, phenylalanine, and valine (Kutay & Guttinger, 2005). A consensus motif for this NES is comprised of 10 to 15 amino acid residues containing several spaced hydrophobic amino acids. These can be ordered as HX2–3HX2–3HXH. In this designation, H is a hydrophobic amino acid (i.e., isoleucine, leucine, methionine, phenylalanine, or valine), X is any amino acid, and the subscripts indicate the potential number of repeats (Turner, et al., 2012). Together, these amino acids form an alpha-helix-loop and/or all loop structure that can bind the hydrophobic groove of CRM1 (Dong, Biswas, & Chook, 2009; Dong et al., 2009).
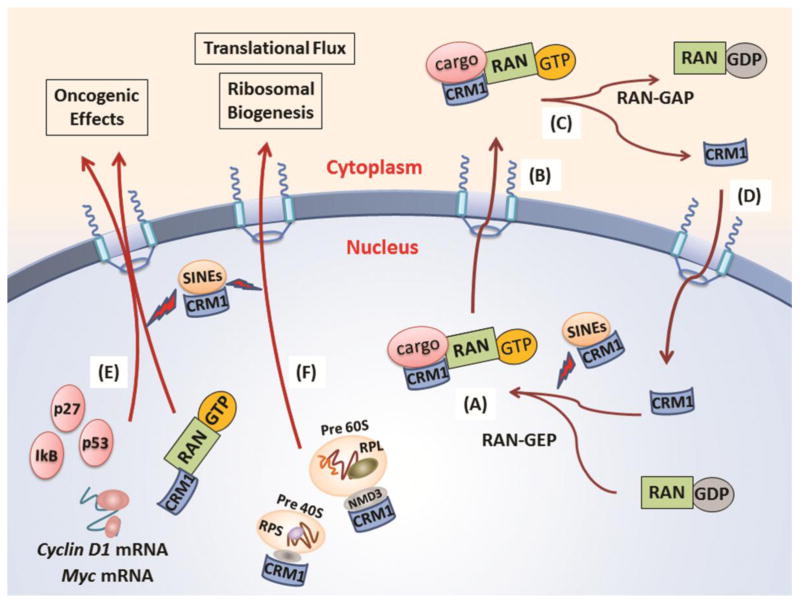
Three-dimensional conformational changes in the cargo protein’s NES, caused by protein phosphorylation, dephosphorylation, or mutation, can regulate CRM1 binding (Craig, Zhang, Davies, & Kalpana, 2002; Vogt, Jiang, & Aoki, 2005). Additional protein modifications such as sumoylation, ubiquitination, acetylation, and/or the binding of protein-specific co-factors can also influence the affinity of the NES for CRM1 binding (Turner, et al., 2012). When CRM1 is bound, it forms a trimer with RanGTP in the nucleus, and exits through the nuclear pore complex into the cytoplasm (Fig. 1A and B). The binding of CRM1 to either RanGTP or to the NES of a cargo protein itself is weak. However, when both RanGTP and the cargo bind to CRM1 simultaneously, affinity to both cargo and Ran GTP is strengthened by 500- to 1000-fold (Dong, Biswas, & Chook, 2009; Dong, Biswas, Suel, et al., 2009; Monecke et al., 2009). In the cytoplasm, the hydrolysis of GTP to GDP by the RanGTP-activating protein promotes the release of the substrate from CRM1 (Kau, et al., 2004) (Fig. 1C). Crystallography has illustrated that this increase in CRM1 affinity is caused by a change in the global conformation of the protein. This change in conformation occurs only when both the cargo substrate and RanGTP bind to CRM1 (Dong, Biswas, & Chook, 2009; Dong, Biswas, Suel, et al., 2009; Monecke, et al., 2009). Ran-GTP and CRM1 are subsequently returned/recycled through the nuclear pore complex back into the nucleus (Turner, et al., 2012) (Fig. 1D).
Figure 1. Pathophysiological functions of CRM1 and inhibition of its function by CRM1 inhibitors.
CRM1 plays its role as nuclear exporter through RanGTP dependent mechanism. A cargo with a leucine-rich nuclear export signal (NES) can bind the hydrophobic groove of CRM1, and once a trimer with RanGTP with these molecules formed in the nucleus (A), these complex exits through nuclear pore complex into the cytoplasm (B). In the cytoplasm, RanGTP-activating protein induces hydrolysis of GTP to GDP, and the cargo is released from CRM1 (C). RanGDP and CRM1 are subsequently recycled through the nuclear pore complex back into the nucleus (D). Over 230 proteins were found as cargo of CRM1 so far, including p53, p27, IkB and so on (E). The 40S and 60S ribosomal subunits are also exported to the cytoplasm in a CRM1 dependent manner, with an adaptor protein NMD3 (F).
Accumulating evidence suggests that CRM1 plays an important role in ribosomal biogenesis. Ribosomal biogenesis involves multiple, coordinated steps including the synthesis and processing of ribosomal RNA (rRNA), the synthesis of ribosomal proteins and their import into the nucleus, the assembly of ribosomal subunits, and the transport of the mature 40S (small subunit) and 60S (large subunit) subunits into the cytoplasm. The first step in rRNA biogenesis is transcription of 47S rRNA precursor by RNA polymerase I, followed by extensive processing, modification and maturation to 18S and 28S rRNAs (Fatica & Tollervey, 2002; Granneman & Baserga, 2005). During synthesis, the rRNAs are assembled into 90S ribosome precursors and processed further to 40S and 60S pre-ribosomes (Granneman & Baserga, 2005).
Nuclear export of the 40S and 60S ribosomal subunits is dependent on CRM1 in eukaryotic cells, and mediated by the binding of the 60S ribosomal export protein NMD3 adaptor protein in RanGTP-dependent manner (Thomas & Kutay, 2003) (Figure 1F). The CRM1-NMD3 complex has been detected in nucleoli, which are membraneless nuclear structures involved in ribosome biogenesis (Bai, et al., 2013). Nucleolar localization of the CRM1-NMD3 complex is induced by inhibition of RNA polymerase I transcription with actinomycin D or by inhibiting RNA polymerase catalytic activity with siRNA (Bai, et al., 2013). On the other hand, leptomycin B, thus CRM1 inhibition, blocked the processing of 28S rRNA. Furthermore, synthesis of pre-47S rRNA was inhibited by the depletion of NMD3 and inhibition of CRM1 (Bai, et al., 2013). However, nucleolar disintegration, a hallmark of RNA polymerase I transcriptional stress (Raska, Koberna, Malinsky, Fidlerova, & Masata, 2004) and pyrimidine depletion (Khutornenko et al., 2010; Linke, Clarkin, Di Leonardo, Tsou, & Wahl, 1996), was not observed by inhibition of NMD3 and CRM1. This would suggest that the CRM1-NMD3 complex functions in pathways of rRNA synthesis and processing, probably by adjusting the rRNA synthesis rate through rRNA processing and export, not by regulating transcription.
It has also been reported that CRM1, as well as other nuclear import receptors like importin-7, is regulated positively by MYC and negatively by p53 (Golomb, et al., 2012), resulting in the modulation of ribosomal biogenesis. This is not surprising, since MYC is a known master regulator of ribosome biogenesis. As such, it can coordinate protein synthesis through the transcriptional control of RNA and the protein components of ribosomes, the gene products required for the processing of ribosomal RNA, the nuclear export of ribosomal subunits, and the initiation of mRNA translation (van Riggelen, Yetil, & Felsher, 2010). MYC seems to transcriptionally upregulate XPO1 as part of a broad range of transcriptional program that also includes numerous ribosomal protein genes (Wu et al., 2008). The burden on the nuclear transport machinery is supposed to be increased once ribosomal protein levels become higher by MYC, so the dual induction of importin-7 and CRM1 presumably plays a pivotal role for a cell to overcome this burden.
Nucleophosmin (NPM1), which also plays a role on the nuclear export of ribosomal proteins and subunits (Maggi et al., 2008), is also a relevant MYC target, presumably through an interaction with CRM1. Conversely, it has been recently reported that specific SINE compounds reduce the levels of MYC in MM cells (Tai, et al., 2014) and MCL (Tabe, et al., 2013). Because the nuclear export of Myc mRNA is mediated by CRM1(Culjkovic, Topisirovic, Skrabanek, Ruiz-Gutierrez, & Borden, 2006), it is certainly possible that CRM1 and MYC maintain their interrelationships in ribosomal biogenesis.
Tabe and Kojima, et al. (Tabe, et al., 2013; Yoshimura, et al., 2014) have very recently analyzed the pro-survival pathways involved in CRM1-dependent nuclear export in MCL cells, using the isobaric tags for relative and absolute quantification (iTRAQ) with two-dimensional-liquid chromatography-tandem mass spectrometry. iTRAQ proteomics of two MCL cell lines revealed that 75 proteins were consistently altered after KPT-185 treatment. Notably, 81% (i.e., 50/62) of the downregulated proteins were ribosomal proteins consisting of both 60S and 40S subunits. A marked suppression of the eukaryotic translation initiation factor 4A1 and eukaryotic elongation factor 2 in these cells suggested that KPT-185 inhibited the CRM1-dependent nuclear export of ribosomal subunits, which led to a defect of ribosomal biogenesis. Recently, the coordination between the net translational activity of ribosomal biogenesis and the transcriptional regulation of heat shock factor 1 has been reported (Santagata et al., 2013). In fact, KPT-185 induced a decrease of heat shock factor 1 targets (e.g., HSP70, FASN, phosphor-HSP90, and EEF1A1), and increased the levels of phosphor-hnRNP D presumably due to the inhibition of translational activity resulting from a decrease in ribosomal protein or RNA. These results indicate that CRM1 may also affect the transcriptional processes that are critical for cellular metabolism and survival (Kohler & Hurt, 2007; Tabe, et al., 2013).
Inhibitors of the CRM1 Protein
Overexpression of CRM1 and its correlation with negative clinical outcomes in various malignancies has recently been reported, and, given this association, the protein is predicted to be a promising therapeutic target in oncology (Huang, et al., 2009; Noske, et al., 2008; Ranganathan, et al., 2012; Shen, et al., 2009; Yao, et al., 2009; Yoshimura, et al., 2014). Indeed, it has been shown that blocking CRM1-mediated nuclear export of any, or all, of these proteins by siRNA or SINE compounds activates apoptotic pathways and increases tumor cell sensitivity to chemotherapeutic drugs such as doxorubicin (Turner et al., 2009), etoposide (Turner, et al., 2009), cisplatin (Takenaka et al., 2004), and imatinib mesylate (Aloisi et al., 2006; Vigneri & Wang, 2001). Based on the structure-function relationship seen in the natural product CRM1 inhibitors leptomycin B, ratjadone (Meissner, et al., 2004), anguinomycin (Bonazzi, et al., 2007), and goniothalamin (Wach, et al., 2010), recently-developed small molecule inhibitors of CRM1, such as N-azolylacrylates (Daelemans, et al., 2002; Van Neck et al., 2008), KOS-2464 (Mutka, et al., 2009), and CBS9106 (Sakakibara, et al., 2011) have clearly demonstrated the requirement of CRM1 nuclear export activity for the growth and survival of cancer cells. The following sections will discuss these agents in detail.
Leptomycin B
Leptomycin B is reportedly the first, and perhaps the most potent (i.e., it is effective at nanomolar concentrations), CRM1 antagonist (Hamamoto, et al., 1985; Komiyama, et al., 1985; Kudo, et al., 1999; Kudo, et al., 1998). Leptomycin B modifies CRM1 by a Michael-type covalent addition reaction at the reactive site (i.e., cysteine 528). In vitro studies have shown that leptomycin B induces acute cytotoxicity at concentration less than 5 μM for 1 h (Mutka, et al., 2009), and the agent was also active in preclinical solid and hematologic tumor models (Mutka, et al., 2009; Shao et al., 2011; Turner, et al., 2009). One of the potential clinical problems associated with leptomycin B that were discovered in preclinical tumor models was its low therapeutic index. Indeed, in mouse studies, the agent induced dose-limiting, adverse gastrointestinal side effects (Newlands, Rustin, & Brampton, 1996).
Nevertheless, leptomycin B entered into a phase I study where the agent produced a similar profound dose-limiting toxicity (Newlands, et al., 1996). In this trial, the target validation of CRM1 inhibition and the etiology of the side effects were not adequately investigated. Even though it was clinically unsuccessful, the preclinical data regarding cytotoxicity to tumor cells promoted the development of new derivatives of leptomycin B that were perhaps more effective as anti-tumor agents, with reduced adverse, off-target effects. Synthetic derivatives of leptomycin B, with improved pharmacologic properties, were shown to have little or no adverse effects in mice. This discovery suggested that at least some of the leptomycin B toxicities were not mediated by its inhibition of CRM1 (Mutka, et al., 2009).
Ratjadone Analogs
Ratjadone was isolated from the myxobacterium Sorangium cellulosum. Ratjadone and its analogs have a chemical structure similar to leptomycin B and identical molecular mechanism of modifying CRM1 at the reactive cysteine 528 site (Meissner, et al., 2004). It has been reported that ratjadone exerts anti-proliferative effects in several human cancer cell lines, via the induction of G1 phase arrest (Burzlaff, Kalesse, Kasper, & Scheper, 2003). The study using the synthetic ratjadone analog C have revealed that this compound sensitizes drug-resistant human myeloma cells to the topoisomerase II inhibitors doxorubicin and etoposide at nanomolar concentrations, by inhibiting nuclear export of topoisomerase IIα (Turner, et al., 2009).
Anguinomycin
The anguinomycins belong to the leptomycin family of natural products and are known as potent antitumor agents that are active at picomolar concentrations (Hayakawa, et al., 1995). Anguinomycin C and D have been reported to be cytotoxic selectively against transformed cell lines, while only inducing cytostatic effects in normal cells. Immunostaining of the Rio2 kinase in HeLa cells showed that anguinomycin C and D inhibit nucleocytoplasmic transport at a concentration of less than 10nM (Bonazzi, et al., 2007; Gademann, 2011). This indicates that these compounds are able to block the CRM1 mediated nuclear export at levels comparable to leptomycin B itself.
Goniothalamin
Goniothalamin is the parent member of a class of styryl lactones that are extracted from different species of plants of the genus Goniothalamus (Gademann, 2011; Jewers, Burbage, Blunden, & Griffin, 1974). In an in vivo nuclear export assay, the immunostaining of Rio2 in HeLa cells demonstrated that goniothalamin is an inhibitor of nucleocytoplasmic transport above 500 nM. The compound also displayed cytotoxicity in breast cancer cells with an IC50 of ~ 1.5 μM (Wach, et al., 2010).
KOS-2464
KOS-2464 is perhaps the most effective analogue among the synthetic leptomycin B derivatives. It has been reported to induce rapid and prolonged inhibition of CRM1-mediated nuclear export and to cause apoptosis at nanomolar concentrations in vitro (Mutka, et al., 2009; Turner, et al., 2012). It also induced cell cycle arrest, but no apoptosis, in normal lung fibroblasts (Mutka, et al., 2009). In vivo, KOS-2464 was reported to have fewer off-target adverse side effects than leptomycin B, while retaining an equivalent anti-tumor cell activity. Indeed, KOS-2464 decreased tumor burden and/or induced growth inhibition in all of the mouse cancer xenograft models tested, including colon, cervical, lung, skin, and CML (Mutka, et al., 2009).
N-azolylacrylate Analogs
N-azolylacrylate analogs are CRM1 inhibitors that were developed to inhibit the nuclear export of the HIV-1 Rev protein (Turner, et al., 2012; Van Neck, et al., 2008). Specifically, the N-azolylacrylate small molecule inhibitor designated PKF050-683 is a reversible and highly specific inhibitor of CRM1 developed for use in HIV therapy (Daelemans, et al., 2002). PKF050-638 also exhibited strict molecular structural requirements, since a trans-enantiomer of the molecule was completely inactive. There are no reports of PKF050-683 being used for cancer treatment.
CBS9106
CBS9106 is a reversible inhibitor of CRM1 that reportedly causes the degradation of CRM1 (Sakakibara, et al., 2011). CBS9106 shows antitumor activity in bladder, breast, colon, CNS, endocrine, lung, kidney, pancreatic, prostate, skin, and hematopoietic malignancies both in vitro and in in vivo (Sakakibara, et al., 2011; Turner, et al., 2012). The covalent CBS9106 binding to CRM1 is reversible, and it occurs at the cysteine 528 residue. A reduction in CRM1 protein levels was observed in tumor cells following CBS9106 treatment. Interestingly, this was not observed with leptomycin B treatment or other SINE compounds, so this effect may be specific to this compound. Nevertheless, the CRM1 degrading ability of CBS9106 shows a strong correlation with the efficacy of this agent in vitro. The CBS9106-induced CRM1 depletion and induction of apoptosis in tumor cells were also confirmed in an in vivo xenograft model using a myeloma cell line (Sakakibara, et al., 2011).
The KPT Series of CRM1 Inhibitors
The SINE series of drugs was generated based on an in silico molecular modeling strategy using a structural model of the NES groove of CRM1 (Lapalombella, et al., 2012). Using this approach, Karyopharm Therapeutics (Karyopharm Therapeutics, Inc., Boston, MA, USA) developed several trans-enantiomers proof-of-concept compounds that are water-soluble, irreversible inhibitors of CRM1. These compounds show no detectable binding to other proteins, including the cysteine proteases which are believed to be the cause of the toxicity by leptomycin B (Lapalombella, et al., 2012).
High-resolution crystal structure analysis of CRM1 bound to KPT-185, -251, -276, and -330 was used to identify the conjugation of these compounds to the cysteine residue (i.e., Cys528) in the groove of CRM1 for a cargo. For example, the 2.2-A crystal structure of KPT-251 binding with a CRM1-Ran-RanBP1 complex reveals that KPT-251 occupies the space commonly for an NES peptide. The groove which the KPT-251-CRM1 complex generates is narrower and deeper than the cargo-bound groove, demonstrating that the CRM1 groove is conformationally plastic. The trifluoromethyl phenyl group of KPT-251 penetrates much deeper into the groove than do the NES side chains, possibly contributing to the potency of the compound in outcompeting the cargo proteins (Etchin, Sanda, et al., 2013; Etchin, Sun, et al., 2013). This evaluation also illustrates the potency of these agents in inhibiting CRM1-cargo interactions and nuclear export (Etchin, Sanda, et al., 2013; Lapalombella, et al., 2012).
While KPT-185 is the most potent CRM1 inhibitor in this series, KPT-251, -276, and -330 are suitable for in vivo use because of their pharmacokinetic and pharmacodynamics properties, including oral bioavailability (Etchin, Sanda, et al., 2013; Lapalombella, et al., 2012; Ranganathan, et al., 2012). Furthermore, in the toxicology studies, the primary adverse effects of the orally administered SINE compounds were dose-dependent reductions in food intake and in body weight. Minimal gastrointestinal symptoms were observed with modest or no gastrointestinal atrophy. These side effects were reduced by giving food supplements to the animals and by reducing the dosing frequency to 2 to 3 times per wk with at least 48 h between dosing. This modified schedule conserved the in vivo activity of these compounds (Etchin, Sun, et al., 2013).
The SINE compounds are relatively non-toxic to normal human cells including normal hematopoietic cells (Etchin, Sanda, et al., 2013; Etchin, Sun, et al., 2013; Kojima, et al., 2013; Lapalombella, et al., 2012). Furthermore, KPT-330 revealed no significant effect on normal immunoglobulin levels in monkeys and rats administered over 4 wk with a 2-wk recovery period, suggesting that the agent had little or no immunotoxic effects (Tai, et al., 2014). Conversely, the KPT compounds induce apoptosis as well as G1 phase cell cycle arrest in renal and pancreatic tumors as well as in melanomas and transformed hematopoietic cells (Etchin, Sanda, et al., 2013; Etchin, Sun, et al., 2013; Kojima, et al., 2013; Lapalombella, et al., 2012; Ranganathan, et al., 2012; Sakakibara, et al., 2011; Schmidt, et al., 2013; Tai, et al., 2014; Walker, et al., 2013; Zhang, et al., 2013). The effectiveness of these agents against hematological malignancies in vitro has emerged recently, and prompted their clinical evaluation. In fact, KPT-330 is currently being evaluated in numerous clinical trials, which are described separately in this review.
CRM1 and Malignancies
Cell cycle regulators and various other anti-neoplastic modulators of cell function that are abnormally exported from the nucleus into the cytoplasm of cancerous cells are, as a result, ancillary targets for CRM1 inhibition (Inoue, et al., 2013; Kojima, et al., 2013; Lapalombella, et al., 2012; Walker, et al., 2013), and include p53 (Inoue, et al., 2013; Kojima, et al., 2013; Lapalombella, et al., 2012; Walker, et al., 2013), p21, p27 (Inoue, et al., 2013; Tai, et al., 2014; van der Watt, et al., 2009), IκB, FOXO (Inoue, et al., 2013; Kojima, et al., 2013; Lapalombella, et al., 2012; Walker, et al., 2013), topoisomerase II (Turner, Engel, Derderian, Jove, & Sullivan, 2004; Turner, et al., 2009), and bcr-abl (Aloisi, et al., 2006; Vigneri & Wang, 2001). As discussed in this section, the elevated expression and/or dysfunction of CRM1 has been reported in various solid tumors and hematological malignancies, and can be correlated with poor disease prognosis and resistance to therapy. This would suggest that alterations in nucleocytoplasmic trafficking, and hence a delocalization of tumor suppressor proteins, cell cycle regulators, pro-apoptotic proteins, etc. could lead to oncogenesis and resistance to chemotherapy.
Ovarian Cancer
The expression of CRM1 was analyzed by immunohistochemistry in 88 ovarian tumors. There was increased nuclear (52.7%) and cytoplasmic (56.8%) expression of CRM1 in 74 carcinomas compared with the expression observed in premalignant or benign lesions. The cytoplasmic CRM1-positive ovarian carcinomas were at a more advanced tumor stage, poorly differentiated, and had a higher mitotic rate. In univariate Kaplan-Meier analysis, the nuclear CRM1 expression was associated with poor overall survival (P = 0.10). Interestingly, this study also demonstrated a positive correlation of CRM1 expression with COX-2 expression, which has been related to poor overall survival [26]. Furthermore, the treatment of cells derived from an ovarian cancer cell line with leptomycin B revealed an anti-proliferative effect and increased apoptosis, as well as reduced COX-2 expression, probably by inhibiting the nuclear export of COX-2 mRNA (Noske, et al., 2008).
Glioma
Immunohistochemical and western blot analysis were performed on samples from 70 glioma patients and 10 normal brain tissue samples (Shen, et al., 2009). The results were categorized according to the percentage of CRM1-positive cells, where 18 patients were strongly positive, 38 patients were moderately positive, and 14 were negative. The survival correlation was analyzed for the same 70 patients who had undergone curative resection, including 32 grade II patients, 26 grade III patients, and 12 grade IV patients. The five-year overall survival was 30.8% in CRM1-positive patients, compared with 55.9% in CRM1-negative patients. Multivariate analysis showed that CRM1 protein expression level was an independent prognostic factor for overall survival (P < 0.001). There was a slight, although not significant, association between higher CRM1 levels and shorter patient survival within the glioma subgroups, The study also showed that CRM1 expression was negatively correlated with p27 expression (P < 0.001) and positively associated with (cytoplasmic) phosphorylated serine 10-p27 expression (P < 0.001). This is compatible with the fact that p27 is supposed to be phosphorylated at serine 10 in order to be exported from the nucleus by CRM1. These results indicated that high levels of CRM1 and increased cytoplasmic p27 are positively correlated with poor clinical prognosis in glioma patients (Shen, et al., 2009).
Pancreatic Cancer
CRM1 expression was examined in pancreatic tissue from nine patients with stage I-II pancreatic cancer as well as tissue from 10 normal subjects. Western blot revealed that, as compared with low CRM1 expression in normal tissue, CRM1 expression in pancreatic cancer was increased (P = 0.0013). Thirty-nine of 69 patients had high CRM1 expression, and none of the 10 normal samples expressed increased CRM1. Increase in CRM1 expression was associated with high serum levels of carcinoembryonic antigen (CEA) (P = 0.002) and carbohydrate antigen 19–9 (CA19-9) (P = 0.005). There was also a correlation between CRM1 expression and tumor size (P = 0.011), lymphadenopathy (P = 0.004), and metastasis (P = 0.041). Increased expression of CRM1 was not correlated with sex, histological grade, or age. High expression of CRM1 was a prognostic indicator of poor overall (P = 0.004) and progression-free (P = 0.011) survival. Multivariate survival analysis showed that high CRM1 expression, lymphadenopathy, tumor size, presence of metastases, and histological grade were related to poor survival (Huang, et al., 2009).
Cervical Cancer
CRM1 gene and protein expression, as well as that of karyopherin-β1 and -α2, in normal (n = 8) and cervical cancer tissues (n = 16) revealed a statistically significantly higher expression of all three in the cancer tissues (van der Watt, et al., 2009). Western blot analysis showed increased CRM1 protein expression also in HPV E6/E7 transformed cells and six cervical cancer cell lines (van der Watt, et al., 2009). The inhibition of CRM1 by leptomycin B or siRNA in vitro showed anti-proliferative effects in CaSki cervical cancer cells. This anti-proliferative effect was associated with an increase in cells in the subG1 cell population and caspase-3/7 activation indicative of apoptosis. Furthermore, CRM1 siRNA-induced apoptosis in these cells was accompanied by an increase in the levels of growth inhibitory proteins, p53, p27, p21, and p18 (van der Watt, et al., 2009).
Osteosarcoma
Protein expression of CRM1 was examined in 57 human osteosarcoma samples and 5 normal cartilage samples. Western blot revealed that expression of CRM1 was significantly increased in osteosarcoma compared with normal tissues. In univariate analysis, there were correlations between CRM1 expression and tumor size (P = 0.014) as well as histological grades of G1 and G2 (P = 0.003). In Kaplan-Meier survival analysis, increased CRM1 protein level was a significant prognostic factor for progression-free (P = 0.016) as well as overall (P = 0.008) survival. Multivariate analysis demonstrated that the expression of CRM1 was one of the clear-cut, independent prognostic indicators for longer overall survival (95% CI, 1.27 – 5.39) for patients with osteosarcoma (Yao, et al., 2009).
Renal Cell Carcinoma
The CRM1 inhibitors KPT-185 and KPT-251 were tested in several renal cell carcinoma cell lines and in a renal cell carcinoma xenograft model (i.e., Caki-1 cells were transplanted subcutaneously into athytic Nu/Nu mice). Compared to sorafenib, the SINE compounds exhibited better tumor growth inhibition and apparent sequestration of p53 and p21 in the nucleus, as well as a decrease in CRM1 protein expression, in vitro and in vivo (Inoue, et al., 2013).
AML
Recently, striking anti-leukemia/anti-lymphoma activity and high selectivity of the SINE class of small molecule CRM1 antagonists has been reported (Etchin, Sanda, et al., 2013; Etchin, Sun, et al., 2013; Kojima, et al., 2013; Ranganathan, et al., 2012; Sakakibara, et al., 2011; Schmidt, et al., 2013; Tai, et al., 2014; Walker, et al., 2013), which provided the basis for initiating the on-going clinical studies with KPT-330. Blockade of CRM1 transport by these inhibitors has been shown to induce cancer cell death, possibly by promoting the forced nuclear retention of tumor suppressor proteins. Kojima et al. (Kojima, et al., 2013) recently reported the profiling of CRM1 protein expression in 511 newly diagnosed AML patients by utilizing reverse-phase protein arrays, and found that CRM1 protein expression has a negative prognostic impact in AML: high CRM1 levels were observed to be an independent predictor of overall survival in both univariate and multivariate analysis (median survival; 66 wk in low expression group, 47.2 wk in the middle third group and 37 wk in highest third group. p < 0.05). Higher levels of CRM1 were significantly associated with higher bone marrow blast percentages, white blood cell counts, peripheral blood blast percentages, and absolute peripheral blood blast counts. CRM1 expression was significantly lower in patients with favorable, compared to intermediate or unfavorable, cytogenetics. The reverse-phase protein array analysis of 207 proteins found that 29 were positively (n = 22) or negatively (n = 7) correlated with CRM1 levels (P < 0.0001). One interesting additional findings was that high CRM1 levels appeared to be associated with activated AKT signaling, including AKT itself, its upstream phospho-PTEN, and downstream phospho-BAK and 14-3–3ζ, which could conceivably contribute to anti-apoptotic properties in AML cells (Kojima, et al., 2013).
It has also been reported that CRM1 expression is higher in AML with FLT3 mutations, which itself is associated with poor overall survival in patients with AML (Whitman et al., 2008; Yanada, Matsuo, Suzuki, Kiyoi, & Naoe, 2005). Another report demonstrated that CRM1 inhibition by SINE compounds downregulated FLT3 post transcriptionally in AML cells (Ranganathan, et al., 2012). Furthermore, considering that FLT3/internal tandem duplication (ITD) AML cells are susceptible to p53-mediated cell death (Long et al., 2010), SINE compounds that indirectly activate p53 may be beneficial in treating FLT3-mutated AML. Indeed, FLT3 mutations were associated with increased sensitivity to KPT-185 in patient samples (Kojima, et al., 2013; Ranganathan, et al., 2012).
Kojima et al. (Kojima, et al., 2013) also reported that the p53 status of cells from AML cell lines and primary AML samples was a major determinant of the apoptotic response of these cells to KPT-185. In fact, apoptosis induction by KPT-185 was much more prominent in p53 wild-type cells (i.e., the effective dose for achieving response in 50% of the treatment population values were < 150 nM in p53 wild-type AML cells and > 1000 nM in the p53 mutant cells). They also observed similarly decreased efficacy of KPT-185 by stable p53 knockdown in three p53 wild-type AML lines. This finding raised the hypothesis that p53 status affects clinical response to SINE compounds; it could be investigated in patients undergoing clinical trials with KPT-330. On the other hand, it has also been reported that KPT-185 exhibits p53-independent anti-leukemic activity in AML cell lines (Etchin, Sun, et al., 2013), and that p53 mutant cells are marginally sensitive to the cytostatic/cytotoxic effects of SINE compounds. There are some discrepancies in the degrees of reported efficacy of these compounds, which is probably due to differences in the assays used to measure cell proliferation and toxicity (Brown & Wilson, 2003; Kojima, et al., 2013).
The correlation of CRM1 protein levels with those of p53 and MDM2 has also been investigated (Kojima, et al., 2013). In a three-way correlation (using distance weighted least squares), there was a clear interaction since p53 levels were highest when CRM1 levels were high and MDM2 levels were low. This indicates that CRM1 possibly plays a role as complementary mechanism of p53 suppression to MDM2. Combination therapy of the MDM2 inhibitor Nutlin-3a and KPT-185 not only increased the cellular p53 levels in AML cells, but it also promoted p53 accumulation in the nucleus, resulting in synergistic apoptosis induction in these cells (Kojima, et al., 2013).
The SINE compounds also provide promising preclinical anti-leukemia results in in vivo experiments. KPT-251 (Etchin, Sun, et al., 2013) (75 mg/kg/day, three times per wk) or KPT-276 (Ranganathan, et al., 2012) (150 mg/kg/day, three times per wk) were strikingly active against human AML cells in a mouse xenograft model (i.e., a luciferase-expressing MV4;11 harboring FLT-ITD mutant that was transplanted into NOD-SCID-IL2Rcγnull [NSG] mice), and the dosing schedule of KPT-251 showed only minimal toxicity to normal tissues, including normal circulating blood and bone marrow cells (Etchin, Sun, et al., 2013).
Perhaps the best-known abnormality of nucleocytoplasmic transport in AML involves the mutant NPM1 protein. NPM1 is a nucleolar tumor suppressor that shuttles between the nucleolus and cytoplasm and regulates the p53-ARF tumor suppressor pathway (Falini et al., 2005; Li & Hann, 2009; Meani & Alcalay, 2009; Ranganathan, et al., 2012). Mutations in exon 12 of NPM1 account for 25% to 35% of AML and are one of the most frequent mutations in this disease. These mutations result in a conformational change of NPM1 protein at the C-terminus, providing a novel and efficient CRM1 biding site, thus resulting in abnormally increased cytoplasmic localization of NPM1. NPM1 mutant protein (NPM1c) can form a heterodimer with wild-type NPM1, which exerts oncogenic effects (Falini et al., 2006). Supporting its oncogenic role of NPM1c, primary AML blasts harboring NPM1 mutations were susceptible to CRM1 inhibition (Ranganathan, et al., 2012). On the other hand, two reports showed anti-leukemia activity of SINE compounds not only against OCI-AML3 cells, the only human cell line with the NPMc, but also against cell lines lacking this mutation (Etchin, Sanda, et al., 2013; Kojima, et al., 2013). From another point of view, nucleoplasmic NPM1 increases p53 stability through interaction with p53 and MDM2, which would indicate that CRM1 inhibition could reactivate p53 and induce apoptosis in a p53 dependent manner in AML cells. Therefore, NPMc is probably dispensable in SINE compounds-induced apoptosis but it may contribute also to the increased sensitivity to these agents. Because patients with NPM1 mutations may also have FLT3-ITD mutations, CRM1 inhibitors also have the potential to target the mutated signaling pathways of both NPM1 and FLT3-ITD (Havelange et al., 2014).
T-cell ALL
Etchin et al. demonstrated that KPT-330 is very active in preclinical models of T-cell ALL as well as AML, with minimal toxicity to normal blood cells both in the peripheral blood and in the bone marrow. Low nanomolar concentrations of SINE compounds induced rapid (within 6 to 13 h) apoptosis and dramatically reduced viability in 13 T-cell ALL cell lines in vitro and suppress growth of human MOLT-4 T-cell ALL cells engrafted into immunocompromised mice. These include the cell lines characterized by TAL1, TLX3, and translocation of MYC-TRA, which control different molecular pathways leading to T-cell ALL, and they also harbor a variety of genetic alterations resulting in abnormal signaling through NOTCH1, NRAS, FLT3, PTEN, and TP53. Interestingly, the most sensitive cell lines to SINE compounds did not display similar patterns of genetic abnormalities, and they were not categorized in the same oncogenic group. This result indicated that common mechanisms or pathways exist, although in different oncogenic subclasses of T-cell ALL, which could presumably reflect differences in the nucleocytoplasmic trafficking of various CRM1 cargoes (Etchin, Sanda, et al., 2013).
CLL
Whole-genome sequencing of CLL has recently identified recurrent mutations of CRM1 (Etchin, Sanda, et al., 2013; Puente et al., 2011), suggesting its role in the pathogenesis of CLL. It has been recently reported that SINE compounds induce apoptosis of CLL cells with a favorable therapeutic index, with effective killing of even genomically high-risk CLL cells that are typically unresponsive to traditional therapies (Lapalombella, et al., 2012).
SINE compounds cause the nuclear retention of key tumor suppressor proteins such as FOXO3a, IkB, and p53 both in vitro and in vivo, a mechanism that potentially plays a partial role in their anti-leukemic effect (Kau, et al., 2004). An unique finding is that KPT-185 induces depletion of Mcl1 message and protein in CLL cells, seemingly correlated with the inactivation of NFκB due to IκB’s nuclear localization, and the sensitivity of patient samples to KPT-185 correlates with the amount of down-modulation of Mcl1 (Lapalombella, et al., 2012). Similarly, a significant reduction of Mcl1 mRNA was also observed by CRM1 downregulation using siRNA knockdown. Because there is no clear consensus about the direct correlation between NFκB and Mcl1, the mechanism of Mcl1 depletion by KPT-185 has not been fully elaborated. High levels of Mcl1 mRNA and protein have been found in CLL, which are inversely correlated with in vitro response to standard chemotherapeutic agents, as well as in CLL patients who fail to respond to standard chemotherapies. The down-regulation of Mcl1 protein expression by antisense oligonucleotides, or indirectly through Mcl1 transcription or translation inhibitors, results in cell death in vitro and in vivo. Therefore, the inhibition of Mcl1 by KPT-185 is anticipated to be one of the critical pathways of apoptosis induction in CLL cells. An in vivo model of CLL also provided a similar promising anti-leukemic effect (Lapalombella, et al., 2012). KPT-251 reduced leukemia cell counts, prevented disease progression, and improved overall survival with minimal weight loss or related toxicities. Similar to the results in vitro, Mcl1, and XIAP mRNA were also down regulated in mice receiving a single dose of KPT-251, although no differences were observed in protein levels. Furthermore, this study illustrated that SINE compounds antagonize pro-survival stimuli from the tumor microenvironment (Lapalombella, et al., 2012). Indeed, soluble factors known to reduce spontaneous apoptosis in CLL cells (e.g., TNF, IL-6 and IL-4) were combined with SINE compounds, which disabled their anti-apoptotic effects. Also in a co-culture system with a human marrow-derived fibroblast cell line, the protective effect of these cells was abrogated by treatment with SINE compounds, and KPT-185-induced apoptosis was enhanced under the co-culture conditions. Therefore, KPT-185 may overcome, at least in part, the protective effects of stromal cells in the CLL cell microenvironment.
Multiple Myeloma (MM)
The inhibition of CRM1 in MM was first demonstrated using leptomycin B, showing its synergistic effect when administered with topoisomerase II inhibitor chemotherapeutic agents (Turner, et al., 2004). The topoisomerase II inhibitors block the binding of topoisomerase IIα during DNA cleavage to produce cleavable complexes, resulting in double-stranded DNA breaks and cell death. For DNA damage to occur, topoisomerase IIα must be in the nucleus and in contact with DNA. The aforementioned study found that MM cells, at high density conditions, are intrinsically resistant to these agents because of cytoplasmic trafficking of topoisomerase IIα, and that the nuclear export of topoisomerase IIα is mediated by CRM1 through its two leucine-rich nuclear NESs. Furthermore this study also proved that leptomycin B maintained topoisomerase IIα in the nucleus and greatly sensitizes MM cells to doxorubicin and etoposide, even in doxorubicin-resistant myeloma cells (Turner, et al., 2004; Turner, et al., 2009).
Recently, a genome-wide RNA interference lethality screening of MM cells (KMS11 cells), using 13984 small interfering RNAs, was performed to identify critical molecular susceptibilities in these cells (Tiedemann et al., 2012). Among the 55 most lethal genes identified, functional testing in human MM and non-MM cell lines indicated that CRM1 is one of the most selectively vulnerable molecules in myeloma. Recently, two studies showed a correlation between CRM1 protein or gene expression levels and the genotypic characteristics of human MM samples (Schmidt, et al., 2013; Tai, et al., 2014). Gene expression analyses demonstrated increased CRM1 expression in newly diagnosed MM cells compared to normal plasma cells (n = 22; p < 0.001), increased levels in MM compared to monoclonal gammopathy of unknown significance (MGUS) (P = 0.007), and in plasma cell leukemias compared to MM (P < 0.01). CRM1 transcripts were also higher in patients unresponsive to bortezomib treatment (P < 0.004). Higher CRM1 was significantly associated with poor outcome (P = 0.024 for event free survival and P = 0.044 for overall survival) and the extent of bone lytic lesion in the “Total Therapy 2” cohort (P = 0.008) (Tai, et al., 2014).
The degree of CRM1 downregulation by shCRM1 lentiviruses targeting CRM1 gene is correlated with the levels of spontaneous MM cell apoptosis (Tai, et al., 2014), and the induction of G1 phase arrest and apoptosis in these cells in vitro and in vivo following SINEs (i.e., KPT-185, -251, -276 and -330) (Schmidt, et al., 2013; Tai, et al., 2014). Importantly, these agents have anti-tumor effects even in MM cells co-cultured with bone marrow stromal cells or osteoclasts. Additionally, KPT-185 and KPT-330 inhibited NFκB activity, through the nuclear localization of IκB and inhibiting pIκB, in both MM and osteoclast cells of the microenvironment. NFκB is known to be generally activated in MM and to contribute to tumor growth, survival and drug resistance (Hideshima, Mitsiades, Tonon, Richardson, & Anderson, 2007), to osteoclast activation and osteolysis. From the point of view of the microenvironment, CRM1 knockdown significantly blocks the key osteoclast differentiation regulator NFATc1 induced by RANKL. By inhibiting both NFκB activity and osteoclast differentiation genes downstream of NFATc1, the SINE compounds prevented adhesion and fusion of preosteoclasts to form functional osteoclast, thereby promising to improve bone lesions of MM. Although they were able to inhibit osteoclastogenesis, the SINE compounds had no impact on osteogenesis in vitro, suggesting that these compounds did not affect new bone formation (Tai, et al., 2014).
Other than NFκB activity, which was compatible with a physiological, anti-cancer role for CRM1, KPT-185 and KPT-330 potently restored the nuclear localization and elevated the levels of p53, FOXO1A, FOXO3a, p27 and PP2A in MM cells. Transcripts of p53 and its downstream targets p21, BAX, PUMA were also induced by KPT-185 treatment. Along with increasing pro-apoptotic PUMA and BAX, KPT-185 also decreased anti-apoptotic proteins MCL-1 and BCL-XL in these cells. Furthermore, the SINE compounds reduced levels of the MM oncogenic growth driver, including MYC. Another group also showed downregulation of two MYC-related genes, CDC25A and BRD4 by KPT-276, as well as MYC itself in MM cells (Schmidt, et al., 2013). BRD4 is known to bind to acetyl-lysine pockets at the IgH enhancer region on the MYC locus, and promotes transcription and activation of MYC downstream targets. Furthermore, a novel BRD4 small-molecule inhibitor JQ1 demonstrated a synergistic effect with KPT-276 against MM cells (Schmidt, et al., 2013). Therefore, SINE compounds may alter MM viability via MYC downregulation by lowering BRD4 levels in MM cells.
In addition, the SINE compounds were active against MM cells harboring mutated p53 (Schmidt, et al., 2013), which is found in over 70% of MM cell lines, and increasingly in the setting of relapsed and refractory disease in humans. A p53-independent mechanism could probably explain the inhibition of NFκB, MYC, MCL1, BCL-XL, or FOXO3a by the SINE compounds. Importantly, these SINE compounds caused minimal apoptosis of stromal cells and osteoblasts even at concentrations that triggered over 80% MM cell apoptosis (Etchin, Sun, et al., 2013; Lapalombella, et al., 2012).
Philadelphia Chromosome Positive (Ph+) Leukemias
Targeting CRM1 in Ph+ leukemia has been reported (Walker, et al., 2013). Using CD34+ progenitor cells from chronic myelogenous leukemia (CML), B cell acute lymphoblastic leukemia (B-ALL), and healthy donors, western blot analysis found that CRM1 protein expression was specifically increased in these diseases. However, CRM1 levels were further increased in Ph− B-ALL, suggesting that BCR-ABL1-dependent and -independent molecular mechanisms may cooperate in increasing CRM1 expression in Ph+ acute leukemias. Walker et al. (Walker, et al., 2013) also showed that KPT-330 (1 μM) induced apoptosis in ~ 75% cells in chronic phase of CML (CML-CP) and ~ 95% in blast crisis of CML (CML-BC) CD34+ BM cells, with no significant induction of apoptosis in KPT-330-treated CD34+ normal bone marrow cells. Similarly, in CD34+/CD19+ leukemic blasts isolated from B-ALL patients, KPT-330 induced apoptosis in ~ 85% of Ph+ and nearly 95% of Ph− leukemic progenitors. KPT-330 exerted anti-leukemia effect in an allograft model of CML-BC in which ICR-SCID mice were intravenously injected with 32D/BCR-ABL cells. As to the mechanisms of action of the SINE compounds, it is unique that they directly inhibit the BCR-ABL1 hnRNP A1 – SET – network thereby restoring its downstream tumor suppressor protein phosphatase 2A (PP2A) that caused selective killing of CML-BC and Ph+ ALL blasts. CRM1 has been shown to directly interact with BCR-ABL1 (Vigneri & Wang, 2001) as well as the two endogenous PP2A inhibitors, SET (Gallouzi, Brennan, & Steitz, 2001) and CIP2A (Lucas et al., 2011), which were also described as BCR-ABL1-regulated factors in Ph+ leukemias (Eiring et al., 2008; Neviani et al., 2005). On the other hand, the nucleocytoplasmic shuttling RNA binding protein hnRNP A1 controls SET mRNA nuclear export (Eiring, et al., 2008; Neviani, et al., 2005). hnRNP A1 expression and nuclear export activity is enhanced in a BCR-ABL1-dependent manner, positively regulating the nuclear export of hnRNP A1 target mRNAs including SET. Thus, hnRNP A1 is an important regulator of BCR-ABL1 leukemogenesis (Eiring, et al., 2008; Neviani, et al., 2005). As expected, KPT-330 caused sequestration of SET and CIP2A proteins in the nucleus, without altering the subcellular localization of PP2Ac and, unexpectedly, caused dysregulated cytoplasmic relocation of hnRNP A1, indicating its deteriorated nuclear export functions. By using BCR-ABL1+ cells engineered to express the PP2A inhibitor small t-antigen, they concluded that PP2A accounts for approximately 50% of the KPT-330-induced apoptosis. Indeed, KPT-330 treatment of BCR-ABL1+ myeloid precursors also expectedly induced nuclear accumulation of the p53, p21, FOXO3a, and IkB, which probably contributes to KPT-330 induced apoptosis (Walker, et al., 2013). Interestingly, the aforementioned study could not confirm a nuclear accumulation of either p210 or p190 BCR-ABL1 unlike previous reports. Instead, both p190 and p210 BCR-ABL1 decreased in amount and activity in the KPT-330-treated CD34+ CML-BC or B-ALL patient cells.
Mantle Cell Lymphoma (MCL)
CRM1 protein expression was evaluated by western blot and it was found to be highly expressed in all of eight MCL cell lines examined as well as in primary samples from MCL patients compared to normal B-lymphocytes (Zhang, et al., 2013). In addition, mRNA expression levels in MCL patient samples were analyzed using Oncomine (Compendia Bioscience, Ann Arbor, MI) (Yoshimura, et al., 2014; Zhang, et al., 2013). Again, the CRM1 expression levels were significantly higher in MCL samples as compared to normal B-cells (P < 0.001). Furthermore, higher CMR1 expression was associated with poor overall prognosis in these MCL patients (i.e., median overall survival was 3.2 years in low CRM1 cases and 1.9 years in high CRM1 expressing cases, P = 0.033). The inhibition of CRM1 by siRNA, KPT-185, or KPT-276 induced significant dose-dependent growth inhibition and apoptosis in MCL cells in vitro. Surprisingly, blastoid-variant MCL cell lines were more sensitive to the SINE compounds than were the typical MCL cell lines (Zhang, et al., 2013).
As mentioned previously, the SINE compounds seem to have both p53-dependent and -independent mechanisms of MCL cell killing, since cytotoxicity of these compounds was independent of p53 status (Tabe, et al., 2013; Zhang, et al., 2013). KPT-185 treatment had little or no effect on BAX and NOXA protein expression, further suggesting that apoptosis induction in these cells might be p53-independent (Zhang, et al., 2013). To investigate the significance of p53-mediated apoptosis further, two wild-type p53 MCL cell lines were infected with lentivirus encoding either negative control shRNA or p53-specific shRNA, and p53 stable knockdown cells were found to be significantly less sensitive to KPT-185 indicating that CRM1 inhibition relies largely on the p53 pathway to induce apoptosis. MCL cells often have cytoplasmic p53 so that wild-type p53-mediated transcription is impaired (Jin et al., 2010). However, KPT-185 induced p53 target genes more potently than the MDM2 inhibitor Nutlin-3a, in spite of higher p53 induction by Nutlin-3a, probably because p53’s cytoplasmic mislocalization was reversed by the SINE (Yoshimura, et al., 2014). As a potential p53-independent mechanism, the CRM1 inhibitor could potently inhibit NFκB activity in MCL cells, by trapping IκBalpha-NFκB complexes within the nucleus (Zhang, et al., 2013). However, specific knockdown of CRM1 has minimal effects on NFκB activity. Several possibilities can account for this result: CRM1 protein must be present for IkBalpha stabilization; there could be other protein exporters shuttling IkBalpha out of the nucleus beside CRM1; or the KPT compounds are not specific for CRM1 inhibition only. It has also been reported that KPT-185 abrogates MCL-related cyclin D1 overexpression and upregulates pro-apoptotic PUMA in a p53-independent manner (Tabe, et al., 2013). In addition, as mentioned previously, KPT-185 can reduce MYC protein levels in MCL cells, and hence CRM1 inhibition could disrupt MYC-related ribosome biogenesis signaling in these cells (Tabe, et al., 2013), which may contribute to p53-independent anti-lymphoma activities of SINE compounds. Conversely, it has been also observed that KPT-185 can induce the translocation of CRM1 itself into the nucleus in MCL cell lines, which will eventually down-regulate the expression of the CRM1 protein. This was positively correlated with the induction of apoptosis in MCL cells in vitro. This effect has been observed also in other malignancies such as renal carcinoma and MM, indicating that it is probably a general effect of the SINE compounds. Interestingly, this result was compatible with, and may be explained by, the reduction of MYC or activation of p53 by these agents, because they are known as positive or negative regulators, respectively of CRM1 (Golomb, et al., 2012). Similar anti-lymphoma activity was also observed in an MCL-xenograph SCID mouse model. Orally administered KPT-276, at a dose of 50mg/kg five times weekly, showed marked effects. Under these conditions, KPT-276 was well tolerated, with <10% weight loss and no remarkable side effects, such as myelosuppression, mucositis or diarrhea (Zhang, et al., 2013).
Clinical Trials of CRM1 Inhibitors
KPT-330 clinical trials have been initiated with promising very early results. One trial includes patients with advanced solid tumors whose disease has progressed after at least one prior therapy for metastatic disease (NCT01607905). Another trial includes patients with advanced hematological malignancies including CLL, non-Hodgkin lymphoma, MM, and Waldenstrom macroglobulinaemia whose disease has relapsed after standard therapies (NCT01607892). Yet another phase IB trial includes patients whose soft tissue or bone sarcomas relapsed, or are progressing, with at least one prior therapy for metastatic disease (NCT01896505). Patients with AML have been also eligible very recently in four on-going clinical trials (NCT02091245, NCT01607892, NCT02088541, NCT02093403), and the trials including patients with glioblastoma or gynecologic malignancies have also been initiated (ClinicalTrials.gov). Early responses include complete remissions in large cell lymphomas, chronic lymphocytic leukemias (Kuruvilla et al., 2013), and AMLs (Savona et al., 2013), which promoted a trial in dogs developing lymphomas (London et al., 2014), and several combination trials. It is obviously too early to assess the full clinical potential of these agents, but the early responses in highly chemo-refractory patients are encouraging. Side effects include anorexia, fatigue, weight loss and gastrointestinal symptoms, which can be substantial (London, et al., 2014).
Conclusion and Future Perspectives
Recently, the role of CRM1 in oncogenesis has provided a, novel focus for the development of therapeutic anti-tumor strategies, and the discovery of the SINE inhibitors may allow the translation of these findings into effective clinical treatments. Clinical trials of SINE compounds are progressing and promising, and mechanism-based combination therapies are under development.
At present, the mechanisms of action of SINE compounds via CRM1 inhibition are gradually being elucidated, specifically in some hematological malignancies and evolving mechanisms of resistance against this therapeutic strategy remain to be better understood and resolved. Because targeting CRM1 causes various anti-tumor pathways to be activated, even in p53 mutant cells, many opportunities for combinatorial therapies with SINE inhibitors exist that are anticipated to overcome evolving resistance mechanisms. In this regard, ribosomal biogenesis, FOXO, and the NFκB signaling pathway can potentially function as co-targets. As described in CLL and MM, it may be also rewarding to focus on the role of CRM1 in the relationship between malignant cell survival and their microenvironment.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (CA49639, 100632, CA136411, and CA16672), the Paul and Mary Haas Chair in Genetics (all to M.A.), and the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad Award (to J. I.)
List of Abbreviations
- CRM
chromosome region maintenance 1
- SINE
CRM1-selective inhibitors of nuclear export
- AML
acute myeloid leukemia
- ALL
acute lymphoid leukemia
- CML
chronic myeloid leukemia
- CLL
chronic lymphoid leukemia
- MCL
mantle cell lymphoma
- MM
multiple myeloma
- rRNA
ribosomal RNA
- NPM1
nucleophosmin1
- iTRAQ
isobaric tags for relative and absolute quantification
Footnotes
Conflict of Interest
M.A. has received research support from Karyopharm Therapeutics. The other authors have no conflict of interest for this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. Journal of Cell Biology. 1989;108(4):1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisi A, Di Gregorio S, Stagno F, Guglielmo P, Mannino F, Sormani MP, et al. BCR-ABL nuclear entrapment kills human CML cells: ex vivo study on 35 patients with the combination of imatinib mesylate and leptomycin B. Blood. 2006;107(4):1591–1598. doi: 10.1182/blood-2005-05-2123. [DOI] [PubMed] [Google Scholar]
- Bai B, Moore HM, Laiho M. CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis. Nucleus. 2013;4(4):315–325. doi: 10.4161/nucl.25342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi S, Guttinger S, Zemp I, Kutay U, Gademann K. Total synthesis, configuration, and biological evaluation of anguinomycin C. Angew Chem Int Ed Engl. 2007;46(45):8707–8710. doi: 10.1002/anie.200703134. [DOI] [PubMed] [Google Scholar]
- Brown JM, Wilson G. Apoptosis genes and resistance to cancer therapy: what does the experimental and clinical data tell us? Cancer Biol Ther. 2003;2(5):477–490. doi: 10.4161/cbt.2.5.450. [DOI] [PubMed] [Google Scholar]
- Burzlaff A, Kalesse M, Kasper C, Scheper T. Multi parameter in vitro testing of ratjadone using flow cytometry. Appl Microbiol Biotechnol. 2003;62(2–3):174–179. doi: 10.1007/s00253-003-1300-0. [DOI] [PubMed] [Google Scholar]
- Craig E, Zhang ZK, Davies KP, Kalpana GV. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: implications for tumorigenesis. Embo Journal. 2002;21(1–2):31–42. doi: 10.1093/emboj/21.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D, Afonina E, Nilsson J, Werner G, Kjems J, De Clercq E, et al. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc Natl Acad Sci U S A. 2002;99(22):14440–14445. doi: 10.1073/pnas.212285299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16(5):558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458(7242):1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring AM, Neviani P, Santhanam R, Oaks JJ, Chang JS, Notari M, et al. Identification of novel posttranscriptional targets of the BCR/ABL oncoprotein by ribonomics: requirement of E2F3 for BCR/ABL leukemogenesis. Blood. 2008;111(2):816–828. doi: 10.1182/blood-2007-05-090472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161(1):117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchin J, Sun Q, Kentsis A, Farmer A, Zhang ZC, Sanda T, et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia. 2013;27(1):66–74. doi: 10.1038/leu.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107(11):4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14(3):313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390(6657):308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gademann K. Controlling protein transport by small molecules. Curr Drug Targets. 2011;12(11):1574–1580. doi: 10.2174/138945011798109446. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7(9):1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, et al. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell. 2012;45(2):222–232. doi: 10.1016/j.molcel.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17(3):281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Hamamoto T, Uozumi T, Beppu T. Leptomycins A and B, new antifungal antibiotics. III. Mode of action of leptomycin B on Schizosaccharomyces pombe. J Antibiot (Tokyo) 1985;38(11):1573–1580. doi: 10.7164/antibiotics.38.1573. [DOI] [PubMed] [Google Scholar]
- Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. [Research Support, N I H, Extramural Research Support, Non-U S Gov’t] Blood. 2014;123(15):2412–2415. doi: 10.1182/blood-2013-10-532374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y, Sohda KY, Shin-Ya K, Hidaka T, Seto H. Anguinomycins C and D, new antitumor antibiotics with selective cytotoxicity against transformed cells. J Antibiot (Tokyo) 1995;48(9):954–961. doi: 10.7164/antibiotics.48.954. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH, Sun YJ. Prognostic value of CRM1 in pancreas cancer. Clin Invest Med. 2009;32(6):E315. [PubMed] [Google Scholar]
- Inoue H, Kauffman M, Shacham S, Landesman Y, Yang J, Evans CP, et al. CRM1 blockade by selective inhibitors of nuclear export attenuates kidney cancer growth. J Urol. 2013;189(6):2317–2326. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewers K, Burbage MB, Blunden G, Griffin WJ. Brisbagenin and brisbenone, two new spirostanes from Cordyline species. Steroids. 1974;24(2):203–208. doi: 10.1016/0039-128x(74)90103-2. [DOI] [PubMed] [Google Scholar]
- Jin L, Tabe Y, Kojima K, Zhou Y, Pittaluga S, Konopleva M, et al. MDM2 antagonist Nutlin-3 enhances bortezomib-mediated mitochondrial apoptosis in TP53-mutated mantle cell lymphoma. Cancer Lett. 2010;299(2):161–170. doi: 10.1016/j.canlet.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4(2):106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci U S A. 2010;107(29):12828–12833. doi: 10.1073/pnas.0910885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8(10):761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, et al. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121(20):4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama K, Okada K, Tomisaka S, Umezawa I, Hamamoto T, Beppu T. Antitumor activity of leptomycin B. J Antibiot (Tokyo) 1985;38(3):427–429. doi: 10.7164/antibiotics.38.427. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A. 1999;96(16):9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242(2):540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Kuruvilla J, Gutierrez M, Shah BD, Gabrail NY, de Nully Brown P, MSR Preliminary Evidence Of Anti Tumor Activity Of Selinexor (KPT-330) In a Phase I Trial Ofa First-In-Class Oral Selective Inhibitor Of Nuclear Export (SINE) In Patients (pts) With Relapsed/Refractory Non Hodgkin’s Lymphoma (NHL) and Chronic Lymphocytic Leukemia (CLL) Blood. 2013;122(21):Abstract 90. [Google Scholar]
- Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15(3):121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120(23):4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hann SR. The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle. 2009;8(17):2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10(8):934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- London CA, Bernabe LF, Barnard S, Kisseberth WC, Borgatti A, Henson M, et al. Preclinical evaluation of the novel, orally bioavailable Selective Inhibitor of Nuclear Export (SINE) KPT-335 in spontaneous canine cancer: results of a phase I study. PLoS One. 2014;9(2):e87585. doi: 10.1371/journal.pone.0087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116(1):71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CM, Harris RJ, Giannoudis A, Copland M, Slupsky JR, Clark RE. Cancerous inhibitor of PP2A (CIP2A) at diagnosis of chronic myeloid leukemia is a critical determinant of disease progression. Blood. 2011;117(24):6660–6668. doi: 10.1182/blood-2010-08-304477. [DOI] [PubMed] [Google Scholar]
- Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Molecular and Cellular Biology. 2008;28(23):7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meani N, Alcalay M. Role of nucleophosmin in acute myeloid leukemia. Expert Rev Anticancer Ther. 2009;9(9):1283–1294. doi: 10.1586/era.09.84. [DOI] [PubMed] [Google Scholar]
- Meissner T, Krause E, Vinkemeier U. Ratjadone and leptomycin B block CRM1-dependent nuclear export by identical mechanisms. FEBS Lett. 2004;576(1–2):27–30. doi: 10.1016/j.febslet.2004.08.056. [DOI] [PubMed] [Google Scholar]
- Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324(5930):1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69(2):510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8(5):355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74(4):648–649. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noske A, Weichert W, Niesporek S, Roske A, Buckendahl AC, Koch I, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112(8):1733–1743. doi: 10.1002/cncr.23354. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278(5335):141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120(9):1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Koberna K, Malinsky J, Fidlerova H, Masata M. The nucleolus and transcription of ribosomal genes. Biol Cell. 2004;96(8):579–594. doi: 10.1016/j.biolcel.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM, et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118(14):3922–3931. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341(6143):1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savona M, Garzon R, de Nully Brown P, Yee K, Lancet JE, Gutierrez M. Phase I Trial of Selinexor (KPT-330), A First-In-Class Oral Selective Inhibitor Of Nuclear Export (SINE) In Patients (pts) With Advanced Acute Myelogenous Leukemia (AML) Blood. 2013;122(21):Abstract 1440. [Google Scholar]
- Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS, et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia. 2013 doi: 10.1038/leu.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Lu C, Chen L, Koty PP, Cobos E, Gao W. p53-Dependent anticancer effects of leptomycin B on lung adenocarcinoma. Cancer Chemother Pharmacol. 2011;67(6):1369–1380. doi: 10.1007/s00280-010-1434-6. [DOI] [PubMed] [Google Scholar]
- Shen A, Wang Y, Zhao Y, Zou L, Sun L, Cheng C. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65(1):153–159. doi: 10.1227/01.NEU.0000348550.47441.4B. discussion 159–160. [DOI] [PubMed] [Google Scholar]
- Tabe Y, Kojima K, Jin L, Iwanami H, Matsushita H, Kazuno S, et al. Molecular mechanisms of inhibition of ribosomal biogenesis and translational flux by the Selective Inhibitor of Nuclear Export (SINE) XPO1/CRM1 antagonist KPT-185 in mantle cell lymphoma. Blood. 2013;122:Abstract 820. [Google Scholar]
- Tai YT, Landesman Y, Acharya C, Calle Y, Zhong MY, Cea M, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2014;28(1):155–165. doi: 10.1038/leu.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka Y, Fukumori T, Yoshii T, Oka N, Inohara H, Kim HR, et al. Nuclear export of phosphorylated galectin-3 regulates its antiapoptotic activity in response to chemotherapeutic drugs. Molecular and Cellular Biology. 2004;24(10):4395–4406. doi: 10.1128/MCB.24.10.4395-4406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci. 2003;116(Pt 12):2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Tiedemann RE, Zhu YX, Schmidt J, Shi CX, Sereduk C, Yin H, et al. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012;72(3):757–768. doi: 10.1158/0008-5472.CAN-11-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83(8):1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Engel R, Derderian JA, Jove R, Sullivan DM. Human topoisomerase IIalpha nuclear export is mediated by two CRM-1-dependent nuclear export signals. J Cell Sci. 2004;117(Pt 14):3061–3071. doi: 10.1242/jcs.01147. [DOI] [PubMed] [Google Scholar]
- Turner JG, Marchion DC, Dawson JL, Emmons MF, Hazlehurst LA, Washausen P, et al. Human multiple myeloma cells are sensitized to topoisomerase II inhibitors by CRM1 inhibition. Cancer Res. 2009;69(17):6899–6905. doi: 10.1158/0008-5472.CAN-09-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, et al. The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124(8):1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Neck T, Pannecouque C, Vanstreels E, Stevens M, Dehaen W, Daelemans D. Inhibition of the CRM1-mediated nucleocytoplasmic transport by N-azolylacrylates: structure-activity relationship and mechanism of action. Bioorg Med Chem. 2008;16(21):9487–9497. doi: 10.1016/j.bmc.2008.09.051. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Vigneri P, Wang JY. Induction of apoptosis in chronic myelogenous leukemia cells through nuclear entrapment of BCR-ABL tyrosine kinase. Nat Med. 2001;7(2):228–234. doi: 10.1038/84683. [DOI] [PubMed] [Google Scholar]
- Vogt PK, Jiang H, Aoki M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle. 2005;4(7):908–913. doi: 10.4161/cc.4.7.1796. [DOI] [PubMed] [Google Scholar]
- Wach JY, Guttinger S, Kutay U, Gademann K. The cytotoxic styryl lactone goniothalamin is an inhibitor of nucleocytoplasmic transport. Bioorg Med Chem Lett. 2010;20(9):2843–2846. doi: 10.1016/j.bmcl.2010.03.049. [DOI] [PubMed] [Google Scholar]
- Walker CJ, Oaks JJ, Santhanam R, Neviani P, Harb JG, Ferenchak G, et al. Preclinical and clinical efficacy of XPO1/CRM1 inhibition by the karyopherin inhibitor KPT-330 in Ph+ leukemias. Blood. 2013 doi: 10.1182/blood-2013-04-495374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82(3):463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Whitman SP, Ruppert AS, Radmacher MD, Mrozek K, Paschka P, Langer C, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111(3):1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW. Combined analysis of murine and human microarrays and ChIP analysis reveals genes associated with the ability of MYC to maintain tumorigenesis. PLoS Genet. 2008;4(6):e1000090. doi: 10.1371/journal.pgen.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Grishin NV, Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell. 2012;23(18):3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanada M, Matsuo K, Suzuki T, Kiyoi H, Naoe T. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: a meta-analysis. Leukemia. 2005;19(8):1345–1349. doi: 10.1038/sj.leu.2403838. [DOI] [PubMed] [Google Scholar]
- Yao Y, Dong Y, Lin F, Zhao H, Shen Z, Chen P, et al. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol Rep. 2009;21(1):229–235. [PubMed] [Google Scholar]
- Yoshimura M, Ishizawa J, Ruvolo V, Dilip A, Quintas-Cardama A, McDonnell TJ, et al. Induction of p53-mediated transcription and apoptosis by Exportin-1 (XPO1) inhibition in mantle cell lymphoma. Cancer Sci. 2014 doi: 10.1111/cas.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wang M, Tamayo AT, Shacham S, Kauffman M, Lee J, et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp Hematol. 2013;41(1):67–78. e64. doi: 10.1016/j.exphem.2012.09.002. [DOI] [PubMed] [Google Scholar]